
A series of mono- and di-Schiff bases formed between benzil dihydrazone {BDH; systematic name: (1
Z)-[(2
E)-2-hydrazinylidene-1,2-diphenylethylidene]hydrazine} and three isomeric chlorobenzaldehydes were designed and synthesized to be used as model compounds to help to explain the reaction mechanisms for the formation of Schiff bases. These compounds are 1-(2-chlorobenzylidene)-2-{2-[2-(2-chlorobenzylidene)hydrazin-1-ylidene]-1,2-diphenylethylidene}hydrazine (BDHOCB), and the 3-chloro (BDHMCB) and 4-chloro (BDHPCB) analogues, all having the formula C
28H
20Cl
2N
4. Surprisingly, only di-Schiff bases were obtained; our attempts to push the reaction in favour of the mono-Schiff bases all failed. Density functional theory (DFT) calculations were performed to explain the trend in the experimental results. In the case of the systems studied, the type of Schiff base produced exhibits a clear dependence on the HOMO–LUMO energy gaps (Δ
EHOMO–LUMO),
i.e. the product is mainly governed by its stability. The compounds were characterized by single-crystal X-ray diffractometry, elemental analysis, melting point,
1H NMR and
13C NMR spectroscopy. The structural features of the three new Schiff bases are similar. For instance, they have the same chemical formula, all the molecules have a symmetrical double helix structure, with each Ph—C=N—N=C—Ph arm exhibiting an
anti conformation, and their supramolecular interactions include intermolecular π–π and weak C—H

π stacking interactions. The crystal systems are different, however,
viz. triclinic (space group
P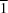
) for BDHPCB, monoclinic (space group
P2
1/
n) for BDHOCB and orthorhombic (space group
Pnna) for BDHMCB.
Supporting information
CCDC references: 1404627; 1404626; 1404625
For all compounds, data collection: SMART (Bruker, 2001). Cell refinement: SMART (Bruker, 2001) for BDHOCB, BDHPCB; SMART for BDHMCB. For all compounds, data reduction: SAINT (Bruker, 2001); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008); molecular graphics: SHELXTL (Bruker, 2001), PLATON (Spek, 2009) and DIAMOND (Brandenburg & Putz, 1999); software used to prepare material for publication: SHELXL97 (Sheldrick, 2008).
(BDHOCB) 1-(2-Chlorobenzylidene)-2-{2-[2-(2-chlorobenzylidene)hydrazin-1-ylidene]-1,2-diphenylethylidene}hydrazine
top
Crystal data top
| C28H20Cl2N4 | F(000) = 1000 |
| Mr = 483.38 | Dx = 1.293 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 380 reflections |
| a = 9.271 (3) Å | θ = 2.5–26.0° |
| b = 15.786 (5) Å | µ = 0.29 mm−1 |
| c = 17.035 (6) Å | T = 298 K |
| β = 95.322 (5)° | Block, yellow |
| V = 2482.4 (14) Å3 | 0.55 × 0.50 × 0.32 mm |
| Z = 4 | |
Data collection top
Bruker SMART CCD area-detector
diffractometer | 5492 independent reflections |
| Radiation source: fine-focus sealed tube | 3106 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.039 |
| phi and ω scans | θmax = 27.5°, θmin = 1.8° |
Absorption correction: multi-scan
(SADABS; Bruker, 2001) | h = −11→10 |
| Tmin = 0.852, Tmax = 0.915 | k = −19→20 |
| 14572 measured reflections | l = −21→21 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.050 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.150 | All H-atom parameters refined |
| S = 0.95 | w = 1/[σ2(Fo2) + (0.0828P)2]
where P = (Fo2 + 2Fc2)/3 |
| 5492 reflections | (Δ/σ)max = 0.001 |
| 387 parameters | Δρmax = 0.27 e Å−3 |
| 0 restraints | Δρmin = −0.27 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.2031 (3) | 0.23878 (17) | 0.70015 (14) | 0.0692 (6) | |
| C2 | 0.1559 (3) | 0.2309 (2) | 0.62107 (16) | 0.0864 (8) | |
| C3 | 0.0460 (3) | 0.1775 (2) | 0.59691 (19) | 0.0877 (9) | |
| C4 | −0.0187 (3) | 0.13020 (19) | 0.65161 (18) | 0.0836 (8) | |
| C5 | 0.0258 (3) | 0.13836 (15) | 0.73091 (16) | 0.0669 (6) | |
| C6 | 0.13700 (19) | 0.19350 (12) | 0.75645 (12) | 0.0519 (5) | |
| C7 | 0.18144 (18) | 0.20286 (11) | 0.84089 (12) | 0.0472 (5) | |
| C8 | 0.0868 (2) | 0.14054 (13) | 1.01586 (13) | 0.0541 (5) | |
| C9 | 0.1350 (2) | 0.13633 (11) | 1.09981 (12) | 0.0517 (5) | |
| C10 | 0.0487 (2) | 0.10266 (13) | 1.15489 (15) | 0.0659 (6) | |
| C11 | 0.0999 (4) | 0.09847 (19) | 1.23475 (17) | 0.0883 (8) | |
| C12 | 0.2363 (4) | 0.1268 (2) | 1.25811 (19) | 0.0932 (9) | |
| C13 | 0.3234 (3) | 0.15985 (17) | 1.20556 (17) | 0.0817 (7) | |
| C14 | 0.2727 (3) | 0.16413 (14) | 1.12724 (15) | 0.0636 (6) | |
| C15 | 0.4309 (3) | 0.09363 (15) | 0.88704 (17) | 0.0744 (7) | |
| C16 | 0.5477 (3) | 0.03943 (18) | 0.9015 (2) | 0.0942 (9) | |
| C17 | 0.6852 (3) | 0.07083 (19) | 0.90641 (19) | 0.0926 (9) | |
| C18 | 0.7087 (3) | 0.1552 (2) | 0.89774 (18) | 0.0878 (8) | |
| C19 | 0.5926 (2) | 0.20920 (16) | 0.88542 (15) | 0.0667 (6) | |
| C20 | 0.45228 (19) | 0.17922 (11) | 0.87971 (11) | 0.0480 (5) | |
| C21 | 0.32974 (18) | 0.23870 (11) | 0.86653 (11) | 0.0444 (4) | |
| C22 | 0.2490 (2) | 0.43873 (12) | 0.89966 (13) | 0.0546 (5) | |
| C23 | 0.1406 (2) | 0.50573 (12) | 0.89255 (13) | 0.0558 (5) | |
| C24 | 0.1317 (3) | 0.56503 (13) | 0.95207 (15) | 0.0709 (7) | |
| C25 | 0.0276 (4) | 0.62802 (18) | 0.9463 (3) | 0.1018 (10) | |
| C26 | −0.0639 (4) | 0.63357 (19) | 0.8797 (3) | 0.1193 (15) | |
| C27 | −0.0564 (3) | 0.57727 (19) | 0.8184 (3) | 0.1014 (11) | |
| C28 | 0.0447 (3) | 0.51328 (16) | 0.82488 (17) | 0.0744 (7) | |
| N1 | 0.09605 (16) | 0.17574 (11) | 0.89066 (10) | 0.0550 (4) | |
| N2 | 0.15540 (17) | 0.18352 (11) | 0.96920 (10) | 0.0575 (4) | |
| N3 | 0.35533 (16) | 0.31738 (10) | 0.87708 (10) | 0.0519 (4) | |
| N4 | 0.23339 (17) | 0.37024 (10) | 0.86230 (10) | 0.0566 (4) | |
| Cl1 | −0.12354 (7) | 0.06504 (5) | 1.12484 (5) | 0.0983 (3) | |
| Cl2 | 0.25019 (11) | 0.56054 (5) | 1.03667 (4) | 0.1100 (3) | |
| H1 | 0.281 (3) | 0.2780 (14) | 0.7172 (13) | 0.071 (7)* | |
| H2 | 0.200 (3) | 0.267 (2) | 0.5844 (19) | 0.123 (11)* | |
| H3 | 0.010 (3) | 0.1689 (17) | 0.5460 (16) | 0.096 (9)* | |
| H4 | −0.091 (3) | 0.0920 (17) | 0.6367 (16) | 0.096 (9)* | |
| H5 | −0.017 (2) | 0.1095 (15) | 0.7678 (14) | 0.069 (7)* | |
| H6 | −0.001 (3) | 0.1085 (15) | 0.9945 (14) | 0.081 (7)* | |
| H7 | 0.035 (3) | 0.0692 (18) | 1.2690 (19) | 0.111 (10)* | |
| H8 | 0.279 (3) | 0.1251 (19) | 1.313 (2) | 0.123 (11)* | |
| H9 | 0.432 (3) | 0.1795 (17) | 1.2252 (15) | 0.097 (8)* | |
| H10 | 0.334 (3) | 0.1880 (15) | 1.0871 (14) | 0.082 (7)* | |
| H11 | 0.343 (3) | 0.0724 (14) | 0.8842 (14) | 0.080 (8)* | |
| H12 | 0.526 (3) | −0.019 (2) | 0.9092 (16) | 0.109 (9)* | |
| H13 | 0.770 (3) | 0.0319 (18) | 0.9167 (15) | 0.106 (8)* | |
| H14 | 0.804 (3) | 0.1780 (19) | 0.8968 (16) | 0.114 (10)* | |
| H15 | 0.607 (2) | 0.2663 (15) | 0.8792 (13) | 0.074 (7)* | |
| H16 | 0.330 (2) | 0.4493 (13) | 0.9328 (13) | 0.071 (7)* | |
| H17 | 0.030 (3) | 0.6658 (19) | 0.9909 (18) | 0.105 (9)* | |
| H18 | −0.132 (4) | 0.677 (2) | 0.8754 (19) | 0.138 (12)* | |
| H19 | −0.119 (4) | 0.5799 (18) | 0.7749 (19) | 0.108 (11)* | |
| H20 | 0.056 (2) | 0.4698 (17) | 0.7830 (13) | 0.082 (7)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1 | 0.0622 (14) | 0.0893 (17) | 0.0556 (15) | −0.0065 (13) | 0.0024 (11) | −0.0042 (12) |
| C2 | 0.0882 (19) | 0.112 (2) | 0.0582 (17) | −0.0042 (17) | 0.0051 (14) | −0.0057 (16) |
| C3 | 0.0851 (19) | 0.116 (2) | 0.0590 (18) | 0.0143 (17) | −0.0114 (15) | −0.0235 (17) |
| C4 | 0.0725 (17) | 0.0904 (19) | 0.083 (2) | −0.0022 (15) | −0.0179 (15) | −0.0250 (16) |
| C5 | 0.0625 (14) | 0.0688 (15) | 0.0676 (16) | −0.0061 (12) | −0.0031 (12) | −0.0073 (13) |
| C6 | 0.0434 (10) | 0.0565 (12) | 0.0546 (13) | 0.0075 (9) | −0.0012 (9) | −0.0068 (9) |
| C7 | 0.0389 (9) | 0.0462 (10) | 0.0561 (12) | 0.0064 (8) | 0.0027 (9) | −0.0030 (9) |
| C8 | 0.0463 (11) | 0.0497 (12) | 0.0666 (14) | 0.0023 (9) | 0.0069 (10) | 0.0018 (10) |
| C9 | 0.0576 (12) | 0.0398 (10) | 0.0586 (13) | 0.0066 (9) | 0.0099 (10) | 0.0033 (9) |
| C10 | 0.0746 (14) | 0.0505 (12) | 0.0751 (17) | 0.0061 (10) | 0.0206 (12) | 0.0015 (11) |
| C11 | 0.120 (2) | 0.0798 (18) | 0.0700 (19) | 0.0135 (17) | 0.0343 (19) | 0.0127 (14) |
| C12 | 0.119 (3) | 0.092 (2) | 0.066 (2) | 0.0101 (19) | −0.0030 (19) | −0.0006 (15) |
| C13 | 0.097 (2) | 0.0730 (16) | 0.0724 (19) | 0.0054 (15) | −0.0081 (16) | 0.0014 (14) |
| C14 | 0.0701 (14) | 0.0553 (13) | 0.0646 (16) | 0.0027 (11) | 0.0021 (12) | 0.0047 (11) |
| C15 | 0.0574 (14) | 0.0561 (14) | 0.110 (2) | 0.0061 (11) | 0.0121 (13) | −0.0006 (13) |
| C16 | 0.087 (2) | 0.0569 (16) | 0.139 (3) | 0.0230 (15) | 0.0140 (18) | 0.0067 (16) |
| C17 | 0.0669 (17) | 0.083 (2) | 0.124 (3) | 0.0342 (15) | −0.0071 (16) | −0.0052 (16) |
| C18 | 0.0472 (14) | 0.090 (2) | 0.123 (2) | 0.0150 (13) | −0.0089 (14) | −0.0095 (16) |
| C19 | 0.0466 (12) | 0.0604 (14) | 0.0915 (18) | 0.0038 (10) | −0.0019 (11) | −0.0042 (12) |
| C20 | 0.0429 (10) | 0.0475 (11) | 0.0533 (12) | 0.0056 (8) | 0.0025 (8) | −0.0015 (8) |
| C21 | 0.0392 (9) | 0.0485 (11) | 0.0453 (11) | 0.0029 (8) | 0.0019 (8) | −0.0030 (8) |
| C22 | 0.0567 (12) | 0.0504 (12) | 0.0560 (13) | 0.0011 (10) | 0.0016 (10) | −0.0038 (10) |
| C23 | 0.0587 (12) | 0.0423 (11) | 0.0675 (14) | 0.0023 (9) | 0.0125 (11) | 0.0006 (9) |
| C24 | 0.0846 (16) | 0.0482 (12) | 0.0834 (18) | −0.0020 (11) | 0.0259 (13) | −0.0074 (11) |
| C25 | 0.100 (2) | 0.0599 (17) | 0.151 (3) | 0.0070 (16) | 0.044 (2) | −0.0247 (19) |
| C26 | 0.080 (2) | 0.0546 (18) | 0.225 (5) | 0.0196 (16) | 0.026 (3) | −0.001 (2) |
| C27 | 0.0746 (18) | 0.0697 (19) | 0.155 (3) | 0.0167 (14) | −0.014 (2) | 0.015 (2) |
| C28 | 0.0703 (15) | 0.0615 (15) | 0.0896 (19) | 0.0109 (12) | −0.0023 (13) | 0.0016 (14) |
| N1 | 0.0413 (8) | 0.0669 (11) | 0.0565 (11) | −0.0005 (8) | 0.0024 (8) | −0.0014 (8) |
| N2 | 0.0518 (9) | 0.0649 (11) | 0.0561 (11) | −0.0053 (8) | 0.0063 (8) | 0.0000 (9) |
| N3 | 0.0450 (9) | 0.0476 (9) | 0.0621 (11) | 0.0068 (7) | −0.0001 (7) | −0.0033 (8) |
| N4 | 0.0521 (9) | 0.0492 (10) | 0.0669 (12) | 0.0105 (7) | −0.0030 (8) | −0.0061 (8) |
| Cl1 | 0.0823 (5) | 0.0966 (5) | 0.1213 (7) | −0.0196 (3) | 0.0373 (4) | 0.0068 (4) |
| Cl2 | 0.1651 (8) | 0.0968 (5) | 0.0677 (5) | −0.0074 (5) | 0.0083 (5) | −0.0249 (4) |
Geometric parameters (Å, º) top
| C1—C2 | 1.383 (3) | C15—C16 | 1.384 (3) |
| C1—C6 | 1.384 (3) | C15—H11 | 0.88 (2) |
| C1—H1 | 0.97 (2) | C16—C17 | 1.363 (4) |
| C2—C3 | 1.357 (4) | C16—H12 | 0.96 (3) |
| C2—H2 | 0.97 (3) | C17—C18 | 1.359 (4) |
| C3—C4 | 1.375 (4) | C17—H13 | 1.00 (3) |
| C3—H3 | 0.91 (3) | C18—C19 | 1.374 (3) |
| C4—C5 | 1.381 (4) | C18—H14 | 0.96 (3) |
| C4—H4 | 0.92 (3) | C19—C20 | 1.379 (3) |
| C5—C6 | 1.388 (3) | C19—H15 | 0.92 (2) |
| C5—H5 | 0.90 (2) | C20—C21 | 1.475 (2) |
| C6—C7 | 1.467 (3) | C21—N3 | 1.274 (2) |
| C7—N1 | 1.286 (2) | C22—N4 | 1.256 (2) |
| C7—C21 | 1.513 (2) | C22—C23 | 1.456 (3) |
| C8—N2 | 1.261 (3) | C22—H16 | 0.91 (2) |
| C8—C9 | 1.460 (3) | C23—C24 | 1.388 (3) |
| C8—H6 | 1.00 (2) | C23—C28 | 1.394 (3) |
| C9—C14 | 1.389 (3) | C24—C25 | 1.383 (4) |
| C9—C10 | 1.394 (3) | C24—Cl2 | 1.731 (3) |
| C10—C11 | 1.400 (4) | C25—C26 | 1.354 (5) |
| C10—Cl1 | 1.736 (2) | C25—H17 | 0.96 (3) |
| C11—C12 | 1.366 (4) | C26—C27 | 1.378 (5) |
| C11—H7 | 0.99 (3) | C26—H18 | 0.93 (4) |
| C12—C13 | 1.364 (4) | C27—C28 | 1.376 (4) |
| C12—H8 | 0.99 (3) | C27—H19 | 0.90 (3) |
| C13—C14 | 1.375 (3) | C28—H20 | 1.00 (2) |
| C13—H9 | 1.08 (3) | N1—N2 | 1.404 (2) |
| C14—H10 | 1.00 (3) | N3—N4 | 1.409 (2) |
| C15—C20 | 1.373 (3) | | |
| | | |
| C2—C1—C6 | 120.6 (2) | C16—C15—H11 | 118.8 (15) |
| C2—C1—H1 | 120.4 (13) | C17—C16—C15 | 119.9 (3) |
| C6—C1—H1 | 119.0 (13) | C17—C16—H12 | 123.3 (16) |
| C3—C2—C1 | 120.8 (3) | C15—C16—H12 | 116.8 (17) |
| C3—C2—H2 | 121.7 (19) | C18—C17—C16 | 120.5 (2) |
| C1—C2—H2 | 117.5 (19) | C18—C17—H13 | 119.2 (16) |
| C2—C3—C4 | 119.7 (3) | C16—C17—H13 | 120.3 (16) |
| C2—C3—H3 | 125.3 (17) | C17—C18—C19 | 119.5 (3) |
| C4—C3—H3 | 115.0 (17) | C17—C18—H14 | 121.9 (18) |
| C3—C4—C5 | 120.2 (3) | C19—C18—H14 | 118.4 (18) |
| C3—C4—H4 | 121.4 (18) | C18—C19—C20 | 121.2 (2) |
| C5—C4—H4 | 118.4 (18) | C18—C19—H15 | 120.3 (14) |
| C4—C5—C6 | 120.7 (3) | C20—C19—H15 | 118.5 (14) |
| C4—C5—H5 | 121.8 (15) | C15—C20—C19 | 118.31 (19) |
| C6—C5—H5 | 117.4 (15) | C15—C20—C21 | 121.66 (18) |
| C1—C6—C5 | 118.1 (2) | C19—C20—C21 | 120.03 (18) |
| C1—C6—C7 | 121.70 (18) | N3—C21—C20 | 117.93 (15) |
| C5—C6—C7 | 120.2 (2) | N3—C21—C7 | 123.89 (15) |
| N1—C7—C6 | 118.62 (17) | C20—C21—C7 | 118.18 (15) |
| N1—C7—C21 | 122.19 (17) | N4—C22—C23 | 122.5 (2) |
| C6—C7—C21 | 119.07 (16) | N4—C22—H16 | 121.3 (13) |
| N2—C8—C9 | 121.0 (2) | C23—C22—H16 | 116.2 (13) |
| N2—C8—H6 | 119.1 (14) | C24—C23—C28 | 118.0 (2) |
| C9—C8—H6 | 119.9 (14) | C24—C23—C22 | 121.1 (2) |
| C14—C9—C10 | 117.4 (2) | C28—C23—C22 | 120.9 (2) |
| C14—C9—C8 | 120.23 (19) | C25—C24—C23 | 121.4 (3) |
| C10—C9—C8 | 122.3 (2) | C25—C24—Cl2 | 118.3 (2) |
| C9—C10—C11 | 120.6 (2) | C23—C24—Cl2 | 120.28 (18) |
| C9—C10—Cl1 | 120.11 (19) | C26—C25—C24 | 119.0 (3) |
| C11—C10—Cl1 | 119.3 (2) | C26—C25—H17 | 125.8 (18) |
| C12—C11—C10 | 119.3 (3) | C24—C25—H17 | 115.2 (18) |
| C12—C11—H7 | 125.3 (18) | C25—C26—C27 | 121.4 (3) |
| C10—C11—H7 | 115.2 (19) | C25—C26—H18 | 119 (2) |
| C13—C12—C11 | 121.5 (3) | C27—C26—H18 | 120 (2) |
| C13—C12—H8 | 115.1 (19) | C28—C27—C26 | 119.7 (4) |
| C11—C12—H8 | 123.4 (19) | C28—C27—H19 | 119 (2) |
| C12—C13—C14 | 119.1 (3) | C26—C27—H19 | 121.4 (19) |
| C12—C13—H9 | 120.0 (14) | C27—C28—C23 | 120.4 (3) |
| C14—C13—H9 | 120.9 (14) | C27—C28—H20 | 124.2 (14) |
| C13—C14—C9 | 122.1 (3) | C23—C28—H20 | 115.3 (13) |
| C13—C14—H10 | 121.2 (13) | C7—N1—N2 | 112.85 (16) |
| C9—C14—H10 | 116.7 (13) | C8—N2—N1 | 112.18 (17) |
| C20—C15—C16 | 120.5 (2) | C21—N3—N4 | 114.60 (15) |
| C20—C15—H11 | 120.7 (15) | C22—N4—N3 | 111.50 (16) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C12—H8···Cl2i | 0.99 (3) | 2.79 (3) | 3.640 (4) | 145 (2) |
| C5—H5···N1 | 0.90 (2) | 2.48 (2) | 2.802 (3) | 101.5 (17) |
| C8—H6···Cl1 | 1.00 (2) | 2.67 (2) | 3.056 (2) | 102.9 (15) |
| C19—H15···N3 | 0.92 (2) | 2.47 (2) | 2.778 (3) | 99.9 (16) |
| C22—H16···Cl2 | 0.91 (2) | 2.65 (2) | 3.023 (2) | 105.6 (16) |
| Symmetry code: (i) −x+1/2, y−1/2, −z+5/2. |
(BDHMCB) 1-(3-Chlorobenzylidene)-2-{2-[2-(2-chlorobenzylidene)hydrazin-1-ylidene]-1,2-diphenylethylidene}hydrazine
top
Crystal data top
| C28H20Cl2N4 | F(000) = 1000 |
| Mr = 483.38 | Dx = 1.301 Mg m−3 |
| Orthorhombic, Pnna | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2a 2bc | Cell parameters from 380 reflections |
| a = 11.619 (4) Å | θ = 2.5–26.0° |
| b = 23.173 (8) Å | µ = 0.29 mm−1 |
| c = 9.165 (3) Å | T = 298 K |
| V = 2467.6 (15) Å3 | Block, yellow |
| Z = 4 | 0.38 × 0.32 × 0.25 mm |
Data collection top
Bruker SMART CCD area detector
diffractometer | 2835 independent reflections |
| Radiation source: fine-focus sealed tube | 1277 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.034 |
| phi and ω scans | θmax = 27.6°, θmin = 1.8° |
Absorption correction: multi-scan
(SADABS; Bruker, 2001) | h = −11→14 |
| Tmin = 0.898, Tmax = 0.932 | k = −29→29 |
| 13835 measured reflections | l = −11→11 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.049 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.151 | All H-atom parameters refined |
| S = 0.98 | w = 1/[σ2(Fo2) + (0.0694P)2 + 0.0138P]
where P = (Fo2 + 2Fc2)/3 |
| 2835 reflections | (Δ/σ)max = 0.001 |
| 194 parameters | Δρmax = 0.15 e Å−3 |
| 0 restraints | Δρmin = −0.16 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.6399 (2) | 0.88360 (10) | 0.5185 (3) | 0.0882 (7) | |
| C2 | 0.6557 (3) | 0.83900 (11) | 0.6152 (3) | 0.1012 (9) | |
| C3 | 0.7494 (3) | 0.83695 (11) | 0.7039 (4) | 0.0960 (8) | |
| C4 | 0.8303 (2) | 0.87958 (10) | 0.6940 (3) | 0.0930 (7) | |
| C5 | 0.81619 (19) | 0.92379 (9) | 0.5972 (3) | 0.0798 (7) | |
| C6 | 0.72127 (16) | 0.92730 (7) | 0.5087 (2) | 0.0667 (6) | |
| C7 | 0.70496 (16) | 0.97683 (8) | 0.4118 (2) | 0.0671 (5) | |
| C8 | 0.5128 (2) | 1.03802 (12) | 0.1865 (3) | 0.0901 (7) | |
| C9 | 0.4866 (2) | 1.08818 (14) | 0.0995 (3) | 0.0954 (8) | |
| C10 | 0.5512 (3) | 1.13736 (14) | 0.1112 (3) | 0.1024 (8) | |
| C11 | 0.5269 (3) | 1.18549 (16) | 0.0282 (4) | 0.1400 (13) | |
| C12 | 0.4386 (5) | 1.1843 (3) | −0.0685 (6) | 0.177 (3) | |
| C13 | 0.3718 (5) | 1.1357 (4) | −0.0809 (5) | 0.174 (2) | |
| C14 | 0.3945 (3) | 1.0876 (2) | 0.0006 (4) | 0.1273 (12) | |
| N1 | 0.61133 (14) | 0.98128 (7) | 0.3382 (2) | 0.0797 (5) | |
| N2 | 0.60586 (14) | 1.03291 (7) | 0.2578 (2) | 0.0833 (6) | |
| Cl1 | 0.60920 (10) | 1.24635 (4) | 0.04030 (15) | 0.2034 (7) | |
| H1 | 0.581 (2) | 0.8856 (9) | 0.449 (2) | 0.098 (8)* | |
| H2 | 0.600 (2) | 0.8119 (10) | 0.619 (3) | 0.116 (9)* | |
| H3 | 0.7602 (17) | 0.8073 (10) | 0.774 (2) | 0.094 (7)* | |
| H4 | 0.896 (2) | 0.8770 (10) | 0.758 (2) | 0.107 (8)* | |
| H5 | 0.8736 (16) | 0.9520 (8) | 0.590 (2) | 0.079 (6)* | |
| H6 | 0.4641 (19) | 1.0070 (10) | 0.190 (2) | 0.098 (8)* | |
| H7 | 0.6109 (19) | 1.1424 (9) | 0.187 (2) | 0.091 (7)* | |
| H8 | 0.414 (4) | 1.2125 (18) | −0.145 (5) | 0.217 (16)* | |
| H9 | 0.313 (3) | 1.1369 (13) | −0.153 (4) | 0.170 (13)* | |
| H10 | 0.357 (2) | 1.0502 (13) | 0.008 (3) | 0.123 (12)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1 | 0.0783 (16) | 0.0754 (16) | 0.111 (2) | −0.0185 (12) | −0.0027 (15) | −0.0001 (14) |
| C2 | 0.101 (2) | 0.0711 (16) | 0.132 (2) | −0.0261 (15) | 0.0133 (18) | 0.0009 (16) |
| C3 | 0.114 (2) | 0.0660 (15) | 0.108 (2) | 0.0040 (15) | 0.0182 (17) | 0.0098 (15) |
| C4 | 0.0870 (17) | 0.0835 (17) | 0.108 (2) | 0.0022 (14) | −0.0081 (16) | 0.0108 (15) |
| C5 | 0.0677 (13) | 0.0662 (13) | 0.1053 (18) | −0.0094 (11) | −0.0017 (13) | 0.0058 (13) |
| C6 | 0.0559 (11) | 0.0585 (12) | 0.0859 (15) | −0.0072 (9) | 0.0050 (11) | −0.0086 (10) |
| C7 | 0.0529 (11) | 0.0633 (12) | 0.0852 (15) | −0.0055 (8) | 0.0018 (11) | −0.0057 (10) |
| C8 | 0.0620 (14) | 0.104 (2) | 0.104 (2) | −0.0012 (14) | −0.0035 (13) | −0.0037 (16) |
| C9 | 0.0630 (14) | 0.130 (2) | 0.0933 (18) | 0.0251 (15) | 0.0000 (13) | 0.0050 (16) |
| C10 | 0.0757 (17) | 0.121 (2) | 0.111 (2) | 0.0317 (16) | 0.0156 (16) | 0.0249 (18) |
| C11 | 0.116 (3) | 0.165 (3) | 0.139 (3) | 0.076 (2) | 0.052 (2) | 0.051 (2) |
| C12 | 0.163 (5) | 0.232 (6) | 0.137 (4) | 0.122 (4) | 0.059 (4) | 0.066 (4) |
| C13 | 0.124 (4) | 0.296 (7) | 0.101 (3) | 0.121 (4) | −0.006 (3) | 0.027 (4) |
| C14 | 0.081 (2) | 0.189 (4) | 0.112 (3) | 0.048 (2) | −0.0007 (19) | −0.008 (2) |
| N1 | 0.0632 (11) | 0.0739 (11) | 0.1021 (14) | −0.0084 (8) | −0.0074 (10) | 0.0002 (10) |
| N2 | 0.0634 (11) | 0.0833 (13) | 0.1030 (15) | −0.0015 (9) | −0.0123 (10) | 0.0032 (10) |
| Cl1 | 0.1921 (10) | 0.1272 (8) | 0.2911 (15) | 0.0564 (7) | 0.1174 (10) | 0.0850 (8) |
Geometric parameters (Å, º) top
| C1—C2 | 1.374 (4) | C8—C9 | 1.442 (4) |
| C1—C6 | 1.388 (3) | C8—H6 | 0.91 (2) |
| C1—H1 | 0.94 (2) | C9—C10 | 1.369 (4) |
| C2—C3 | 1.359 (4) | C9—C14 | 1.403 (4) |
| C2—H2 | 0.90 (2) | C10—C11 | 1.380 (4) |
| C3—C4 | 1.367 (3) | C10—H7 | 0.99 (2) |
| C3—H3 | 0.95 (2) | C11—C12 | 1.356 (7) |
| C4—C5 | 1.365 (3) | C11—Cl1 | 1.708 (4) |
| C4—H4 | 0.96 (2) | C12—C13 | 1.372 (7) |
| C5—C6 | 1.371 (3) | C12—H8 | 1.00 (4) |
| C5—H5 | 0.936 (19) | C13—C14 | 1.366 (7) |
| C6—C7 | 1.464 (3) | C13—H9 | 0.96 (3) |
| C7—N1 | 1.284 (2) | C14—H10 | 0.97 (3) |
| C7—C7i | 1.500 (3) | N1—N2 | 1.407 (2) |
| C8—N2 | 1.269 (3) | | |
| | | |
| C2—C1—C6 | 120.0 (3) | N2—C8—H6 | 115.9 (15) |
| C2—C1—H1 | 125.0 (14) | C9—C8—H6 | 121.3 (15) |
| C6—C1—H1 | 114.7 (14) | C10—C9—C14 | 118.5 (3) |
| C3—C2—C1 | 121.3 (3) | C10—C9—C8 | 120.8 (2) |
| C3—C2—H2 | 122.1 (16) | C14—C9—C8 | 120.7 (3) |
| C1—C2—H2 | 116.6 (16) | C9—C10—C11 | 121.2 (3) |
| C2—C3—C4 | 119.1 (3) | C9—C10—H7 | 122.5 (13) |
| C2—C3—H3 | 122.5 (13) | C11—C10—H7 | 115.9 (13) |
| C4—C3—H3 | 118.5 (13) | C12—C11—C10 | 119.9 (5) |
| C5—C4—C3 | 120.2 (3) | C12—C11—Cl1 | 118.9 (4) |
| C5—C4—H4 | 122.6 (15) | C10—C11—Cl1 | 121.1 (3) |
| C3—C4—H4 | 117.2 (15) | C11—C12—C13 | 119.9 (5) |
| C4—C5—C6 | 121.7 (2) | C11—C12—H8 | 132 (3) |
| C4—C5—H5 | 119.0 (12) | C13—C12—H8 | 108 (3) |
| C6—C5—H5 | 119.3 (12) | C14—C13—C12 | 121.0 (5) |
| C5—C6—C1 | 117.8 (2) | C14—C13—H9 | 123 (2) |
| C5—C6—C7 | 120.63 (17) | C12—C13—H9 | 116 (2) |
| C1—C6—C7 | 121.6 (2) | C13—C14—C9 | 119.5 (5) |
| N1—C7—C6 | 119.43 (16) | C13—C14—H10 | 132.6 (18) |
| N1—C7—C7i | 122.27 (18) | C9—C14—H10 | 107.9 (18) |
| C6—C7—C7i | 118.11 (16) | C7—N1—N2 | 112.43 (16) |
| N2—C8—C9 | 122.6 (2) | C8—N2—N1 | 112.78 (19) |
| Symmetry code: (i) −x+3/2, −y+2, z. |
(BDHPCB) 1-(4-Chlorobenzylidene)-2-{2-[2-(2-chlorobenzylidene)hydrazin-1-ylidene]-1,2-diphenylethylidene}hydrazine
top
Crystal data top
| C28H20Cl2N4 | Z = 2 |
| Mr = 483.38 | F(000) = 500 |
| Triclinic, P1 | Dx = 1.293 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.661 (5) Å | Cell parameters from 380 reflections |
| b = 11.492 (6) Å | θ = 2.5–26.0° |
| c = 13.018 (7) Å | µ = 0.29 mm−1 |
| α = 111.040 (7)° | T = 298 K |
| β = 109.532 (7)° | Block, yellow |
| γ = 93.954 (7)° | 0.38 × 0.26 × 0.18 mm |
| V = 1241.7 (11) Å3 | |
Data collection top
Bruker SMART CCD area-detector
diffractometer | 4757 independent reflections |
| Radiation source: fine-focus sealed tube | 2699 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.052 |
| phi and ω scans | θmax = 26.0°, θmin = 1.8° |
Absorption correction: multi-scan
(SADABS; Bruker, 2001) | h = −10→11 |
| Tmin = 0.914, Tmax = 0.954 | k = −14→14 |
| 6793 measured reflections | l = −15→16 |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.058 | All H-atom parameters refined |
| wR(F2) = 0.158 | w = 1/[σ2(Fo2) + (0.0918P)2]
where P = (Fo2 + 2Fc2)/3 |
| S = 0.91 | (Δ/σ)max < 0.001 |
| 4757 reflections | Δρmax = 0.30 e Å−3 |
| 388 parameters | Δρmin = −0.44 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.033 (4) |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.3977 (3) | 0.5324 (3) | 0.2655 (3) | 0.0692 (8) | |
| C2 | 0.4360 (4) | 0.5111 (3) | 0.1689 (3) | 0.0900 (10) | |
| C3 | 0.4965 (5) | 0.6119 (4) | 0.1527 (3) | 0.0978 (11) | |
| C4 | 0.5180 (4) | 0.7344 (3) | 0.2321 (3) | 0.0856 (10) | |
| C5 | 0.4801 (3) | 0.7565 (3) | 0.3290 (3) | 0.0631 (7) | |
| C6 | 0.4206 (2) | 0.6563 (2) | 0.3487 (2) | 0.0488 (6) | |
| C7 | 0.3868 (2) | 0.6822 (2) | 0.4556 (2) | 0.0456 (5) | |
| C8 | 0.2211 (3) | 0.5467 (3) | 0.5866 (3) | 0.0590 (6) | |
| C9 | 0.1943 (3) | 0.5707 (2) | 0.6951 (2) | 0.0559 (6) | |
| C10 | 0.2739 (3) | 0.6789 (3) | 0.7981 (3) | 0.0621 (7) | |
| C11 | 0.2449 (3) | 0.7056 (3) | 0.8989 (3) | 0.0698 (8) | |
| C12 | 0.1313 (3) | 0.6229 (3) | 0.8966 (3) | 0.0707 (8) | |
| C13 | 0.0518 (4) | 0.5145 (3) | 0.7972 (3) | 0.0718 (8) | |
| C14 | 0.0830 (3) | 0.4876 (3) | 0.6963 (3) | 0.0671 (7) | |
| C15 | 0.6769 (3) | 0.7390 (3) | 0.6461 (2) | 0.0584 (7) | |
| C16 | 0.8145 (3) | 0.7579 (3) | 0.7349 (3) | 0.0703 (8) | |
| C17 | 0.8721 (3) | 0.8713 (3) | 0.8311 (3) | 0.0743 (8) | |
| C18 | 0.7947 (4) | 0.9675 (3) | 0.8383 (3) | 0.0824 (9) | |
| C19 | 0.6583 (3) | 0.9498 (3) | 0.7502 (2) | 0.0695 (8) | |
| C20 | 0.5964 (2) | 0.8344 (2) | 0.65252 (19) | 0.0446 (5) | |
| C21 | 0.4506 (2) | 0.8128 (2) | 0.55597 (19) | 0.0444 (5) | |
| C22 | 0.1910 (3) | 0.9688 (3) | 0.4592 (2) | 0.0555 (6) | |
| C23 | 0.0526 (3) | 0.9583 (2) | 0.3630 (2) | 0.0565 (6) | |
| C24 | −0.0154 (3) | 1.0615 (3) | 0.3738 (3) | 0.0680 (8) | |
| C25 | −0.1458 (4) | 1.0557 (4) | 0.2843 (3) | 0.0778 (9) | |
| C26 | −0.2066 (3) | 0.9459 (4) | 0.1840 (3) | 0.0771 (9) | |
| C27 | −0.1423 (3) | 0.8417 (4) | 0.1700 (3) | 0.0853 (9) | |
| C28 | −0.0126 (3) | 0.8486 (3) | 0.2605 (3) | 0.0734 (8) | |
| N1 | 0.3121 (2) | 0.59119 (18) | 0.46430 (17) | 0.0548 (5) | |
| N2 | 0.2974 (2) | 0.63244 (19) | 0.57530 (18) | 0.0589 (5) | |
| N3 | 0.3862 (2) | 0.90753 (18) | 0.55885 (17) | 0.0521 (5) | |
| N4 | 0.2515 (2) | 0.87315 (19) | 0.45753 (18) | 0.0564 (5) | |
| Cl1 | 0.08981 (13) | 0.66030 (9) | 1.02296 (9) | 0.1160 (4) | |
| Cl2 | −0.36990 (9) | 0.93629 (11) | 0.06886 (10) | 0.1202 (4) | |
| H1 | 0.354 (3) | 0.465 (3) | 0.280 (3) | 0.092 (10)* | |
| H2 | 0.422 (3) | 0.424 (3) | 0.121 (3) | 0.082 (9)* | |
| H3 | 0.528 (4) | 0.596 (4) | 0.089 (4) | 0.134 (13)* | |
| H4 | 0.566 (3) | 0.810 (3) | 0.231 (3) | 0.098 (10)* | |
| H5 | 0.496 (3) | 0.839 (3) | 0.388 (3) | 0.074 (8)* | |
| H6 | 0.177 (3) | 0.467 (2) | 0.522 (2) | 0.056 (7)* | |
| H7 | 0.354 (3) | 0.735 (2) | 0.801 (2) | 0.068 (7)* | |
| H8 | 0.298 (3) | 0.778 (3) | 0.974 (3) | 0.074 (8)* | |
| H9 | −0.034 (3) | 0.457 (3) | 0.790 (3) | 0.093 (9)* | |
| H10 | 0.029 (3) | 0.413 (3) | 0.629 (2) | 0.070 (8)* | |
| H11 | 0.640 (2) | 0.659 (2) | 0.583 (2) | 0.056 (7)* | |
| H12 | 0.866 (3) | 0.690 (3) | 0.732 (2) | 0.084 (9)* | |
| H13 | 0.965 (3) | 0.882 (3) | 0.888 (3) | 0.082 (9)* | |
| H14 | 0.830 (3) | 1.050 (3) | 0.906 (3) | 0.090 (9)* | |
| H15 | 0.604 (3) | 1.015 (3) | 0.756 (2) | 0.088 (9)* | |
| H16 | 0.238 (3) | 1.050 (2) | 0.528 (2) | 0.058 (7)* | |
| H17 | 0.031 (3) | 1.139 (3) | 0.443 (2) | 0.070 (8)* | |
| H18 | −0.184 (3) | 1.134 (3) | 0.295 (3) | 0.087 (9)* | |
| H19 | −0.185 (4) | 0.761 (3) | 0.102 (3) | 0.101 (11)* | |
| H20 | 0.027 (3) | 0.783 (3) | 0.250 (3) | 0.080 (9)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1 | 0.089 (2) | 0.0533 (17) | 0.0569 (17) | 0.0143 (14) | 0.0273 (15) | 0.0142 (14) |
| C2 | 0.134 (3) | 0.062 (2) | 0.063 (2) | 0.026 (2) | 0.046 (2) | 0.0068 (17) |
| C3 | 0.141 (3) | 0.094 (3) | 0.074 (2) | 0.031 (2) | 0.067 (2) | 0.027 (2) |
| C4 | 0.119 (3) | 0.072 (2) | 0.075 (2) | 0.0142 (19) | 0.055 (2) | 0.0256 (18) |
| C5 | 0.0763 (18) | 0.0564 (17) | 0.0563 (17) | 0.0118 (14) | 0.0311 (14) | 0.0180 (14) |
| C6 | 0.0467 (13) | 0.0489 (14) | 0.0431 (13) | 0.0126 (10) | 0.0101 (10) | 0.0166 (11) |
| C7 | 0.0404 (12) | 0.0458 (13) | 0.0432 (13) | 0.0114 (10) | 0.0090 (10) | 0.0163 (11) |
| C8 | 0.0561 (15) | 0.0528 (16) | 0.0597 (17) | 0.0056 (12) | 0.0159 (13) | 0.0209 (14) |
| C9 | 0.0559 (15) | 0.0539 (15) | 0.0607 (16) | 0.0132 (12) | 0.0199 (12) | 0.0284 (13) |
| C10 | 0.0585 (16) | 0.0643 (17) | 0.0680 (19) | 0.0074 (14) | 0.0222 (14) | 0.0349 (15) |
| C11 | 0.0784 (19) | 0.0629 (18) | 0.066 (2) | 0.0116 (15) | 0.0271 (16) | 0.0259 (16) |
| C12 | 0.0841 (19) | 0.0664 (19) | 0.084 (2) | 0.0249 (15) | 0.0444 (17) | 0.0419 (17) |
| C13 | 0.078 (2) | 0.0632 (19) | 0.090 (2) | 0.0133 (16) | 0.0417 (18) | 0.0410 (18) |
| C14 | 0.0681 (18) | 0.0548 (17) | 0.077 (2) | 0.0055 (14) | 0.0265 (16) | 0.0283 (16) |
| C15 | 0.0575 (15) | 0.0477 (15) | 0.0548 (16) | 0.0124 (12) | 0.0080 (12) | 0.0166 (13) |
| C16 | 0.0593 (16) | 0.0659 (18) | 0.075 (2) | 0.0227 (15) | 0.0084 (15) | 0.0310 (16) |
| C17 | 0.0604 (18) | 0.082 (2) | 0.0608 (19) | 0.0111 (16) | −0.0025 (15) | 0.0315 (17) |
| C18 | 0.086 (2) | 0.068 (2) | 0.0517 (18) | 0.0124 (17) | −0.0041 (15) | 0.0082 (16) |
| C19 | 0.0747 (18) | 0.0571 (16) | 0.0527 (17) | 0.0233 (14) | 0.0080 (14) | 0.0096 (13) |
| C20 | 0.0454 (12) | 0.0438 (13) | 0.0415 (13) | 0.0078 (10) | 0.0130 (10) | 0.0177 (11) |
| C21 | 0.0439 (12) | 0.0467 (13) | 0.0431 (13) | 0.0130 (10) | 0.0165 (10) | 0.0186 (11) |
| C22 | 0.0551 (15) | 0.0557 (16) | 0.0539 (16) | 0.0190 (13) | 0.0183 (12) | 0.0216 (14) |
| C23 | 0.0514 (14) | 0.0690 (17) | 0.0611 (17) | 0.0246 (13) | 0.0234 (12) | 0.0358 (15) |
| C24 | 0.0714 (18) | 0.0689 (19) | 0.073 (2) | 0.0309 (16) | 0.0272 (16) | 0.0368 (17) |
| C25 | 0.074 (2) | 0.089 (2) | 0.096 (3) | 0.0395 (18) | 0.0331 (18) | 0.060 (2) |
| C26 | 0.0473 (15) | 0.115 (3) | 0.093 (2) | 0.0262 (17) | 0.0220 (15) | 0.071 (2) |
| C27 | 0.0668 (19) | 0.091 (2) | 0.076 (2) | 0.0125 (18) | 0.0062 (17) | 0.030 (2) |
| C28 | 0.0675 (18) | 0.073 (2) | 0.068 (2) | 0.0283 (16) | 0.0146 (15) | 0.0237 (17) |
| N1 | 0.0557 (12) | 0.0519 (12) | 0.0493 (12) | 0.0060 (9) | 0.0175 (10) | 0.0161 (10) |
| N2 | 0.0607 (13) | 0.0587 (13) | 0.0552 (13) | 0.0051 (10) | 0.0222 (10) | 0.0222 (11) |
| N3 | 0.0510 (11) | 0.0515 (12) | 0.0478 (12) | 0.0179 (9) | 0.0129 (9) | 0.0179 (9) |
| N4 | 0.0517 (12) | 0.0581 (13) | 0.0512 (12) | 0.0218 (10) | 0.0117 (10) | 0.0189 (10) |
| Cl1 | 0.1693 (10) | 0.0998 (7) | 0.1126 (8) | 0.0236 (6) | 0.0946 (8) | 0.0437 (6) |
| Cl2 | 0.0670 (5) | 0.1721 (10) | 0.1308 (9) | 0.0229 (6) | 0.0031 (5) | 0.1043 (8) |
Geometric parameters (Å, º) top
| C1—C2 | 1.372 (4) | C15—C20 | 1.382 (3) |
| C1—C6 | 1.392 (3) | C15—H11 | 0.93 (2) |
| C1—H1 | 0.97 (3) | C16—C17 | 1.356 (4) |
| C2—C3 | 1.375 (5) | C16—H12 | 0.95 (3) |
| C2—H2 | 0.94 (3) | C17—C18 | 1.370 (4) |
| C3—C4 | 1.365 (5) | C17—H13 | 0.92 (3) |
| C3—H3 | 0.94 (4) | C18—C19 | 1.371 (4) |
| C4—C5 | 1.373 (4) | C18—H14 | 0.97 (3) |
| C4—H4 | 0.97 (3) | C19—C20 | 1.380 (3) |
| C5—C6 | 1.392 (3) | C19—H15 | 0.94 (3) |
| C5—H5 | 0.94 (3) | C20—C21 | 1.473 (3) |
| C6—C7 | 1.467 (3) | C21—N3 | 1.286 (3) |
| C7—N1 | 1.290 (3) | C22—N4 | 1.276 (3) |
| C7—C21 | 1.505 (3) | C22—C23 | 1.457 (3) |
| C8—N2 | 1.269 (3) | C22—H16 | 0.97 (3) |
| C8—C9 | 1.454 (4) | C23—C28 | 1.374 (4) |
| C8—H6 | 0.94 (3) | C23—C24 | 1.381 (4) |
| C9—C10 | 1.384 (4) | C24—C25 | 1.378 (4) |
| C9—C14 | 1.397 (4) | C24—H17 | 0.96 (3) |
| C10—C11 | 1.365 (4) | C25—C26 | 1.358 (5) |
| C10—H7 | 0.95 (3) | C25—H18 | 0.98 (3) |
| C11—C12 | 1.387 (4) | C26—C27 | 1.369 (5) |
| C11—H8 | 0.96 (3) | C26—Cl2 | 1.739 (3) |
| C12—C13 | 1.364 (4) | C27—C28 | 1.377 (4) |
| C12—Cl1 | 1.734 (3) | C27—H19 | 0.97 (3) |
| C13—C14 | 1.377 (4) | C28—H20 | 0.85 (3) |
| C13—H9 | 0.98 (3) | N1—N2 | 1.409 (3) |
| C14—H10 | 0.93 (3) | N3—N4 | 1.413 (3) |
| C15—C16 | 1.380 (4) | | |
| | | |
| C2—C1—C6 | 120.3 (3) | C20—C15—H11 | 122.2 (14) |
| C2—C1—H1 | 123.2 (18) | C17—C16—C15 | 120.0 (3) |
| C6—C1—H1 | 116.5 (18) | C17—C16—H12 | 119.5 (18) |
| C1—C2—C3 | 120.4 (3) | C15—C16—H12 | 120.4 (18) |
| C1—C2—H2 | 113.9 (17) | C16—C17—C18 | 119.7 (3) |
| C3—C2—H2 | 125.6 (17) | C16—C17—H13 | 118.4 (18) |
| C4—C3—C2 | 120.4 (3) | C18—C17—H13 | 121.9 (18) |
| C4—C3—H3 | 120 (2) | C17—C18—C19 | 120.7 (3) |
| C2—C3—H3 | 120 (2) | C17—C18—H14 | 122.9 (17) |
| C3—C4—C5 | 119.6 (3) | C19—C18—H14 | 116.3 (17) |
| C3—C4—H4 | 126.1 (18) | C18—C19—C20 | 120.5 (3) |
| C5—C4—H4 | 114.2 (18) | C18—C19—H15 | 120.8 (18) |
| C4—C5—C6 | 121.3 (3) | C20—C19—H15 | 118.7 (18) |
| C4—C5—H5 | 122.8 (16) | C19—C20—C15 | 118.0 (2) |
| C6—C5—H5 | 115.8 (16) | C19—C20—C21 | 121.7 (2) |
| C1—C6—C5 | 118.0 (2) | C15—C20—C21 | 120.3 (2) |
| C1—C6—C7 | 121.7 (2) | N3—C21—C20 | 118.5 (2) |
| C5—C6—C7 | 120.3 (2) | N3—C21—C7 | 122.9 (2) |
| N1—C7—C6 | 119.5 (2) | C20—C21—C7 | 118.55 (18) |
| N1—C7—C21 | 121.9 (2) | N4—C22—C23 | 121.8 (3) |
| C6—C7—C21 | 118.4 (2) | N4—C22—H16 | 118.8 (13) |
| N2—C8—C9 | 121.7 (3) | C23—C22—H16 | 119.5 (13) |
| N2—C8—H6 | 119.6 (15) | C28—C23—C24 | 118.5 (3) |
| C9—C8—H6 | 118.6 (15) | C28—C23—C22 | 122.0 (2) |
| C10—C9—C14 | 118.4 (3) | C24—C23—C22 | 119.5 (3) |
| C10—C9—C8 | 120.9 (2) | C25—C24—C23 | 121.2 (3) |
| C14—C9—C8 | 120.6 (3) | C25—C24—H17 | 119.6 (15) |
| C11—C10—C9 | 121.5 (3) | C23—C24—H17 | 119.1 (16) |
| C11—C10—H7 | 118.4 (16) | C26—C25—C24 | 118.6 (3) |
| C9—C10—H7 | 120.1 (16) | C26—C25—H18 | 123.8 (17) |
| C10—C11—C12 | 118.8 (3) | C24—C25—H18 | 117.3 (18) |
| C10—C11—H8 | 125.3 (16) | C25—C26—C27 | 121.8 (3) |
| C12—C11—H8 | 115.9 (16) | C25—C26—Cl2 | 119.7 (2) |
| C13—C12—C11 | 121.3 (3) | C27—C26—Cl2 | 118.5 (3) |
| C13—C12—Cl1 | 120.0 (2) | C26—C27—C28 | 118.9 (3) |
| C11—C12—Cl1 | 118.7 (3) | C26—C27—H19 | 124.0 (19) |
| C12—C13—C14 | 119.5 (3) | C28—C27—H19 | 116.9 (19) |
| C12—C13—H9 | 123.6 (18) | C23—C28—C27 | 120.9 (3) |
| C14—C13—H9 | 116.6 (18) | C23—C28—H20 | 121 (2) |
| C13—C14—C9 | 120.4 (3) | C27—C28—H20 | 118 (2) |
| C13—C14—H10 | 119.4 (16) | C7—N1—N2 | 111.31 (19) |
| C9—C14—H10 | 120.2 (16) | C8—N2—N1 | 113.5 (2) |
| C16—C15—C20 | 121.1 (3) | C21—N3—N4 | 111.75 (19) |
| C16—C15—H11 | 116.6 (14) | C22—N4—N3 | 111.3 (2) |
Calculated reaction enthalpy changes (ΔH) (kJ mol-1) top| Schiff base | ΔHgas | ΔHcpcm |
| BDHMOCB | -2.588743 | -8.217815 |
| BDHOCB | -6.597881501 | -12.513133 |
| BDHMMCB | -0.855913 | -6.4561045 |
| BDHMCB | -5.159107501 | -10.6569045 |
| BDHMPCB | -0.2179165 | -5.854865 |
| BDHPCB | -4.7285255 | -10.0267845 |
Calculated reaction Gibbs free energy changes (ΔG) (kJ mol-1) top| Schiff base | ΔGgas | ΔGcpcm |
| BDHMOCB | 5.6527015 | 0.0918925 |
| BDHOCB | 3.549676 | -4.1509155 |
| BDHMMCB | 6.810547 | 0.5172235 |
| BDHMCB | 4.4607245 | 0.168032 |
| BDHMPCB | 7.82399 | 2.118778499 |
| BDHPCB | 3.62319 | -1.714451501 |
Energy gaps between HOMO and LUMO (ΔE) (kJ mol-1). top| Schiff base | ΔEgas | ΔEcpcm |
| BDHMOCB | 327.2685251 | 340.6848281 |
| BDHOCB | 377.8619024 | 375.1576378 |
| BDHMMCB | 322.1750559 | 343.6516426 |
| BDHMCB | 368.6989088 | 380.749952 |
| BDHMPCB | 322.9889608 | 338.6369384 |
| BDHPCB | 375.7352478 | 376.7329376 |
Selected π–π interaction geometry for BDHOCB. top| d is the centroid–centroid distance (Å),
α is the dihedral angle (α/°),
β is the slippage angle between the centroid–centroid distance
and the normal to plane I (°),
γ is the slippage angle between the centroid–centroid distance
and the normal to plane J (°),
e is the perpendicular distance of centroid I to
plane J (Å) and f is the perpendicular distance
of centroid J to plane I (Å). |
| π–π interactions (I→J) | d | α | β | γ | e | f |
| Cg1→Cg2i | 4.754 (2) | 66.28 | 10.69 | 75.49 | 1.191 | 4.671 |
| Cg1→Cg4ii | 4.108 (2) | 20.78 | 22.72 | 16.21 | 3.945 | 3.789 |
| Cg2→Cg3iii | 4.851 (2) | 72.63 | 9.82 | 65.95 | 1.977 | 4.780 |
| Cg2→Cg3iv | 5.467 (2) | 85.33 | 32.92 | 53.32 | 3.265 | 4.589 |
| Cg2→Cg4v | 5.222 (2) | 77.27 | 20.83 | 68.43 | 1.919 | 4.880 |
| Cg2→Cg4vi | 5.560 (3) | 29.58 | 50.81 | 63.42 | 2.488 | 3.514 |
| Cg3→Cg1vi | 5.673 (3) | 86.14 | 25.42 | 72.85 | 1.673 | 5.124 |
| Cg3→Cg3iii | 5.595 (3) | 0.02 | 53.86 | 53.86 | 3.299 | 3.299 |
| Cg3→Cg4ii | 4.834 (3) | 56.21 | 13.58 | 64.57 | 2.076 | 4.699 |
| Cg4→Cg4v | 4.595 (3) | 0.00 | 37.35 | 37.35 | 3.653 | 3.652 |
| Cg1 is the centroid of the C1–C6 ring,
Cg2 is the centroid of the C9–C14 ring,
Cg3 is the centroid of the C15–C20 ring and
Cg4 is the centroid of the C23–C28 ring.
Symmetry codes: (i) x-1/2, -y+1/2, z-1/2;
(ii) -x+3/2, y+1/2, -z+1/2;
(iii) -x+2, -y+1, -z+1;
(iv) x-1/2, -y+1/2, z+1/2;
(v) -x+1, -y, -z+1;
(vi) x+1/2, -y+1/2, z+1/2. |
Selected C—H···π interaction geometry for BDHOCB. top| H···Cg is the distance between a H atom and the centroid of the benzene
ring (Å), H-Perp is the perpendicular distance of a H atom to the benzene
plane (Å), γ is the slippage angle between the C—H distance and the normal
to the benzene plane (°), C—H···Cg is the angle of C—H with the
centroid (°) and C···Cg is the C–centroid distance (Å). |
| C—H···π interactions | H···Cg | H-Perp | γ | C—H···Cg | C···Cg |
| C13—H9···Cg1i | 2.77 (3) | 2.688 | 14.33 | 129.6 (18) | 3.559 (3) |
| C17—H13···Cg2ii | 3.11 (3) | 2.807 | 25.35 | 123.8 (18) | 3.756 (3) |
| C26—H18···Cg2iii | 3.20 (3) | 2.777 | 29.63 | 144 (3) | 3.984 (4) |
| C27—H19···Cg3iv | 3.02 (3) | 2.975 | 10.01 | 130 (3) | 3.668 (5) |
| Cg1 is the centroid of the C1–C6 ring,
Cg2 is the centroid of the C9–C14 ring and
Cg3 is the centroid of the C15–C20 ring.
Symmetry codes: (i) x+1/2, -y+1/2, z+1/2;
(ii) -x+2, -y+1,-z+1;
(iii) -x+1, -y, -z+1;
(iv) -x+3/2, y-1/2, -z+1/2. |
Selected π–π interaction geometry for BDHMCB. top| d is the centroid–centroid distance (Å),
α is the dihedral angle (α/°),
β is the slippage angle between the centroid–centroid distance
and the normal to plane I (°),
γ is the slippage angle between the centroid–centroid distance
and the normal to plane J (°),
e is the perpendicular distance of centroid I to
plane J (Å) and f is the perpendicular distance
of centroid J to plane I (Å). |
| π–π interactions (I→J) | d | α | β | γ | e | f |
| Cg1→Cg2i | 4.180 (2) | 12.79 | 37.25 | 26.49 | 3.741 | 3.327 |
| Cg2→Cg1ii | 5.158 (3) | 88.54 | 15.30 | 82.33 | 0.688 | 4.975 |
| Cg2→Cg2iii | 5.816 (3) | 88.77 | 54.70 | 49.80 | 3.754 | 3.361 |
| Cg1 is the centroid of the C1–C6 ring and
Cg2 is the centroid of the C9–C14 ring.
Symmetry codes: (i) -x, -y, -z+1;
(ii) -x+1/2, -y, z+1;
(iii) x-1/2, y, -z+2. |
Selected C—H···π interaction geometry for BDHMCB. top| H···Cg is the distance between a H atom and the centroid of the benzene
ring (Å), H-Perp is the perpendicular distance of a H atom to the benzene
plane (Å), γ is the slippage angle between the C—H distance and the normal
to the benzene plane (°), C—H···Cg is the angle of C—H with the
centroid (°) and C···Cg is the C–centroid distance (Å). |
| C—H···π interactions | H···Cg | H-Perp | γ | C—H···Cg | C···Cg |
| C4—H4···Cg2i | 2.90 (2) | 2.802 | 14.60 | 162.5 (17) | 3.826 (3) |
| Cg2 is the centroid of the C9—C14 ring.
Symmetry code: (i) -x+1/2, -y, z-1. |
Selected π–π interaction geometry for BDHPCB. top| d is the centroid–centroid distance (Å),
α is the dihedral angle (α/°),
β is the slippage angle between the centroid–centroid distance
and the normal to plane I (°),
γ is the slippage angle between the centroid–centroid distance
and the normal to plane J (°),
e is the perpendicular distance of centroid I to
plane J (Å) and f is the perpendicular distance
of centroid J to plane I (Å). |
| π–π interactions (I→J) | d | α | β | γ | e | f |
| Cg1→Cg2i | 5.459 (3) | 28.10 | 44.56 | 51.41 | 3.405 | 3.890 |
| Cg1→Cg2ii | 4.736 (3) | 28.10 | 39.65 | 29.26 | 4.132 | 3.646 |
| Cg2→Cg3iii | 5.244 (3) | 83.00 | 14.35 | 83.85 | 0.562 | 5.081 |
| Cg2→Cg4iv | 5.645 (4) | 87.63 | 34.55 | 61.16 | 2.723 | 4.649 |
| Cg2→Cg4v | 5.995 (4) | 87.63 | 50.21 | 59.96 | 3.001 | 3.836 |
| Cg3→Cg1ii | 5.828 (4) | 83.79 | 42.57 | 85.50 | 0.457 | 4.292 |
| Cg3→Cg4vi | 3.966 (3) | 5.48 | 26.81 | 22.14 | 3.674 | 3.540 |
| Cg4→Cg1iii | 5.437 (4) | 78.37 | 27.42 | 63.67 | 2.412 | 4.827 |
| Cg4→Cg4iv | 5.258 (3) | 0.00 | 53.25 | 53.25 | 3.146 | 3.146 |
Cg1 is the centroid of the C1–C6 ring,
Cg2 is the centroid of the C9–C14 ring,
Cg3 is the centroid of the C15–C20 ring and
Cg4 is the centroid of the C23–C28 ring.
Symmetry codes: (i) x, y, z+1;
(ii) -x+1, -y+1, -z+1;
(iii) x+1, y, z;
(iv) -x+2, -y, -z+1;
(v) -x+2, -y+1, -z+1;
(vi) -x+1, -y, -z+1. |
Selected C—H···π interaction geometry for BDHPCB. top| H···Cg is the distance between a H atom and the centroid of the benzene
ring
(Å), H-Perp is the perpendicular distance of a H atom to the benzene plane
(Å), γ is the slippage angle between the C—H distance and the normal to the
benzene plane (°), C—H···Cg is the angle of C—H with the centroid
(°) and
C···Cg is the C–centroid distance (Å). |
| C—H···π interactions | H···Cg | H-Perp | γ | C—H···Cg | C···Cg |
| C16—H12···Cg2i | 3.09 (3) | 2.920 | 19.19 | 150 (2) | 3.943 (4) |
| Cg2 is the centroid of the C9–C14 ring.
Symmetry code: (i) z-1, y, z. |
 π stacking interactions. The crystal systems are different, however, viz. triclinic (space group P
π stacking interactions. The crystal systems are different, however, viz. triclinic (space group P
 journal menu
journal menu











