The polymorphic study of 3-(3-phenyl-1
H-1,2,4-triazol-5-yl)-2
H-1-benzopyran-2-one, C
17H
11N
3O
2, was performed due to its potential biological activity and revealed three polymorphic modifications in the triclinic space group
P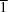
, the monoclinic space group
P2
1 and the orthorhombic space group
Pbca. These polymorphs have a one-column layered type of crystal organization. The strongest interactions between the molecules of the studied structures is stacking between π-systems, while N—H

N and C—H

O hydrogen bonds link stacked columns forming layers as a secondary basic structural motif. C—H

π hydrogen bonds were observed between neighbouring layers and their role is the least significant in the formation of the crystal structure. Packing differences between the polymorphic modifications are minor and can be identified only using an analysis based on a comparison of the pairwise interaction energies.
Supporting information
CCDC references: 1896591; 1896590; 1896589
For all structures, data collection: CrysAlis PRO (Rigaku OD, 2018); cell refinement: CrysAlis PRO (Rigaku OD, 2018); data reduction: CrysAlis PRO (Rigaku OD, 2018); program(s) used to solve structure: SHELXT2014 (Sheldrick, 2015a); program(s) used to refine structure: SHELXL2016 (Sheldrick, 2015b); software used to prepare material for publication: SHELXL2016 (Sheldrick, 2015b).
3-(3-Phenyl-1
H-1,2,4-triazol-5-yl)-2
H-1-benzopyran-2-one (1t)
top
Crystal data top
| C17H11N3O2 | Z = 4 |
| Mr = 289.29 | F(000) = 600 |
| Triclinic, P1 | Dx = 1.391 Mg m−3 |
| a = 6.8415 (7) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 10.8931 (12) Å | Cell parameters from 1138 reflections |
| c = 20.163 (2) Å | θ = 3.3–21.2° |
| α = 74.965 (9)° | µ = 0.09 mm−1 |
| β = 83.815 (9)° | T = 293 K |
| γ = 72.183 (10)° | Plate, colorless |
| V = 1380.9 (3) Å3 | 0.20 × 0.10 × 0.06 mm |
Data collection top
Rigaku Xcalibur Sapphire3
diffractometer | 4841 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2514 reflections with I > 2σ(I) |
| Detector resolution: 16.1827 pixels mm-1 | Rint = 0.057 |
| ω scans | θmax = 25.0°, θmin = 3.1° |
Absorption correction: multi-scan
(CrysAlis PRO; Rigaku OD, 2018) | h = −7→8 |
| Tmin = 0.347, Tmax = 1.000 | k = −8→12 |
| 10538 measured reflections | l = −22→23 |
Refinement top
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.058 | w = 1/[σ2(Fo2) + (0.0307P)2]
where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.136 | (Δ/σ)max < 0.001 |
| S = 0.97 | Δρmax = 0.17 e Å−3 |
| 4841 reflections | Δρmin = −0.16 e Å−3 |
| 398 parameters | Extinction correction: SHELXL2016 (Sheldrick, 2015b), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.0023 (6) |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1A | 0.4008 (3) | 1.0215 (2) | −0.11353 (10) | 0.0755 (6) | |
| O2A | 0.3009 (3) | 0.8450 (2) | −0.06352 (12) | 0.0844 (7) | |
| N1A | 0.5679 (3) | 0.6233 (2) | 0.01294 (12) | 0.0669 (7) | |
| H1NA | 0.452422 | 0.643077 | −0.006324 | 0.080* | |
| N2A | 0.6610 (4) | 0.5033 (2) | 0.05329 (12) | 0.0695 (7) | |
| N3A | 0.8452 (4) | 0.6478 (2) | 0.04421 (11) | 0.0609 (6) | |
| C1A | 0.5422 (5) | 1.0932 (3) | −0.12559 (15) | 0.0672 (8) | |
| C2A | 0.4889 (6) | 1.2166 (3) | −0.17011 (18) | 0.0882 (10) | |
| H2A | 0.362055 | 1.249931 | −0.190797 | 0.106* | |
| C3A | 0.6255 (6) | 1.2893 (3) | −0.18339 (18) | 0.0954 (11) | |
| H3A | 0.591601 | 1.373311 | −0.213079 | 0.115* | |
| C4A | 0.8149 (6) | 1.2384 (3) | −0.15272 (19) | 0.0914 (11) | |
| H4A | 0.908863 | 1.287358 | −0.162915 | 0.110* | |
| C5A | 0.8643 (5) | 1.1164 (3) | −0.10757 (16) | 0.0775 (9) | |
| H5A | 0.989783 | 1.084682 | −0.086106 | 0.093* | |
| C6A | 0.7292 (5) | 1.0394 (3) | −0.09335 (15) | 0.0633 (8) | |
| C7A | 0.7706 (4) | 0.9107 (3) | −0.04805 (14) | 0.0612 (8) | |
| H7A | 0.895536 | 0.873615 | −0.026228 | 0.073* | |
| C8A | 0.6323 (4) | 0.8416 (3) | −0.03624 (14) | 0.0564 (7) | |
| C9A | 0.4363 (5) | 0.8975 (3) | −0.06980 (15) | 0.0648 (8) | |
| C10A | 0.6806 (4) | 0.7074 (3) | 0.00711 (14) | 0.0592 (7) | |
| C11A | 0.8277 (4) | 0.5232 (3) | 0.07162 (14) | 0.0610 (7) | |
| C12A | 0.9789 (4) | 0.4179 (3) | 0.11695 (14) | 0.0613 (8) | |
| C13A | 1.1602 (5) | 0.4372 (3) | 0.12948 (15) | 0.0707 (9) | |
| H13A | 1.187706 | 0.517019 | 0.108194 | 0.085* | |
| C14A | 1.3004 (5) | 0.3405 (4) | 0.17286 (17) | 0.0862 (10) | |
| H14A | 1.421697 | 0.355065 | 0.181079 | 0.103* | |
| C15A | 1.2616 (6) | 0.2224 (4) | 0.20406 (17) | 0.0924 (11) | |
| H15A | 1.355660 | 0.156618 | 0.233803 | 0.111* | |
| C16A | 1.0815 (6) | 0.2017 (3) | 0.19100 (17) | 0.0872 (10) | |
| H16A | 1.055369 | 0.121255 | 0.211950 | 0.105* | |
| C17A | 0.9412 (5) | 0.2974 (3) | 0.14779 (15) | 0.0710 (8) | |
| H17A | 0.821098 | 0.282036 | 0.139096 | 0.085* | |
| O1B | 1.1426 (3) | 0.07088 (19) | 0.38985 (10) | 0.0714 (6) | |
| O2B | 1.2224 (3) | 0.2095 (2) | 0.43673 (12) | 0.0863 (7) | |
| N1B | 0.9317 (4) | 0.3641 (2) | 0.51047 (12) | 0.0688 (7) | |
| H1NB | 1.050368 | 0.364303 | 0.491496 | 0.083* | |
| N2B | 0.8274 (4) | 0.4445 (2) | 0.55156 (13) | 0.0713 (7) | |
| N3B | 0.6506 (3) | 0.3076 (2) | 0.54006 (11) | 0.0604 (6) | |
| C1B | 1.0030 (5) | 0.0137 (3) | 0.37562 (15) | 0.0644 (8) | |
| C2B | 1.0713 (5) | −0.0736 (3) | 0.33325 (16) | 0.0780 (9) | |
| H2B | 1.206841 | −0.094192 | 0.316857 | 0.094* | |
| C3B | 0.9352 (6) | −0.1288 (3) | 0.31596 (18) | 0.0934 (11) | |
| H3B | 0.978834 | −0.188075 | 0.287777 | 0.112* | |
| C4B | 0.7326 (6) | −0.0971 (4) | 0.3401 (2) | 0.0991 (12) | |
| H4B | 0.639632 | −0.132390 | 0.326751 | 0.119* | |
| C5B | 0.6702 (5) | −0.0141 (3) | 0.38361 (17) | 0.0852 (10) | |
| H5B | 0.535829 | 0.003963 | 0.401188 | 0.102* | |
| C6B | 0.8052 (5) | 0.0439 (3) | 0.40202 (15) | 0.0648 (8) | |
| C7B | 0.7506 (4) | 0.1332 (3) | 0.44635 (14) | 0.0621 (8) | |
| H7B | 0.618526 | 0.152490 | 0.465850 | 0.074* | |
| C8B | 0.8843 (4) | 0.1899 (3) | 0.46063 (14) | 0.0574 (7) | |
| C9B | 1.0904 (5) | 0.1609 (3) | 0.43024 (15) | 0.0653 (8) | |
| C10B | 0.8242 (4) | 0.2840 (3) | 0.50338 (14) | 0.0600 (7) | |
| C11B | 0.6592 (4) | 0.4040 (3) | 0.56920 (14) | 0.0605 (8) | |
| C12B | 0.4965 (4) | 0.4635 (3) | 0.61495 (14) | 0.0613 (8) | |
| C13B | 0.3188 (5) | 0.4260 (3) | 0.62550 (15) | 0.0720 (9) | |
| H13B | 0.303394 | 0.364430 | 0.603522 | 0.086* | |
| C14B | 0.1630 (5) | 0.4793 (4) | 0.66854 (17) | 0.0835 (10) | |
| H14B | 0.042410 | 0.454503 | 0.674850 | 0.100* | |
| C15B | 0.1858 (5) | 0.5685 (3) | 0.70191 (16) | 0.0843 (10) | |
| H15B | 0.082003 | 0.603024 | 0.731557 | 0.101* | |
| C16B | 0.3613 (5) | 0.6064 (3) | 0.69148 (16) | 0.0784 (9) | |
| H16B | 0.376217 | 0.667153 | 0.714101 | 0.094* | |
| C17B | 0.5166 (5) | 0.5558 (3) | 0.64788 (15) | 0.0688 (8) | |
| H17B | 0.634646 | 0.583486 | 0.640570 | 0.083* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1A | 0.0653 (13) | 0.0665 (14) | 0.0835 (15) | −0.0053 (12) | −0.0173 (11) | −0.0093 (12) |
| O2A | 0.0616 (13) | 0.0792 (15) | 0.1125 (18) | −0.0194 (13) | −0.0164 (13) | −0.0188 (13) |
| N1A | 0.0595 (15) | 0.0604 (16) | 0.0763 (17) | −0.0141 (14) | −0.0160 (13) | −0.0072 (13) |
| N2A | 0.0629 (16) | 0.0641 (16) | 0.0764 (17) | −0.0157 (14) | −0.0156 (14) | −0.0059 (14) |
| N3A | 0.0604 (14) | 0.0582 (15) | 0.0637 (15) | −0.0155 (13) | −0.0056 (12) | −0.0142 (12) |
| C1A | 0.0650 (19) | 0.059 (2) | 0.071 (2) | −0.0097 (17) | 0.0001 (17) | −0.0148 (17) |
| C2A | 0.087 (2) | 0.071 (2) | 0.090 (3) | −0.006 (2) | −0.011 (2) | −0.006 (2) |
| C3A | 0.103 (3) | 0.057 (2) | 0.096 (3) | 0.003 (2) | −0.002 (2) | 0.003 (2) |
| C4A | 0.103 (3) | 0.060 (2) | 0.098 (3) | −0.018 (2) | 0.015 (2) | −0.010 (2) |
| C5A | 0.076 (2) | 0.071 (2) | 0.084 (2) | −0.021 (2) | 0.0015 (19) | −0.0177 (19) |
| C6A | 0.0641 (19) | 0.0538 (18) | 0.0661 (19) | −0.0084 (16) | −0.0031 (16) | −0.0142 (15) |
| C7A | 0.0559 (17) | 0.0601 (19) | 0.0645 (19) | −0.0084 (16) | −0.0059 (15) | −0.0179 (15) |
| C8A | 0.0497 (16) | 0.0532 (17) | 0.0597 (17) | −0.0032 (14) | −0.0088 (14) | −0.0132 (14) |
| C9A | 0.0615 (19) | 0.059 (2) | 0.065 (2) | −0.0039 (17) | −0.0067 (16) | −0.0146 (16) |
| C10A | 0.0541 (17) | 0.0612 (19) | 0.0636 (18) | −0.0142 (16) | −0.0032 (15) | −0.0199 (15) |
| C11A | 0.0585 (18) | 0.066 (2) | 0.0578 (18) | −0.0162 (16) | −0.0007 (15) | −0.0156 (15) |
| C12A | 0.0608 (18) | 0.065 (2) | 0.0572 (18) | −0.0134 (17) | −0.0044 (15) | −0.0181 (15) |
| C13A | 0.070 (2) | 0.077 (2) | 0.063 (2) | −0.0149 (19) | −0.0051 (17) | −0.0178 (17) |
| C14A | 0.074 (2) | 0.102 (3) | 0.075 (2) | −0.008 (2) | −0.017 (2) | −0.024 (2) |
| C15A | 0.093 (3) | 0.084 (3) | 0.074 (2) | 0.012 (2) | −0.014 (2) | −0.015 (2) |
| C16A | 0.114 (3) | 0.065 (2) | 0.072 (2) | −0.014 (2) | −0.005 (2) | −0.0108 (18) |
| C17A | 0.080 (2) | 0.063 (2) | 0.066 (2) | −0.0149 (19) | −0.0029 (18) | −0.0142 (17) |
| O1B | 0.0631 (13) | 0.0677 (14) | 0.0782 (14) | −0.0154 (12) | 0.0082 (11) | −0.0171 (12) |
| O2B | 0.0588 (13) | 0.0865 (16) | 0.1194 (19) | −0.0226 (13) | 0.0081 (13) | −0.0373 (14) |
| N1B | 0.0552 (14) | 0.0686 (17) | 0.0802 (18) | −0.0156 (14) | 0.0028 (13) | −0.0186 (14) |
| N2B | 0.0624 (16) | 0.0705 (18) | 0.0827 (18) | −0.0167 (15) | 0.0039 (14) | −0.0266 (15) |
| N3B | 0.0555 (15) | 0.0575 (15) | 0.0655 (15) | −0.0142 (13) | 0.0002 (13) | −0.0135 (13) |
| C1B | 0.0649 (19) | 0.0537 (19) | 0.068 (2) | −0.0117 (17) | −0.0022 (16) | −0.0096 (16) |
| C2B | 0.088 (2) | 0.061 (2) | 0.075 (2) | −0.010 (2) | 0.0060 (19) | −0.0140 (17) |
| C3B | 0.117 (3) | 0.067 (2) | 0.096 (3) | −0.015 (2) | −0.008 (3) | −0.030 (2) |
| C4B | 0.094 (3) | 0.086 (3) | 0.126 (3) | −0.016 (2) | −0.018 (3) | −0.047 (3) |
| C5B | 0.076 (2) | 0.082 (2) | 0.105 (3) | −0.020 (2) | −0.007 (2) | −0.036 (2) |
| C6B | 0.0608 (19) | 0.0558 (19) | 0.070 (2) | −0.0073 (16) | −0.0064 (16) | −0.0114 (16) |
| C7B | 0.0531 (17) | 0.0566 (18) | 0.0660 (19) | −0.0069 (15) | 0.0000 (15) | −0.0081 (15) |
| C8B | 0.0465 (15) | 0.0540 (18) | 0.0627 (18) | −0.0092 (15) | 0.0001 (14) | −0.0058 (15) |
| C9B | 0.0627 (19) | 0.0520 (19) | 0.074 (2) | −0.0093 (17) | −0.0034 (17) | −0.0116 (16) |
| C10B | 0.0537 (17) | 0.0551 (18) | 0.0673 (19) | −0.0115 (15) | −0.0049 (16) | −0.0118 (15) |
| C11B | 0.0555 (17) | 0.0567 (19) | 0.0643 (19) | −0.0133 (16) | −0.0113 (15) | −0.0059 (15) |
| C12B | 0.0582 (18) | 0.0625 (19) | 0.0560 (17) | −0.0108 (16) | −0.0017 (15) | −0.0100 (15) |
| C13B | 0.070 (2) | 0.076 (2) | 0.071 (2) | −0.0217 (19) | 0.0016 (17) | −0.0193 (17) |
| C14B | 0.068 (2) | 0.100 (3) | 0.080 (2) | −0.024 (2) | 0.0085 (19) | −0.022 (2) |
| C15B | 0.073 (2) | 0.098 (3) | 0.067 (2) | −0.001 (2) | 0.0042 (19) | −0.025 (2) |
| C16B | 0.086 (2) | 0.074 (2) | 0.069 (2) | −0.004 (2) | −0.010 (2) | −0.0251 (18) |
| C17B | 0.070 (2) | 0.061 (2) | 0.070 (2) | −0.0127 (18) | −0.0129 (18) | −0.0106 (17) |
Geometric parameters (Å, º) top
| O1A—C9A | 1.374 (3) | O1B—C9B | 1.374 (3) |
| O1A—C1A | 1.386 (3) | O1B—C1B | 1.379 (3) |
| O2A—C9A | 1.209 (3) | O2B—C9B | 1.212 (3) |
| N1A—C10A | 1.343 (3) | N1B—C10B | 1.342 (3) |
| N1A—N2A | 1.351 (3) | N1B—N2B | 1.352 (3) |
| N1A—H1NA | 0.8600 | N1B—H1NB | 0.8600 |
| N2A—C11A | 1.329 (3) | N2B—C11B | 1.336 (3) |
| N3A—C10A | 1.316 (3) | N3B—C10B | 1.321 (3) |
| N3A—C11A | 1.363 (3) | N3B—C11B | 1.347 (3) |
| C1A—C2A | 1.375 (4) | C1B—C6B | 1.374 (4) |
| C1A—C6A | 1.388 (4) | C1B—C2B | 1.384 (4) |
| C2A—C3A | 1.365 (4) | C2B—C3B | 1.368 (4) |
| C2A—H2A | 0.9300 | C2B—H2B | 0.9300 |
| C3A—C4A | 1.387 (4) | C3B—C4B | 1.388 (5) |
| C3A—H3A | 0.9300 | C3B—H3B | 0.9300 |
| C4A—C5A | 1.368 (4) | C4B—C5B | 1.365 (4) |
| C4A—H4A | 0.9300 | C4B—H4B | 0.9300 |
| C5A—C6A | 1.390 (3) | C5B—C6B | 1.394 (3) |
| C5A—H5A | 0.9300 | C5B—H5B | 0.9300 |
| C6A—C7A | 1.425 (4) | C6B—C7B | 1.429 (4) |
| C7A—C8A | 1.346 (3) | C7B—C8B | 1.341 (3) |
| C7A—H7A | 0.9300 | C7B—H7B | 0.9300 |
| C8A—C9A | 1.452 (4) | C8B—C10B | 1.445 (4) |
| C8A—C10A | 1.454 (4) | C8B—C9B | 1.451 (4) |
| C11A—C12A | 1.468 (4) | C11B—C12B | 1.469 (4) |
| C12A—C13A | 1.378 (3) | C12B—C13B | 1.376 (3) |
| C12A—C17A | 1.387 (3) | C12B—C17B | 1.386 (4) |
| C13A—C14A | 1.371 (4) | C13B—C14B | 1.382 (4) |
| C13A—H13A | 0.9300 | C13B—H13B | 0.9300 |
| C14A—C15A | 1.369 (4) | C14B—C15B | 1.369 (4) |
| C14A—H14A | 0.9300 | C14B—H14B | 0.9300 |
| C15A—C16A | 1.382 (4) | C15B—C16B | 1.364 (4) |
| C15A—H15A | 0.9300 | C15B—H15B | 0.9300 |
| C16A—C17A | 1.365 (4) | C16B—C17B | 1.378 (4) |
| C16A—H16A | 0.9300 | C16B—H16B | 0.9300 |
| C17A—H17A | 0.9300 | C17B—H17B | 0.9300 |
| | | |
| C9A—O1A—C1A | 122.7 (2) | C9B—O1B—C1B | 121.8 (2) |
| C10A—N1A—N2A | 110.6 (2) | C10B—N1B—N2B | 110.6 (2) |
| C10A—N1A—H1NA | 124.7 | C10B—N1B—H1NB | 124.7 |
| N2A—N1A—H1NA | 124.7 | N2B—N1B—H1NB | 124.7 |
| C11A—N2A—N1A | 102.1 (2) | C11B—N2B—N1B | 101.9 (2) |
| C10A—N3A—C11A | 103.4 (2) | C10B—N3B—C11B | 103.9 (2) |
| C2A—C1A—O1A | 117.4 (3) | C6B—C1B—O1B | 120.9 (3) |
| C2A—C1A—C6A | 122.8 (3) | C6B—C1B—C2B | 122.2 (3) |
| O1A—C1A—C6A | 119.8 (3) | O1B—C1B—C2B | 116.9 (3) |
| C3A—C2A—C1A | 118.7 (3) | C3B—C2B—C1B | 118.6 (3) |
| C3A—C2A—H2A | 120.6 | C3B—C2B—H2B | 120.7 |
| C1A—C2A—H2A | 120.6 | C1B—C2B—H2B | 120.7 |
| C2A—C3A—C4A | 120.2 (3) | C2B—C3B—C4B | 120.6 (4) |
| C2A—C3A—H3A | 119.9 | C2B—C3B—H3B | 119.7 |
| C4A—C3A—H3A | 119.9 | C4B—C3B—H3B | 119.7 |
| C5A—C4A—C3A | 120.4 (3) | C5B—C4B—C3B | 119.8 (3) |
| C5A—C4A—H4A | 119.8 | C5B—C4B—H4B | 120.1 |
| C3A—C4A—H4A | 119.8 | C3B—C4B—H4B | 120.1 |
| C4A—C5A—C6A | 120.8 (3) | C4B—C5B—C6B | 120.9 (3) |
| C4A—C5A—H5A | 119.6 | C4B—C5B—H5B | 119.5 |
| C6A—C5A—H5A | 119.6 | C6B—C5B—H5B | 119.5 |
| C1A—C6A—C5A | 117.0 (3) | C1B—C6B—C5B | 117.9 (3) |
| C1A—C6A—C7A | 118.7 (3) | C1B—C6B—C7B | 118.0 (3) |
| C5A—C6A—C7A | 124.3 (3) | C5B—C6B—C7B | 124.2 (3) |
| C8A—C7A—C6A | 121.2 (3) | C8B—C7B—C6B | 121.9 (3) |
| C8A—C7A—H7A | 119.4 | C8B—C7B—H7B | 119.1 |
| C6A—C7A—H7A | 119.4 | C6B—C7B—H7B | 119.1 |
| C7A—C8A—C9A | 120.3 (3) | C7B—C8B—C10B | 121.2 (3) |
| C7A—C8A—C10A | 120.8 (2) | C7B—C8B—C9B | 119.5 (3) |
| C9A—C8A—C10A | 118.9 (2) | C10B—C8B—C9B | 119.3 (3) |
| O2A—C9A—O1A | 116.4 (3) | O2B—C9B—O1B | 116.2 (3) |
| O2A—C9A—C8A | 126.3 (3) | O2B—C9B—C8B | 126.0 (3) |
| O1A—C9A—C8A | 117.3 (3) | O1B—C9B—C8B | 117.8 (3) |
| N3A—C10A—N1A | 109.6 (2) | N3B—C10B—N1B | 109.3 (3) |
| N3A—C10A—C8A | 125.4 (2) | N3B—C10B—C8B | 125.0 (3) |
| N1A—C10A—C8A | 124.9 (2) | N1B—C10B—C8B | 125.7 (3) |
| N2A—C11A—N3A | 114.2 (3) | N2B—C11B—N3B | 114.3 (3) |
| N2A—C11A—C12A | 122.0 (3) | N2B—C11B—C12B | 122.1 (3) |
| N3A—C11A—C12A | 123.8 (2) | N3B—C11B—C12B | 123.7 (3) |
| C13A—C12A—C17A | 119.1 (3) | C13B—C12B—C17B | 119.1 (3) |
| C13A—C12A—C11A | 120.6 (3) | C13B—C12B—C11B | 118.8 (3) |
| C17A—C12A—C11A | 120.3 (3) | C17B—C12B—C11B | 122.1 (3) |
| C14A—C13A—C12A | 121.0 (3) | C12B—C13B—C14B | 120.4 (3) |
| C14A—C13A—H13A | 119.5 | C12B—C13B—H13B | 119.8 |
| C12A—C13A—H13A | 119.5 | C14B—C13B—H13B | 119.8 |
| C15A—C14A—C13A | 119.9 (3) | C15B—C14B—C13B | 120.1 (3) |
| C15A—C14A—H14A | 120.1 | C15B—C14B—H14B | 119.9 |
| C13A—C14A—H14A | 120.1 | C13B—C14B—H14B | 119.9 |
| C14A—C15A—C16A | 119.5 (3) | C16B—C15B—C14B | 119.7 (3) |
| C14A—C15A—H15A | 120.3 | C16B—C15B—H15B | 120.1 |
| C16A—C15A—H15A | 120.3 | C14B—C15B—H15B | 120.1 |
| C17A—C16A—C15A | 121.0 (3) | C15B—C16B—C17B | 120.8 (3) |
| C17A—C16A—H16A | 119.5 | C15B—C16B—H16B | 119.6 |
| C15A—C16A—H16A | 119.5 | C17B—C16B—H16B | 119.6 |
| C16A—C17A—C12A | 119.6 (3) | C16B—C17B—C12B | 119.8 (3) |
| C16A—C17A—H17A | 120.2 | C16B—C17B—H17B | 120.1 |
| C12A—C17A—H17A | 120.2 | C12B—C17B—H17B | 120.1 |
| | | |
| C10A—N1A—N2A—C11A | 1.6 (3) | C10B—N1B—N2B—C11B | 1.8 (3) |
| C9A—O1A—C1A—C2A | −179.6 (3) | C9B—O1B—C1B—C6B | −0.6 (4) |
| C9A—O1A—C1A—C6A | 0.7 (4) | C9B—O1B—C1B—C2B | 178.7 (2) |
| O1A—C1A—C2A—C3A | −179.6 (3) | C6B—C1B—C2B—C3B | 1.5 (5) |
| C6A—C1A—C2A—C3A | 0.2 (5) | O1B—C1B—C2B—C3B | −177.7 (2) |
| C1A—C2A—C3A—C4A | 0.5 (6) | C1B—C2B—C3B—C4B | 0.4 (5) |
| C2A—C3A—C4A—C5A | −1.7 (6) | C2B—C3B—C4B—C5B | −2.5 (6) |
| C3A—C4A—C5A—C6A | 2.2 (6) | C3B—C4B—C5B—C6B | 2.6 (6) |
| C2A—C1A—C6A—C5A | 0.3 (5) | O1B—C1B—C6B—C5B | 177.8 (2) |
| O1A—C1A—C6A—C5A | −180.0 (3) | C2B—C1B—C6B—C5B | −1.4 (4) |
| C2A—C1A—C6A—C7A | −180.0 (3) | O1B—C1B—C6B—C7B | −1.9 (4) |
| O1A—C1A—C6A—C7A | −0.3 (4) | C2B—C1B—C6B—C7B | 178.8 (3) |
| C4A—C5A—C6A—C1A | −1.5 (5) | C4B—C5B—C6B—C1B | −0.7 (5) |
| C4A—C5A—C6A—C7A | 178.8 (3) | C4B—C5B—C6B—C7B | 179.0 (3) |
| C1A—C6A—C7A—C8A | −0.1 (4) | C1B—C6B—C7B—C8B | 2.1 (4) |
| C5A—C6A—C7A—C8A | 179.6 (3) | C5B—C6B—C7B—C8B | −177.7 (3) |
| C6A—C7A—C8A—C9A | 0.0 (4) | C6B—C7B—C8B—C10B | 177.5 (2) |
| C6A—C7A—C8A—C10A | 176.6 (3) | C6B—C7B—C8B—C9B | 0.2 (4) |
| C1A—O1A—C9A—O2A | 179.9 (3) | C1B—O1B—C9B—O2B | −176.7 (2) |
| C1A—O1A—C9A—C8A | −0.7 (4) | C1B—O1B—C9B—C8B | 2.9 (4) |
| C7A—C8A—C9A—O2A | 179.7 (3) | C7B—C8B—C9B—O2B | 176.8 (3) |
| C10A—C8A—C9A—O2A | 3.0 (5) | C10B—C8B—C9B—O2B | −0.5 (4) |
| C7A—C8A—C9A—O1A | 0.3 (4) | C7B—C8B—C9B—O1B | −2.7 (4) |
| C10A—C8A—C9A—O1A | −176.3 (2) | C10B—C8B—C9B—O1B | 180.0 (2) |
| C11A—N3A—C10A—N1A | 1.0 (3) | C11B—N3B—C10B—N1B | −0.2 (3) |
| C11A—N3A—C10A—C8A | −176.3 (3) | C11B—N3B—C10B—C8B | −178.2 (3) |
| N2A—N1A—C10A—N3A | −1.7 (3) | N2B—N1B—C10B—N3B | −1.1 (3) |
| N2A—N1A—C10A—C8A | 175.6 (3) | N2B—N1B—C10B—C8B | 176.9 (3) |
| C7A—C8A—C10A—N3A | 8.3 (5) | C7B—C8B—C10B—N3B | 10.8 (4) |
| C9A—C8A—C10A—N3A | −175.1 (3) | C9B—C8B—C10B—N3B | −171.9 (3) |
| C7A—C8A—C10A—N1A | −168.6 (3) | C7B—C8B—C10B—N1B | −166.9 (2) |
| C9A—C8A—C10A—N1A | 8.0 (4) | C9B—C8B—C10B—N1B | 10.4 (4) |
| N1A—N2A—C11A—N3A | −1.0 (3) | N1B—N2B—C11B—N3B | −2.0 (3) |
| N1A—N2A—C11A—C12A | 179.7 (3) | N1B—N2B—C11B—C12B | 179.7 (2) |
| C10A—N3A—C11A—N2A | 0.0 (3) | C10B—N3B—C11B—N2B | 1.4 (3) |
| C10A—N3A—C11A—C12A | 179.3 (3) | C10B—N3B—C11B—C12B | 179.7 (3) |
| N2A—C11A—C12A—C13A | 173.4 (3) | N2B—C11B—C12B—C13B | 173.7 (3) |
| N3A—C11A—C12A—C13A | −5.9 (4) | N3B—C11B—C12B—C13B | −4.5 (4) |
| N2A—C11A—C12A—C17A | −6.6 (4) | N2B—C11B—C12B—C17B | −6.5 (4) |
| N3A—C11A—C12A—C17A | 174.1 (3) | N3B—C11B—C12B—C17B | 175.4 (3) |
| C17A—C12A—C13A—C14A | −1.4 (5) | C17B—C12B—C13B—C14B | −0.3 (4) |
| C11A—C12A—C13A—C14A | 178.6 (3) | C11B—C12B—C13B—C14B | 179.6 (3) |
| C12A—C13A—C14A—C15A | 0.4 (5) | C12B—C13B—C14B—C15B | −1.0 (5) |
| C13A—C14A—C15A—C16A | 0.5 (5) | C13B—C14B—C15B—C16B | 1.2 (5) |
| C14A—C15A—C16A—C17A | −0.4 (5) | C14B—C15B—C16B—C17B | −0.2 (5) |
| C15A—C16A—C17A—C12A | −0.6 (5) | C15B—C16B—C17B—C12B | −1.1 (5) |
| C13A—C12A—C17A—C16A | 1.4 (5) | C13B—C12B—C17B—C16B | 1.3 (4) |
| C11A—C12A—C17A—C16A | −178.6 (3) | C11B—C12B—C17B—C16B | −178.5 (3) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1A—H1NA···O2A | 0.86 | 2.19 | 2.742 (3) | 122 |
| N1B—H1NB···O2B | 0.86 | 2.22 | 2.760 (3) | 121 |
3-(3-Phenyl-1
H-1,2,4-triazol-5-yl)-2
H-1-benzopyran-2-one (1m)
top
Crystal data top
| C17H11N3O2 | F(000) = 600 |
| Mr = 289.29 | Dx = 1.391 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.71073 Å |
| a = 6.8355 (11) Å | Cell parameters from 881 reflections |
| b = 19.512 (3) Å | θ = 3.7–22.3° |
| c = 10.881 (2) Å | µ = 0.09 mm−1 |
| β = 107.811 (19)° | T = 293 K |
| V = 1381.7 (4) Å3 | Stick, colorless |
| Z = 4 | 0.30 × 0.05 × 0.05 mm |
Data collection top
Rigaku Xcalibur Sapphire3
diffractometer | 4818 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2351 reflections with I > 2σ(I) |
| Detector resolution: 16.1827 pixels mm-1 | Rint = 0.080 |
| ω scans | θmax = 25.0°, θmin = 3.1° |
Absorption correction: multi-scan
(CrysAlis PRO; Rigaku OD, 2018) | h = −8→7 |
| Tmin = 0.283, Tmax = 1.000 | k = −23→23 |
| 10615 measured reflections | l = −12→10 |
Refinement top
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.093 | w = 1/[σ2(Fo2) + (0.1063P)2]
where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.263 | (Δ/σ)max < 0.001 |
| S = 1.01 | Δρmax = 0.39 e Å−3 |
| 4818 reflections | Δρmin = −0.22 e Å−3 |
| 397 parameters | Absolute structure: Flack x determined using 637 quotients [(I+)-(I-)]/[(I+)+(I-)]
(Parsons et al., 2013) |
| 1 restraint | Absolute structure parameter: −3.5 (10) |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1A | 0.8453 (10) | 0.6138 (4) | 1.0861 (7) | 0.082 (2) | |
| O2A | 0.9366 (11) | 0.5625 (5) | 0.9325 (7) | 0.094 (2) | |
| N1A | 0.6589 (13) | 0.4870 (4) | 0.7457 (8) | 0.074 (2) | |
| H1NA | 0.775813 | 0.506193 | 0.754747 | 0.089* | |
| N2A | 0.5582 (13) | 0.4461 (5) | 0.6445 (8) | 0.072 (2) | |
| N3A | 0.3806 (12) | 0.4562 (4) | 0.7893 (7) | 0.064 (2) | |
| C1A | 0.7060 (17) | 0.6257 (6) | 1.1524 (10) | 0.074 (3) | |
| C2A | 0.758 (2) | 0.6718 (7) | 1.2517 (12) | 0.101 (4) | |
| H2A | 0.886354 | 0.692983 | 1.273921 | 0.122* | |
| C3A | 0.625 (2) | 0.6869 (8) | 1.3184 (13) | 0.108 (5) | |
| H3A | 0.661099 | 0.718378 | 1.385665 | 0.129* | |
| C4A | 0.429 (2) | 0.6544 (8) | 1.2851 (13) | 0.107 (4) | |
| H4A | 0.337575 | 0.663186 | 1.331389 | 0.128* | |
| C5A | 0.3787 (18) | 0.6099 (6) | 1.1832 (10) | 0.079 (3) | |
| H5A | 0.250182 | 0.589048 | 1.157967 | 0.095* | |
| C6A | 0.5152 (15) | 0.5959 (6) | 1.1185 (9) | 0.071 (3) | |
| C7A | 0.4705 (15) | 0.5486 (5) | 1.0072 (9) | 0.069 (3) | |
| H7A | 0.345003 | 0.525819 | 0.982161 | 0.082* | |
| C8A | 0.6021 (14) | 0.5372 (5) | 0.9412 (9) | 0.065 (3) | |
| C9A | 0.8056 (15) | 0.5695 (5) | 0.9840 (10) | 0.069 (3) | |
| C10A | 0.5496 (14) | 0.4934 (5) | 0.8300 (8) | 0.059 (2) | |
| C11A | 0.3914 (15) | 0.4286 (5) | 0.6781 (9) | 0.067 (3) | |
| C12A | 0.2328 (15) | 0.3836 (5) | 0.5939 (9) | 0.063 (3) | |
| C13A | 0.0555 (17) | 0.3729 (7) | 0.6274 (10) | 0.083 (3) | |
| H13A | 0.037278 | 0.394856 | 0.698993 | 0.100* | |
| C14A | −0.0942 (19) | 0.3294 (6) | 0.5537 (14) | 0.091 (4) | |
| H14A | −0.211587 | 0.321071 | 0.577528 | 0.109* | |
| C15A | −0.071 (2) | 0.2981 (7) | 0.4443 (13) | 0.092 (4) | |
| H15A | −0.175179 | 0.270805 | 0.391979 | 0.110* | |
| C16A | 0.1066 (19) | 0.3082 (6) | 0.4152 (10) | 0.083 (3) | |
| H16A | 0.127001 | 0.285713 | 0.344706 | 0.099* | |
| C17A | 0.2558 (18) | 0.3509 (6) | 0.4882 (10) | 0.078 (3) | |
| H17A | 0.374857 | 0.357800 | 0.465432 | 0.093* | |
| O1B | 0.6081 (10) | 0.3891 (4) | 0.1390 (6) | 0.077 (2) | |
| O2B | 0.5156 (11) | 0.4388 (5) | 0.2978 (7) | 0.094 (3) | |
| N1B | 0.7947 (14) | 0.5107 (5) | 0.4835 (8) | 0.077 (3) | |
| H1NB | 0.677837 | 0.491220 | 0.472848 | 0.093* | |
| N2B | 0.8923 (13) | 0.5521 (5) | 0.5859 (9) | 0.079 (3) | |
| N3B | 1.0751 (13) | 0.5425 (5) | 0.4483 (8) | 0.069 (2) | |
| C1B | 0.7509 (16) | 0.3751 (6) | 0.0749 (9) | 0.068 (3) | |
| C2B | 0.6886 (19) | 0.3325 (6) | −0.0306 (10) | 0.079 (3) | |
| H2B | 0.555969 | 0.314737 | −0.058312 | 0.095* | |
| C3B | 0.826 (2) | 0.3173 (6) | −0.0925 (12) | 0.097 (4) | |
| H3B | 0.786503 | 0.289200 | −0.164914 | 0.117* | |
| C4B | 1.027 (2) | 0.3426 (8) | −0.0507 (12) | 0.108 (5) | |
| H4B | 1.121311 | 0.329972 | −0.092802 | 0.130* | |
| C5B | 1.0839 (17) | 0.3862 (7) | 0.0527 (11) | 0.086 (3) | |
| H5B | 1.216051 | 0.404341 | 0.079762 | 0.103* | |
| C6B | 0.9443 (14) | 0.4035 (5) | 0.1172 (9) | 0.066 (3) | |
| C7B | 0.9897 (14) | 0.4482 (6) | 0.2278 (9) | 0.067 (3) | |
| H7B | 1.120153 | 0.467471 | 0.257810 | 0.081* | |
| C8B | 0.8522 (15) | 0.4634 (5) | 0.2897 (8) | 0.061 (3) | |
| C9B | 0.6531 (15) | 0.4311 (6) | 0.2471 (10) | 0.073 (3) | |
| C10B | 0.9055 (14) | 0.5050 (5) | 0.4038 (9) | 0.066 (3) | |
| C11B | 1.0608 (16) | 0.5691 (5) | 0.5578 (10) | 0.066 (3) | |
| C12B | 1.2181 (15) | 0.6149 (5) | 0.6418 (10) | 0.069 (3) | |
| C13B | 1.3979 (16) | 0.6267 (5) | 0.6145 (11) | 0.076 (3) | |
| H13B | 1.423656 | 0.605510 | 0.544530 | 0.091* | |
| C14B | 1.542 (2) | 0.6712 (7) | 0.6938 (14) | 0.097 (4) | |
| H14B | 1.662305 | 0.679778 | 0.673996 | 0.117* | |
| C15B | 1.515 (2) | 0.7014 (8) | 0.7946 (14) | 0.101 (4) | |
| H15B | 1.615628 | 0.730004 | 0.845856 | 0.121* | |
| C16B | 1.332 (2) | 0.6901 (7) | 0.8245 (12) | 0.095 (4) | |
| H16B | 1.310387 | 0.711525 | 0.895479 | 0.114* | |
| C17B | 1.1846 (18) | 0.6466 (6) | 0.7470 (10) | 0.077 (3) | |
| H17B | 1.062792 | 0.638795 | 0.766012 | 0.093* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1A | 0.063 (4) | 0.092 (5) | 0.085 (5) | −0.014 (4) | 0.012 (4) | −0.013 (4) |
| O2A | 0.058 (4) | 0.135 (7) | 0.090 (5) | −0.018 (5) | 0.025 (4) | −0.007 (5) |
| N1A | 0.058 (5) | 0.085 (6) | 0.084 (6) | −0.010 (5) | 0.030 (5) | −0.002 (5) |
| N2A | 0.060 (5) | 0.081 (6) | 0.076 (5) | −0.012 (5) | 0.019 (4) | −0.005 (5) |
| N3A | 0.053 (5) | 0.071 (5) | 0.065 (5) | −0.009 (4) | 0.014 (4) | −0.001 (4) |
| C1A | 0.073 (7) | 0.071 (7) | 0.074 (7) | −0.003 (6) | 0.017 (6) | 0.004 (6) |
| C2A | 0.105 (11) | 0.093 (9) | 0.092 (9) | −0.024 (8) | 0.010 (8) | −0.024 (8) |
| C3A | 0.108 (11) | 0.115 (11) | 0.088 (9) | 0.023 (10) | 0.012 (8) | −0.030 (9) |
| C4A | 0.095 (10) | 0.127 (12) | 0.090 (9) | 0.034 (9) | 0.017 (8) | −0.002 (9) |
| C5A | 0.077 (8) | 0.083 (8) | 0.071 (7) | 0.004 (6) | 0.012 (6) | −0.009 (6) |
| C6A | 0.054 (6) | 0.097 (8) | 0.058 (5) | −0.005 (6) | 0.011 (5) | 0.000 (6) |
| C7A | 0.054 (6) | 0.074 (7) | 0.071 (6) | −0.005 (5) | 0.008 (5) | 0.003 (5) |
| C8A | 0.048 (5) | 0.068 (6) | 0.078 (7) | 0.000 (5) | 0.018 (5) | 0.006 (5) |
| C9A | 0.055 (6) | 0.068 (6) | 0.083 (7) | −0.001 (5) | 0.019 (5) | −0.002 (6) |
| C10A | 0.055 (6) | 0.062 (6) | 0.055 (5) | 0.004 (5) | 0.011 (5) | 0.002 (5) |
| C11A | 0.055 (6) | 0.075 (7) | 0.069 (6) | 0.009 (5) | 0.015 (5) | −0.002 (5) |
| C12A | 0.055 (6) | 0.063 (6) | 0.066 (6) | 0.002 (5) | 0.010 (5) | 0.007 (5) |
| C13A | 0.065 (7) | 0.095 (8) | 0.086 (7) | −0.008 (7) | 0.016 (6) | −0.004 (7) |
| C14A | 0.064 (8) | 0.084 (8) | 0.123 (10) | −0.015 (6) | 0.027 (7) | 0.013 (8) |
| C15A | 0.085 (9) | 0.077 (8) | 0.101 (10) | −0.016 (7) | 0.009 (7) | −0.006 (7) |
| C16A | 0.089 (9) | 0.069 (7) | 0.073 (7) | 0.013 (7) | −0.001 (6) | 0.002 (6) |
| C17A | 0.069 (7) | 0.094 (8) | 0.067 (6) | 0.006 (7) | 0.016 (5) | 0.001 (6) |
| O1B | 0.057 (4) | 0.095 (6) | 0.074 (4) | −0.010 (4) | 0.013 (3) | 0.003 (4) |
| O2B | 0.046 (4) | 0.129 (7) | 0.107 (6) | −0.008 (5) | 0.026 (4) | 0.003 (6) |
| N1B | 0.054 (5) | 0.108 (7) | 0.070 (5) | −0.012 (5) | 0.018 (4) | −0.004 (5) |
| N2B | 0.051 (5) | 0.107 (7) | 0.083 (6) | 0.004 (5) | 0.023 (5) | −0.011 (6) |
| N3B | 0.049 (5) | 0.080 (6) | 0.071 (5) | −0.009 (4) | 0.009 (4) | 0.011 (5) |
| C1B | 0.067 (6) | 0.070 (7) | 0.066 (6) | 0.003 (5) | 0.018 (5) | 0.005 (6) |
| C2B | 0.087 (8) | 0.074 (7) | 0.073 (7) | −0.007 (6) | 0.018 (6) | −0.009 (6) |
| C3B | 0.099 (10) | 0.101 (10) | 0.083 (8) | −0.006 (8) | 0.013 (7) | −0.013 (7) |
| C4B | 0.095 (10) | 0.164 (14) | 0.067 (7) | −0.005 (10) | 0.027 (7) | −0.016 (8) |
| C5B | 0.066 (7) | 0.108 (10) | 0.087 (8) | −0.004 (7) | 0.027 (6) | −0.013 (7) |
| C6B | 0.048 (5) | 0.067 (6) | 0.075 (6) | −0.001 (5) | 0.007 (5) | 0.005 (5) |
| C7B | 0.047 (5) | 0.081 (7) | 0.070 (6) | −0.002 (5) | 0.013 (5) | 0.009 (6) |
| C8B | 0.057 (6) | 0.070 (7) | 0.055 (5) | 0.000 (5) | 0.015 (5) | 0.006 (5) |
| C9B | 0.045 (6) | 0.086 (8) | 0.080 (7) | −0.003 (5) | 0.009 (5) | −0.005 (6) |
| C10B | 0.046 (6) | 0.080 (7) | 0.072 (6) | 0.000 (5) | 0.019 (5) | −0.006 (5) |
| C11B | 0.058 (6) | 0.062 (6) | 0.078 (7) | 0.007 (5) | 0.019 (5) | 0.012 (6) |
| C12B | 0.058 (6) | 0.068 (7) | 0.081 (7) | −0.002 (5) | 0.018 (5) | 0.005 (6) |
| C13B | 0.070 (7) | 0.059 (6) | 0.095 (8) | 0.010 (6) | 0.021 (6) | 0.016 (6) |
| C14B | 0.081 (9) | 0.087 (9) | 0.110 (10) | −0.015 (7) | 0.008 (7) | 0.005 (8) |
| C15B | 0.078 (9) | 0.105 (11) | 0.095 (9) | −0.019 (8) | −0.010 (7) | −0.001 (8) |
| C16B | 0.096 (9) | 0.095 (9) | 0.083 (8) | −0.009 (8) | 0.009 (7) | −0.014 (7) |
| C17B | 0.082 (7) | 0.075 (7) | 0.068 (6) | 0.009 (6) | 0.014 (6) | 0.004 (6) |
Geometric parameters (Å, º) top
| O1A—C9A | 1.367 (12) | O1B—C9B | 1.388 (12) |
| O1A—C1A | 1.379 (13) | O1B—C1B | 1.390 (11) |
| O2A—C9A | 1.201 (12) | O2B—C9B | 1.236 (12) |
| N1A—C10A | 1.354 (11) | N1B—C10B | 1.319 (12) |
| N1A—N2A | 1.364 (11) | N1B—N2B | 1.372 (12) |
| N1A—H1NA | 0.8600 | N1B—H1NB | 0.8600 |
| N2A—C11A | 1.344 (12) | N2B—C11B | 1.322 (13) |
| N3A—C10A | 1.320 (12) | N3B—C10B | 1.330 (13) |
| N3A—C11A | 1.347 (11) | N3B—C11B | 1.331 (12) |
| C1A—C2A | 1.366 (15) | C1B—C2B | 1.375 (13) |
| C1A—C6A | 1.372 (14) | C1B—C6B | 1.377 (13) |
| C2A—C3A | 1.362 (17) | C2B—C3B | 1.346 (15) |
| C2A—H2A | 0.9300 | C2B—H2B | 0.9300 |
| C3A—C4A | 1.424 (19) | C3B—C4B | 1.399 (18) |
| C3A—H3A | 0.9300 | C3B—H3B | 0.9300 |
| C4A—C5A | 1.367 (15) | C4B—C5B | 1.369 (16) |
| C4A—H4A | 0.9300 | C4B—H4B | 0.9300 |
| C5A—C6A | 1.357 (13) | C5B—C6B | 1.388 (13) |
| C5A—H5A | 0.9300 | C5B—H5B | 0.9300 |
| C6A—C7A | 1.478 (14) | C6B—C7B | 1.441 (13) |
| C7A—C8A | 1.329 (12) | C7B—C8B | 1.346 (12) |
| C7A—H7A | 0.9300 | C7B—H7B | 0.9300 |
| C8A—C10A | 1.435 (13) | C8B—C10B | 1.434 (13) |
| C8A—C9A | 1.466 (14) | C8B—C9B | 1.442 (14) |
| C11A—C12A | 1.476 (14) | C11B—C12B | 1.479 (14) |
| C12A—C17A | 1.366 (13) | C12B—C13B | 1.369 (14) |
| C12A—C13A | 1.384 (13) | C12B—C17B | 1.380 (14) |
| C13A—C14A | 1.381 (17) | C13B—C14B | 1.394 (16) |
| C13A—H13A | 0.9300 | C13B—H13B | 0.9300 |
| C14A—C15A | 1.390 (16) | C14B—C15B | 1.306 (17) |
| C14A—H14A | 0.9300 | C14B—H14B | 0.9300 |
| C15A—C16A | 1.361 (16) | C15B—C16B | 1.397 (17) |
| C15A—H15A | 0.9300 | C15B—H15B | 0.9300 |
| C16A—C17A | 1.367 (15) | C16B—C17B | 1.391 (16) |
| C16A—H16A | 0.9300 | C16B—H16B | 0.9300 |
| C17A—H17A | 0.9300 | C17B—H17B | 0.9300 |
| | | |
| C9A—O1A—C1A | 122.3 (8) | C9B—O1B—C1B | 122.2 (8) |
| C10A—N1A—N2A | 110.7 (8) | C10B—N1B—N2B | 110.4 (9) |
| C10A—N1A—H1NA | 124.6 | C10B—N1B—H1NB | 124.8 |
| N2A—N1A—H1NA | 124.6 | N2B—N1B—H1NB | 124.8 |
| C11A—N2A—N1A | 100.9 (7) | C11B—N2B—N1B | 101.7 (8) |
| C10A—N3A—C11A | 103.8 (8) | C10B—N3B—C11B | 104.2 (9) |
| C2A—C1A—C6A | 119.3 (12) | C2B—C1B—C6B | 122.9 (10) |
| C2A—C1A—O1A | 117.9 (11) | C2B—C1B—O1B | 116.9 (10) |
| C6A—C1A—O1A | 122.6 (10) | C6B—C1B—O1B | 120.2 (9) |
| C3A—C2A—C1A | 120.4 (14) | C3B—C2B—C1B | 117.7 (11) |
| C3A—C2A—H2A | 119.8 | C3B—C2B—H2B | 121.1 |
| C1A—C2A—H2A | 119.8 | C1B—C2B—H2B | 121.1 |
| C2A—C3A—C4A | 120.0 (13) | C2B—C3B—C4B | 121.8 (12) |
| C2A—C3A—H3A | 120.0 | C2B—C3B—H3B | 119.1 |
| C4A—C3A—H3A | 120.0 | C4B—C3B—H3B | 119.1 |
| C5A—C4A—C3A | 118.2 (13) | C5B—C4B—C3B | 119.3 (12) |
| C5A—C4A—H4A | 120.9 | C5B—C4B—H4B | 120.3 |
| C3A—C4A—H4A | 120.9 | C3B—C4B—H4B | 120.3 |
| C6A—C5A—C4A | 120.5 (13) | C4B—C5B—C6B | 120.1 (11) |
| C6A—C5A—H5A | 119.8 | C4B—C5B—H5B | 120.0 |
| C4A—C5A—H5A | 119.8 | C6B—C5B—H5B | 120.0 |
| C5A—C6A—C1A | 121.5 (11) | C1B—C6B—C5B | 118.1 (10) |
| C5A—C6A—C7A | 123.2 (10) | C1B—C6B—C7B | 117.6 (9) |
| C1A—C6A—C7A | 115.2 (9) | C5B—C6B—C7B | 124.3 (9) |
| C8A—C7A—C6A | 122.8 (9) | C8B—C7B—C6B | 123.1 (9) |
| C8A—C7A—H7A | 118.6 | C8B—C7B—H7B | 118.5 |
| C6A—C7A—H7A | 118.6 | C6B—C7B—H7B | 118.5 |
| C7A—C8A—C10A | 121.2 (9) | C7B—C8B—C10B | 121.7 (9) |
| C7A—C8A—C9A | 119.3 (10) | C7B—C8B—C9B | 118.5 (10) |
| C10A—C8A—C9A | 119.5 (9) | C10B—C8B—C9B | 119.4 (9) |
| O2A—C9A—O1A | 117.4 (9) | O2B—C9B—O1B | 116.6 (9) |
| O2A—C9A—C8A | 125.0 (10) | O2B—C9B—C8B | 125.1 (10) |
| O1A—C9A—C8A | 117.5 (9) | O1B—C9B—C8B | 118.3 (9) |
| N3A—C10A—N1A | 109.3 (9) | N1B—C10B—N3B | 109.0 (10) |
| N3A—C10A—C8A | 125.3 (9) | N1B—C10B—C8B | 124.9 (10) |
| N1A—C10A—C8A | 125.3 (9) | N3B—C10B—C8B | 126.1 (9) |
| N2A—C11A—N3A | 115.3 (9) | N2B—C11B—N3B | 114.6 (10) |
| N2A—C11A—C12A | 120.4 (9) | N2B—C11B—C12B | 122.0 (10) |
| N3A—C11A—C12A | 124.4 (9) | N3B—C11B—C12B | 123.4 (10) |
| C17A—C12A—C13A | 119.2 (10) | C13B—C12B—C17B | 119.5 (10) |
| C17A—C12A—C11A | 123.3 (9) | C13B—C12B—C11B | 119.9 (10) |
| C13A—C12A—C11A | 117.5 (9) | C17B—C12B—C11B | 120.6 (10) |
| C14A—C13A—C12A | 119.5 (11) | C12B—C13B—C14B | 118.7 (11) |
| C14A—C13A—H13A | 120.3 | C12B—C13B—H13B | 120.7 |
| C12A—C13A—H13A | 120.3 | C14B—C13B—H13B | 120.7 |
| C13A—C14A—C15A | 120.7 (11) | C15B—C14B—C13B | 123.1 (13) |
| C13A—C14A—H14A | 119.7 | C15B—C14B—H14B | 118.4 |
| C15A—C14A—H14A | 119.7 | C13B—C14B—H14B | 118.4 |
| C16A—C15A—C14A | 118.6 (12) | C14B—C15B—C16B | 119.3 (12) |
| C16A—C15A—H15A | 120.7 | C14B—C15B—H15B | 120.4 |
| C14A—C15A—H15A | 120.7 | C16B—C15B—H15B | 120.4 |
| C15A—C16A—C17A | 120.9 (12) | C17B—C16B—C15B | 119.2 (12) |
| C15A—C16A—H16A | 119.5 | C17B—C16B—H16B | 120.4 |
| C17A—C16A—H16A | 119.5 | C15B—C16B—H16B | 120.4 |
| C12A—C17A—C16A | 121.1 (11) | C12B—C17B—C16B | 120.2 (12) |
| C12A—C17A—H17A | 119.5 | C12B—C17B—H17B | 119.9 |
| C16A—C17A—H17A | 119.5 | C16B—C17B—H17B | 119.9 |
| | | |
| C10A—N1A—N2A—C11A | −1.6 (10) | C10B—N1B—N2B—C11B | 0.4 (11) |
| C9A—O1A—C1A—C2A | −178.9 (9) | C9B—O1B—C1B—C2B | 179.7 (9) |
| C9A—O1A—C1A—C6A | −2.2 (16) | C9B—O1B—C1B—C6B | −0.4 (14) |
| C6A—C1A—C2A—C3A | 1.5 (18) | C6B—C1B—C2B—C3B | 1.4 (17) |
| O1A—C1A—C2A—C3A | 178.3 (12) | O1B—C1B—C2B—C3B | −178.8 (9) |
| C1A—C2A—C3A—C4A | 0 (2) | C1B—C2B—C3B—C4B | 1.1 (19) |
| C2A—C3A—C4A—C5A | −2 (2) | C2B—C3B—C4B—C5B | −3 (2) |
| C3A—C4A—C5A—C6A | 1.9 (19) | C3B—C4B—C5B—C6B | 2 (2) |
| C4A—C5A—C6A—C1A | −0.2 (18) | C2B—C1B—C6B—C5B | −2.3 (16) |
| C4A—C5A—C6A—C7A | −179.7 (10) | O1B—C1B—C6B—C5B | 177.9 (9) |
| C2A—C1A—C6A—C5A | −1.5 (17) | C2B—C1B—C6B—C7B | 179.0 (9) |
| O1A—C1A—C6A—C5A | −178.2 (10) | O1B—C1B—C6B—C7B | −0.8 (14) |
| C2A—C1A—C6A—C7A | 178.0 (10) | C4B—C5B—C6B—C1B | 0.6 (17) |
| O1A—C1A—C6A—C7A | 1.3 (15) | C4B—C5B—C6B—C7B | 179.2 (11) |
| C5A—C6A—C7A—C8A | 177.3 (10) | C1B—C6B—C7B—C8B | −0.5 (15) |
| C1A—C6A—C7A—C8A | −2.2 (15) | C5B—C6B—C7B—C8B | −179.1 (10) |
| C6A—C7A—C8A—C10A | −177.1 (9) | C6B—C7B—C8B—C10B | 176.5 (9) |
| C6A—C7A—C8A—C9A | 3.6 (15) | C6B—C7B—C8B—C9B | 2.8 (15) |
| C1A—O1A—C9A—O2A | −180.0 (9) | C1B—O1B—C9B—O2B | −178.6 (9) |
| C1A—O1A—C9A—C8A | 3.4 (14) | C1B—O1B—C9B—C8B | 2.8 (14) |
| C7A—C8A—C9A—O2A | 179.5 (10) | C7B—C8B—C9B—O2B | 177.6 (10) |
| C10A—C8A—C9A—O2A | 0.2 (16) | C10B—C8B—C9B—O2B | 3.8 (16) |
| C7A—C8A—C9A—O1A | −4.2 (14) | C7B—C8B—C9B—O1B | −3.9 (14) |
| C10A—C8A—C9A—O1A | 176.5 (8) | C10B—C8B—C9B—O1B | −177.7 (9) |
| C11A—N3A—C10A—N1A | −0.9 (11) | N2B—N1B—C10B—N3B | −1.0 (12) |
| C11A—N3A—C10A—C8A | 175.3 (9) | N2B—N1B—C10B—C8B | 177.4 (9) |
| N2A—N1A—C10A—N3A | 1.6 (11) | C11B—N3B—C10B—N1B | 1.1 (11) |
| N2A—N1A—C10A—C8A | −174.5 (8) | C11B—N3B—C10B—C8B | −177.3 (9) |
| C7A—C8A—C10A—N3A | −6.3 (15) | C7B—C8B—C10B—N1B | −165.6 (10) |
| C9A—C8A—C10A—N3A | 173.0 (9) | C9B—C8B—C10B—N1B | 8.0 (15) |
| C7A—C8A—C10A—N1A | 169.3 (9) | C7B—C8B—C10B—N3B | 12.5 (16) |
| C9A—C8A—C10A—N1A | −11.4 (14) | C9B—C8B—C10B—N3B | −173.9 (10) |
| N1A—N2A—C11A—N3A | 1.1 (11) | N1B—N2B—C11B—N3B | 0.3 (12) |
| N1A—N2A—C11A—C12A | 179.5 (8) | N1B—N2B—C11B—C12B | 179.6 (9) |
| C10A—N3A—C11A—N2A | −0.2 (12) | C10B—N3B—C11B—N2B | −0.8 (12) |
| C10A—N3A—C11A—C12A | −178.5 (9) | C10B—N3B—C11B—C12B | 179.9 (9) |
| N2A—C11A—C12A—C17A | 8.2 (14) | N2B—C11B—C12B—C13B | 174.7 (10) |
| N3A—C11A—C12A—C17A | −173.5 (10) | N3B—C11B—C12B—C13B | −6.0 (15) |
| N2A—C11A—C12A—C13A | −173.8 (10) | N2B—C11B—C12B—C17B | −6.1 (15) |
| N3A—C11A—C12A—C13A | 4.5 (15) | N3B—C11B—C12B—C17B | 173.2 (10) |
| C17A—C12A—C13A—C14A | 0.1 (16) | C17B—C12B—C13B—C14B | −0.6 (15) |
| C11A—C12A—C13A—C14A | −178.0 (10) | C11B—C12B—C13B—C14B | 178.7 (9) |
| C12A—C13A—C14A—C15A | −2.0 (18) | C12B—C13B—C14B—C15B | 1.2 (18) |
| C13A—C14A—C15A—C16A | 3.5 (19) | C13B—C14B—C15B—C16B | −1 (2) |
| C14A—C15A—C16A—C17A | −3.2 (18) | C14B—C15B—C16B—C17B | 1 (2) |
| C13A—C12A—C17A—C16A | 0.2 (15) | C13B—C12B—C17B—C16B | 0.0 (15) |
| C11A—C12A—C17A—C16A | 178.2 (10) | C11B—C12B—C17B—C16B | −179.3 (10) |
| C15A—C16A—C17A—C12A | 1.4 (17) | C15B—C16B—C17B—C12B | 0.0 (18) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1A—H1NA···O2A | 0.86 | 2.20 | 2.746 (12) | 121 |
| N1B—H1NB···O2B | 0.86 | 2.15 | 2.708 (12) | 123 |
| N1A—H1NA···N2B | 0.86 | 2.39 | 2.979 (12) | 126 |
| N1B—H1NB···N2A | 0.86 | 2.42 | 3.000 (12) | 125 |
3-(3-Phenyl-1
H-1,2,4-triazol-5-yl)-2
H-1-benzopyran-2-one (1r)
top
Crystal data top
| C17H11N3O2 | Dx = 1.389 Mg m−3 |
| Mr = 289.29 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pbca | Cell parameters from 1802 reflections |
| a = 13.0369 (8) Å | θ = 3.4–24.6° |
| b = 13.0804 (7) Å | µ = 0.09 mm−1 |
| c = 16.2223 (8) Å | T = 293 K |
| V = 2766.4 (3) Å3 | Block, colorless |
| Z = 8 | 0.30 × 0.12 × 0.12 mm |
| F(000) = 1200 | |
Data collection top
Rigaku Xcalibur Sapphire3
diffractometer | 2434 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 1687 reflections with I > 2σ(I) |
| Detector resolution: 16.1827 pixels mm-1 | Rint = 0.086 |
| ω scans | θmax = 25.0°, θmin = 3.0° |
Absorption correction: multi-scan
(CrysAlis PRO; Rigaku OD, 2018) | h = −14→15 |
| Tmin = 0.109, Tmax = 1.000 | k = −10→15 |
| 18857 measured reflections | l = −19→19 |
Refinement top
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.053 | w = 1/[σ2(Fo2) + (0.0911P)2]
where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.170 | (Δ/σ)max < 0.001 |
| S = 1.07 | Δρmax = 0.22 e Å−3 |
| 2434 reflections | Δρmin = −0.17 e Å−3 |
| 200 parameters | Extinction correction: SHELXL2016 (Sheldrick, 2015b), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.0028 (9) |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.55275 (12) | 0.82349 (12) | 0.30139 (10) | 0.0626 (5) | |
| O2 | 0.47671 (13) | 0.68927 (14) | 0.35456 (11) | 0.0777 (6) | |
| N1 | 0.58664 (15) | 0.54585 (13) | 0.43886 (11) | 0.0588 (5) | |
| H1N | 0.523389 | 0.562957 | 0.432834 | 0.071* | |
| N2 | 0.61898 (15) | 0.46386 (14) | 0.48195 (12) | 0.0609 (5) | |
| N3 | 0.75265 (13) | 0.54970 (13) | 0.42686 (10) | 0.0517 (5) | |
| C1 | 0.63897 (17) | 0.87601 (17) | 0.27645 (13) | 0.0514 (6) | |
| C2 | 0.6240 (2) | 0.96771 (17) | 0.23729 (14) | 0.0621 (7) | |
| H2 | 0.558333 | 0.994046 | 0.230321 | 0.075* | |
| C3 | 0.7083 (2) | 1.01985 (19) | 0.20855 (14) | 0.0640 (7) | |
| H3 | 0.699646 | 1.081995 | 0.181589 | 0.077* | |
| C4 | 0.8057 (2) | 0.98078 (19) | 0.21935 (15) | 0.0673 (7) | |
| H4 | 0.862025 | 1.016417 | 0.199012 | 0.081* | |
| C5 | 0.82020 (18) | 0.89003 (17) | 0.25975 (14) | 0.0593 (6) | |
| H5 | 0.886215 | 0.864805 | 0.267378 | 0.071* | |
| C6 | 0.73571 (17) | 0.83501 (17) | 0.28965 (12) | 0.0507 (6) | |
| C7 | 0.74293 (17) | 0.74110 (17) | 0.33302 (12) | 0.0521 (6) | |
| H7 | 0.807366 | 0.713629 | 0.343698 | 0.062* | |
| C8 | 0.65868 (17) | 0.69054 (17) | 0.35921 (12) | 0.0501 (6) | |
| C9 | 0.55756 (17) | 0.73097 (18) | 0.33958 (14) | 0.0559 (6) | |
| C10 | 0.66619 (16) | 0.59730 (16) | 0.40658 (13) | 0.0500 (6) | |
| C11 | 0.71992 (18) | 0.46861 (16) | 0.47263 (12) | 0.0517 (6) | |
| C12 | 0.79027 (18) | 0.39262 (17) | 0.50711 (13) | 0.0537 (6) | |
| C13 | 0.8946 (2) | 0.40662 (19) | 0.50197 (18) | 0.0727 (7) | |
| H13 | 0.920177 | 0.464708 | 0.476085 | 0.087* | |
| C14 | 0.9620 (2) | 0.3361 (2) | 0.5345 (2) | 0.0872 (9) | |
| H14 | 1.032352 | 0.346288 | 0.530106 | 0.105* | |
| C15 | 0.9244 (3) | 0.2504 (2) | 0.57352 (17) | 0.0802 (8) | |
| H15 | 0.969472 | 0.203175 | 0.596502 | 0.096* | |
| C16 | 0.8207 (2) | 0.23475 (19) | 0.57844 (15) | 0.0708 (8) | |
| H16 | 0.795369 | 0.176365 | 0.604017 | 0.085* | |
| C17 | 0.7538 (2) | 0.30573 (18) | 0.54541 (13) | 0.0595 (6) | |
| H17 | 0.683409 | 0.294814 | 0.549041 | 0.071* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0430 (10) | 0.0652 (11) | 0.0795 (10) | −0.0015 (7) | −0.0040 (8) | 0.0199 (8) |
| O2 | 0.0457 (11) | 0.0822 (13) | 0.1051 (14) | −0.0108 (8) | −0.0042 (9) | 0.0286 (10) |
| N1 | 0.0471 (11) | 0.0557 (12) | 0.0737 (12) | −0.0017 (9) | 0.0032 (9) | 0.0109 (9) |
| N2 | 0.0538 (13) | 0.0561 (12) | 0.0729 (12) | −0.0009 (9) | 0.0025 (10) | 0.0136 (9) |
| N3 | 0.0489 (11) | 0.0491 (11) | 0.0571 (10) | 0.0011 (8) | 0.0023 (8) | 0.0019 (9) |
| C1 | 0.0469 (13) | 0.0555 (14) | 0.0519 (12) | −0.0040 (10) | −0.0009 (10) | 0.0049 (10) |
| C2 | 0.0601 (16) | 0.0584 (16) | 0.0677 (14) | 0.0026 (11) | −0.0016 (12) | 0.0119 (12) |
| C3 | 0.0718 (18) | 0.0596 (15) | 0.0607 (14) | −0.0033 (12) | −0.0008 (12) | 0.0106 (11) |
| C4 | 0.0672 (18) | 0.0649 (16) | 0.0698 (14) | −0.0142 (12) | 0.0097 (13) | 0.0067 (13) |
| C5 | 0.0491 (14) | 0.0609 (15) | 0.0680 (14) | −0.0021 (10) | 0.0071 (11) | 0.0027 (12) |
| C6 | 0.0501 (13) | 0.0513 (13) | 0.0506 (11) | −0.0030 (10) | 0.0017 (10) | 0.0016 (10) |
| C7 | 0.0436 (13) | 0.0555 (14) | 0.0571 (12) | 0.0044 (10) | 0.0009 (10) | −0.0009 (10) |
| C8 | 0.0461 (13) | 0.0500 (13) | 0.0544 (12) | −0.0009 (9) | 0.0004 (10) | 0.0016 (10) |
| C9 | 0.0458 (14) | 0.0594 (15) | 0.0626 (12) | −0.0047 (11) | −0.0021 (10) | 0.0070 (11) |
| C10 | 0.0467 (13) | 0.0465 (13) | 0.0567 (12) | −0.0026 (9) | 0.0026 (10) | −0.0006 (10) |
| C11 | 0.0540 (14) | 0.0467 (13) | 0.0543 (12) | −0.0017 (10) | 0.0036 (11) | −0.0001 (10) |
| C12 | 0.0587 (15) | 0.0483 (13) | 0.0540 (11) | 0.0033 (10) | 0.0015 (11) | −0.0007 (10) |
| C13 | 0.0650 (18) | 0.0559 (15) | 0.0974 (18) | 0.0023 (12) | −0.0029 (14) | 0.0127 (13) |
| C14 | 0.0673 (19) | 0.077 (2) | 0.117 (2) | 0.0114 (15) | −0.0104 (17) | 0.0132 (17) |
| C15 | 0.089 (2) | 0.0666 (19) | 0.0854 (17) | 0.0213 (15) | −0.0084 (16) | 0.0095 (14) |
| C16 | 0.091 (2) | 0.0574 (16) | 0.0636 (14) | 0.0080 (14) | 0.0065 (14) | 0.0057 (12) |
| C17 | 0.0660 (16) | 0.0562 (14) | 0.0562 (12) | 0.0027 (11) | 0.0081 (11) | −0.0005 (11) |
Geometric parameters (Å, º) top
| O1—C9 | 1.361 (3) | C5—H5 | 0.9300 |
| O1—C1 | 1.378 (3) | C6—C7 | 1.419 (3) |
| O2—C9 | 1.211 (3) | C7—C8 | 1.351 (3) |
| N1—C10 | 1.343 (3) | C7—H7 | 0.9300 |
| N1—N2 | 1.348 (2) | C8—C10 | 1.445 (3) |
| N1—H1N | 0.8600 | C8—C9 | 1.456 (3) |
| N2—C11 | 1.326 (3) | C11—C12 | 1.464 (3) |
| N3—C10 | 1.329 (3) | C12—C13 | 1.375 (4) |
| N3—C11 | 1.363 (3) | C12—C17 | 1.380 (3) |
| C1—C2 | 1.371 (3) | C13—C14 | 1.379 (4) |
| C1—C6 | 1.387 (3) | C13—H13 | 0.9300 |
| C2—C3 | 1.375 (3) | C14—C15 | 1.377 (4) |
| C2—H2 | 0.9300 | C14—H14 | 0.9300 |
| C3—C4 | 1.380 (3) | C15—C16 | 1.370 (4) |
| C3—H3 | 0.9300 | C15—H15 | 0.9300 |
| C4—C5 | 1.369 (3) | C16—C17 | 1.383 (3) |
| C4—H4 | 0.9300 | C16—H16 | 0.9300 |
| C5—C6 | 1.402 (3) | C17—H17 | 0.9300 |
| | | |
| C9—O1—C1 | 122.64 (17) | C10—C8—C9 | 119.0 (2) |
| C10—N1—N2 | 111.07 (19) | O2—C9—O1 | 116.8 (2) |
| C10—N1—H1N | 124.5 | O2—C9—C8 | 125.5 (2) |
| N2—N1—H1N | 124.5 | O1—C9—C8 | 117.67 (19) |
| C11—N2—N1 | 102.35 (17) | N3—C10—N1 | 108.89 (19) |
| C10—N3—C11 | 103.52 (18) | N3—C10—C8 | 125.77 (19) |
| C2—C1—O1 | 117.1 (2) | N1—C10—C8 | 125.3 (2) |
| C2—C1—C6 | 122.6 (2) | N2—C11—N3 | 114.15 (19) |
| O1—C1—C6 | 120.24 (19) | N2—C11—C12 | 123.13 (19) |
| C1—C2—C3 | 118.5 (2) | N3—C11—C12 | 122.7 (2) |
| C1—C2—H2 | 120.7 | C13—C12—C17 | 118.6 (2) |
| C3—C2—H2 | 120.7 | C13—C12—C11 | 120.4 (2) |
| C2—C3—C4 | 120.6 (2) | C17—C12—C11 | 121.0 (2) |
| C2—C3—H3 | 119.7 | C12—C13—C14 | 121.2 (3) |
| C4—C3—H3 | 119.7 | C12—C13—H13 | 119.4 |
| C5—C4—C3 | 120.6 (2) | C14—C13—H13 | 119.4 |
| C5—C4—H4 | 119.7 | C15—C14—C13 | 119.6 (3) |
| C3—C4—H4 | 119.7 | C15—C14—H14 | 120.2 |
| C4—C5—C6 | 120.2 (2) | C13—C14—H14 | 120.2 |
| C4—C5—H5 | 119.9 | C16—C15—C14 | 119.9 (3) |
| C6—C5—H5 | 119.9 | C16—C15—H15 | 120.0 |
| C1—C6—C5 | 117.5 (2) | C14—C15—H15 | 120.0 |
| C1—C6—C7 | 118.14 (19) | C15—C16—C17 | 120.0 (2) |
| C5—C6—C7 | 124.3 (2) | C15—C16—H16 | 120.0 |
| C8—C7—C6 | 121.7 (2) | C17—C16—H16 | 120.0 |
| C8—C7—H7 | 119.1 | C12—C17—C16 | 120.6 (3) |
| C6—C7—H7 | 119.1 | C12—C17—H17 | 119.7 |
| C7—C8—C10 | 121.7 (2) | C16—C17—H17 | 119.7 |
| C7—C8—C9 | 119.3 (2) | | |
| | | |
| C10—N1—N2—C11 | −0.8 (2) | C11—N3—C10—N1 | −0.3 (2) |
| C9—O1—C1—C2 | 178.85 (19) | C11—N3—C10—C8 | 178.39 (19) |
| C9—O1—C1—C6 | 0.2 (3) | N2—N1—C10—N3 | 0.7 (2) |
| O1—C1—C2—C3 | −177.3 (2) | N2—N1—C10—C8 | −177.93 (19) |
| C6—C1—C2—C3 | 1.4 (3) | C7—C8—C10—N3 | −2.5 (3) |
| C1—C2—C3—C4 | −0.3 (4) | C9—C8—C10—N3 | 177.7 (2) |
| C2—C3—C4—C5 | −0.8 (4) | C7—C8—C10—N1 | 175.9 (2) |
| C3—C4—C5—C6 | 0.9 (4) | C9—C8—C10—N1 | −3.9 (3) |
| C2—C1—C6—C5 | −1.3 (3) | N1—N2—C11—N3 | 0.7 (2) |
| O1—C1—C6—C5 | 177.34 (19) | N1—N2—C11—C12 | −178.26 (19) |
| C2—C1—C6—C7 | 178.12 (19) | C10—N3—C11—N2 | −0.3 (2) |
| O1—C1—C6—C7 | −3.3 (3) | C10—N3—C11—C12 | 178.67 (19) |
| C4—C5—C6—C1 | 0.1 (3) | N2—C11—C12—C13 | −173.9 (2) |
| C4—C5—C6—C7 | −179.2 (2) | N3—C11—C12—C13 | 7.2 (3) |
| C1—C6—C7—C8 | 1.8 (3) | N2—C11—C12—C17 | 6.0 (3) |
| C5—C6—C7—C8 | −178.9 (2) | N3—C11—C12—C17 | −172.87 (19) |
| C6—C7—C8—C10 | −177.17 (18) | C17—C12—C13—C14 | −0.3 (4) |
| C6—C7—C8—C9 | 2.6 (3) | C11—C12—C13—C14 | 179.7 (2) |
| C1—O1—C9—O2 | −176.0 (2) | C12—C13—C14—C15 | −0.6 (4) |
| C1—O1—C9—C8 | 4.2 (3) | C13—C14—C15—C16 | 1.2 (4) |
| C7—C8—C9—O2 | 174.6 (2) | C14—C15—C16—C17 | −1.0 (4) |
| C10—C8—C9—O2 | −5.6 (4) | C13—C12—C17—C16 | 0.5 (3) |
| C7—C8—C9—O1 | −5.6 (3) | C11—C12—C17—C16 | −179.4 (2) |
| C10—C8—C9—O1 | 174.22 (18) | C15—C16—C17—C12 | 0.1 (4) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O2 | 0.86 | 2.17 | 2.728 (3) | 122 |
| N1—H1N···N2i | 0.86 | 2.34 | 2.975 (3) | 131 |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Results of crystallization from different solvents and equimolar
two-component mixtures top| Solvent | Crystal form | Structure |
| Methylene chloride | Plate-like | 1t |
| Methanol | Plate-like | 1t |
| Ethanol | Plate-like | 1t |
| Isopropanol/ethyl acetate | Plate-like | 1t |
| Chloroform/tetrahydrofuran | Plate-like | 1t |
| Tetrahydrofuran/acetone | Stick-like | 1m+1t |
| Acetonitrile | Prismatic | 1r |
| Acetonitrile/tetrahydrofuran | Prismatic | 1r |
Selected geometric parameters for molecules 1 in different polymorphic
forms top| Parameter | Polymorph 1t | | Polymorph 1m | | Polymorph 1r |
| 1t_A | 1t_B | 1m_A | 1m_B | |
| The accuracy of the molecular mean-square plane (Å) | | | | | |
| 0.07 | 0.09 | 0.08 | 0.08 | 0.06 |
| Bond lengths (Å) | | | | | |
| C9—O2 | 1.209 (4) | 1.211 (5) | 1.201 (12) | 1.236 (12) | 1.211 (3) |
| C8—C10 | 1.454 (4) | 1.445 (5) | 1.435 (13) | 1.434 (13) | 1.445 (3) |
| C11—C12 | 1.468 (3) | 1.469 (4) | 1.476 (14) | 1.479 (14) | 1.464 (3) |
| | | | | |
| Torsion angles (°) | | | | | |
| C9—C8—C10—N1 | 8.0 (4) | 10.4 (5) | -11.4 (14) | 8.0 (15) | -3.9 (3) |
| N3—C11—C12—C13 | -5.9 (5) | -4.5 (4) | 4.5 (15) | -6.0 (15) | 7.2 (3) |
| | | | | |
| N1—H···O2 intramolecular hydrogen bond | | | | | |
| H···O2 | 2.19 | 2.22 | 2.17 | 2.15 | 2.17 |
| N1—H···O2 | 122 | 121 | 121 | 122 | 122 |
| | | | | |
| Intramolecular short contacts (Å) | | | | | |
| H7···N3 | 2.61 | 2.60 | 2.57 | 2.63 | 2.63 |
| H13···N3 | 2.62 | 2.55 | 2.55 | 2.60 | 2.58 |
| H17···N2 | 2.58 | 2.63 | 2.62 | 2.58 | 2.60 |
Geometric characteristics (Å, °of the intermolecular hydrogen bonds and
stacking interactions in different polymorphic crystals of compound 1 top| Interaction | Symmetry operation | Geometric characteristics | |
| | H···A | D—H···A |
| Polymorph 1t | | | |
| N1A—H···N2A | -x+1, -y+1, -z | 2.40 | 125 |
| N1B—H···N2B | -x+2, -y+1, -z+1 | 2.40 | 126 |
| C3A—H···C16B | x, y+1, z | 2.89 | 160 |
| C16A—H···C2B | x, y, z | 2.78 | 145 |
| C5B—H···O1B | x-1, y, z | 2.58 | 153 |
| C15B—H···C16A | -x+1, -y+1, -z+1 | 2.85 | 147 |
| C15B—H···C17A | -x+1, -y+1, -z+1 | 2.88 | 136 |
| C7A···C16A (π–π) | -x+2, -y+1, -z | Distance between the atoms is 3.357 (4) Å | |
| C7B···C16B (π–π) | -x+1, -y+1, -z+1 | Distance between the atoms is 3.371 (3) Å | |
| C9A···C7A (π–π) | -x+1, -y+2, -z | Distance between the atoms is 3.474 (4) Å | |
| C9B···C7B (π–π) | -x+2, -y, -z+1 | Distance between the atoms is 3.459 (3) Å | |
| | | |
| Polymorph 1m | | | |
| N1A—H···N2B | x, y, z | 2.39 | 126 |
| N1B—H···N2A | x, y, z | 2.42 | 125 |
| C15A—H···C16B | -x+1, y-1/2, -z+1 | 2.76 | 151 |
| C15A—H···C17B | -x+1, y-1/2, -z+1 | 2.85 | 134 |
| C3A—H···C16A | -x+1, y+1/2, -z+2 | 2.86 | 163 |
| C3A—H···C17A | -x+1, y+1/2, -z+2 | 2.89 | 159 |
| C5B—H···O1B | x+1, y, z | 2.58 | 151 |
| C16B—H···C2B | -x+2, y+1/2, -z+1 | 2.78 | 148 |
| C9B···C7A (π–π) | x, y, z-1 | Distance between the atoms is 3.41 (1) Å | |
| C7B···C16A (π–π) | x+1, y, z | Distance between the atoms is 3.36 (1) Å | |
| | | |
| Polymorph 1r | | | |
| N1—H···N2 | -x+1, -y+1, -z+1 | 2.34 | 131 |
| C3—H···C17 | x, -y+3/2, z-1/2 | 2.75 | 145 |
| C17—H···O2 | -x+1, -y+1, -z+1 | 2.62 | 144 |
| C5—H···O1 | x+1/2, y, -z+1/2 | 2.50 | 145 |
| C10···C16 (π–π) | -x+3/2, y+1/2, z | Distance between the atoms is 3.321 (3) Å | |
| C10···C4 (π–π) | -x+3/2, y-1/2, z | Distance between the atoms is 3.419 (3) Å | |
Symmetry codes, interaction energies of the basic molecule with neighbouring
molecules (Eint, kcal mol-1) with the highest values (more than
5% of total interaction energy) and the contribution of this energy to the
total interaction energy (%) in 1t crystals (for full list of dimers,
see Tables S1 in the supporting information) top| Dimer | Molecules | Symmetry operation | Eint | The contribution to the total interaction energy (%) | Type of interaction |
| Molecule 1t_A | | | | | |
| 1t_d1 | A–A | -x+2, -y+1, -z | -17.6 | 23.6 | Stacking |
| 1t_d2 | A–A | -x+1, -y+2, -z | -15.9 | 21.3 | Stacking |
| 1t_d3 | A–A | -x+2, -y+2, -z | -7.4 | 9.9 | Nonspecific |
| 1t_d4 | A–A | -x+1, -y+1, -z | -7.3 | 9.9 | N—H···N |
| 1t_d5 | A–A | x+1, y, z | -5.4 | 7.2 | C—H···O |
| 1t_d6 | A–A | -x+1, y, z | -5.4 | 7.2 | C—H···O |
| 1t_d7 | A–B | x, y+1, z-1 | -4.2 | 5.6 | C—H···π |
| | | | | |
| Molecule 1t_B | | | | | |
| 1t_d14 | B–B | -x+1, -y+1, -z+1 | -16.6 | 22.4 | Stacking |
| 1t_d15 | B–B | -x+2, -y, -z+1 | -16.4 | 22.1 | Stacking |
| 1t_d16 | B–B | -x+2, -y+1, -z+1 | -7.8 | 10.5 | N—H···N |
| 1t_d17 | B–B | -x+1, -y, -z+1 | -7.1 | 9.6 | Nonspecific |
| 1t_d18 | B–B | x+1, y, z | -5.4 | 7.2 | C—H···O |
| 1t_d19 | B–B | -x+1, y, z | -5.4 | 7.2 | C—H···O |
| 1t_d20 | B–A | x, y-1, z+1 | -4.2 | 5.6 | C—H···π |
Symmetry codes, interaction energies of the basic molecule with neighbouring
molecules (Eint, kcal mol-1) with the highest values (more than 5%
of total interaction energy) and the contribution of this energy to the total
interaction energy (%) in 1m crystals (for full list of dimers, see
Table S2 in the supporting information) top| Dimer | Molecules | Symmetry operation | Eint | The contribution to the total interaction energy (%) | Type of interaction |
| Molecule 1m_A | | | | | |
| 1m_d1 | A–B | x+1, y, z | -17.23 | 23.6 | Stacking |
| 1m_d2 | A–B | x, y, z-1 | -15.74 | 21.6 | Stacking |
| 1m_d3 | A–B | x+1, y, z-1 | -7.58 | 10.4 | Nonspecific |
| 1m_d4 | A–B | x, y, z | -7.24 | 9.9 | N—H···N |
| 1m_d5 | A–A | x+1, y, z | -5.15 | 7.1 | C—H···O |
| 1m_d6 | A–A | -x+1, y, z | -5.15 | 7.1 | C—H···O |
| | | | | |
| Molecule 1m_B | | | | | |
| 1m_d15 | B–A | -x+1, y, z | -17.23 | 23.2 | Stacking |
| 1m_d16 | B–A | x, y, z+1 | -15.74 | 21.2 | Stacking |
| 1m_d17 | B–A | -x+1, y, z+1 | -7.58 | 10.2 | Nonspecific |
| 1m_d18 | B–A | x, y, z | -7.24 | 9.8 | N—H···N |
| 1m_d19 | B–B | x+1, y, z | -5.28 | 7.1 | C—H···O |
| 1m_d20 | B–B | -x+1, y, z | -5.28 | 7.1 | C—H···O |
| 1m_d21 | B–B | -x+1, y+1/2, -z+2 | -4.31 | 5.8 | C—H···π |
Symmetry codes, interaction energies of the basic molecule with neighbouring
molecules (Eint, kcal mol-1) with the highest values (more than
5% of total interaction energy) and the contribution of this energy to the
total interaction energy (%) in 1r crystals (for full list of dimers,
see Table S3 in the supporting information) top| Dimer | Symmetry code | Eint | The contribution to the total interaction energy (%) | Type of interaction |
| 1r_d1 | -x+3/2, y+1/2, z | -14.02 | 18.3 | Stacking |
| 1r_d2 | -x+3/2, -y+1/2, z | -14.02 | 18.3 | Stacking |
| 1r_d3 | -x+1, -y+1, -z+1 | -10.02 | 13.1 | N—H···N |
| 1r_d4 | -x+2, -y+1, -z+1 | -5.21 | 6.8 | Nonspecific |
| 1r_d5 | x, -y+3/2, z+1/2 | -4.81 | 6.3 | C—H···π |
| 1r_d6 | x, -y+3/2, -z+1/2 | -4.81 | 6.3 | C—H···π |
| 1r_d7 | -x+3/2, -y+1, z+1/2 | -4.56 | 6.0 | C—H···π |
| 1r_d8 | -x+3/2, -y+1, -z+1/2 | -4.56 | 6.0 | C—H···π |
 N and C—H
N and C—H O hydrogen bonds link stacked columns forming layers as a secondary basic structural motif. C—H
O hydrogen bonds link stacked columns forming layers as a secondary basic structural motif. C—H π hydrogen bonds were observed between neighbouring layers and their role is the least significant in the formation of the crystal structure. Packing differences between the polymorphic modifications are minor and can be identified only using an analysis based on a comparison of the pairwise interaction energies.
π hydrogen bonds were observed between neighbouring layers and their role is the least significant in the formation of the crystal structure. Packing differences between the polymorphic modifications are minor and can be identified only using an analysis based on a comparison of the pairwise interaction energies.
 journal menu
journal menu











