
Thermal analysis, X-ray diffraction and temperature-dependent IR spectroscopy were used to study the dehydration process of crystalline
DL-phenylglycinium trifluoromethanesulfonate monohydrate (PGTFH), C
8H
10NO
2+·CF
3SO
3−·H
2O. PGTFH dehydrates in one step centred at 353 K and crystallizes in the monoclinic space group
C2/
c, whereas the anhydrous compound (PGTF) crystallizes in the triclinic space group
P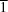
. The dehydration process in PGTFH is preceded by a weakening of both the noncovalent aromatic–aromatic interactions and the packing contacts. This process is accompanied by the breakage of medium-strength O—H

O hydrogen bonds between ions inside layers and a reorganization of the ions within the layers. This reorganization results in the formation of two different ion pairs (
DL-phenylglycinium trifluoromethanesulfonate) and the formation of a new hydrogen-bond network. The dehydration process does not destroy the nature of the crystal structure. Both crystals,
i.e. hydrated and anhydrous, have a layered structure, although the layers of each crystal are arranged somewhat differently.
Supporting information
CCDC references: 1902197; 1902198
For both structures, data collection: CrysAlis PRO (Rigaku OD, 2018); cell refinement: CrysAlis PRO (Rigaku OD, 2018); data reduction: CrysAlis PRO (Rigaku OD, 2018); program(s) used to solve structure: SHELXT (Sheldrick, 2015a); program(s) used to refine structure: SHELXL2018 (Sheldrick, 2015b); molecular graphics: OLEX2 (Dolomanov et al., 2009); software used to prepare material for publication: OLEX2 (Dolomanov et al., 2009).
DL-Phenylglycinium trifluoromethanesulfonate monohydrate (ptfh_abs_twin1_hklf4)
top
Crystal data top
| C8H10NO2+·CF3O3S−·H2O | F(000) = 1312 |
| Mr = 319.26 | Dx = 1.589 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 18.804 (5) Å | Cell parameters from 3549 reflections |
| b = 5.792 (2) Å | θ = 3.7–29.0° |
| c = 24.523 (6) Å | µ = 0.30 mm−1 |
| β = 92.57 (3)° | T = 100 K |
| V = 2668.2 (13) Å3 | Plate, colourless |
| Z = 8 | 0.42 × 0.37 × 0.08 mm |
Data collection top
Rigaku Xcalibur Atlas
diffractometer | 3085 independent reflections |
| Radiation source: fine-focus sealed X-ray tube, Enhance (Mo) X-ray Source | 2316 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.072 |
| Detector resolution: 10.6249 pixels mm-1 | θmax = 29.4°, θmin = 3.7° |
| ω scans | h = −24→24 |
Absorption correction: analytical
(CrysAlis PRO; Rigaku OD, 2018) | k = −5→7 |
| Tmin = 0.891, Tmax = 0.974 | l = −32→28 |
| 8083 measured reflections | |
Refinement top
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.056 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.153 | w = 1/[σ2(Fo2) + (0.0884P)2 + 2.4562P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max < 0.001 |
| 3085 reflections | Δρmax = 0.52 e Å−3 |
| 191 parameters | Δρmin = −0.53 e Å−3 |
| 0 restraints | |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. The single-crystal XRD measurements for hydrated form PGTFH and anhydrous
form PGTF were performed on an Xcalibur CCD diffractometer (Mo Kα
radiation, λ = 0.71073 nm). The crystallographic data for PGTFH were
collected at 100 K. CrysAlis CCD and CrysAlis RED were used for data
collection and reduction (Rigaku OD, 2015). The crystal structure was solved
and refined using the SHELXT (Sheldrick, 2015a) and SHELXL (Sheldrick, 2015b)
software packages. Non-hydrogen bonded atoms were refined with anisotropic
displacement parameters. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| S1A | 0.49397 (3) | 0.20005 (11) | 0.41586 (2) | 0.01486 (18) | |
| O1A | 0.55588 (10) | 0.0539 (3) | 0.42069 (7) | 0.0241 (4) | |
| O2A | 0.43939 (10) | 0.1506 (3) | 0.45376 (7) | 0.0230 (4) | |
| O3A | 0.50941 (9) | 0.4451 (3) | 0.41086 (7) | 0.0186 (4) | |
| C1A | 0.45288 (15) | 0.1260 (5) | 0.34933 (11) | 0.0248 (6) | |
| F1A | 0.43419 (10) | −0.0963 (3) | 0.34787 (6) | 0.0343 (5) | |
| F2A | 0.39490 (11) | 0.2523 (4) | 0.33923 (8) | 0.0467 (6) | |
| F3A | 0.49720 (11) | 0.1635 (4) | 0.30971 (7) | 0.0443 (6) | |
| O1B | 0.28684 (9) | 0.8192 (3) | 0.41034 (7) | 0.0192 (4) | |
| H1B | 0.325796 | 0.782411 | 0.426084 | 0.029* | |
| O2B | 0.26903 (9) | 1.0031 (3) | 0.48952 (7) | 0.0170 (4) | |
| C1B | 0.24969 (13) | 0.9442 (4) | 0.44343 (9) | 0.0137 (5) | |
| C2B | 0.17706 (12) | 1.0075 (4) | 0.41768 (9) | 0.0145 (5) | |
| H2B | 0.149778 | 0.862043 | 0.410004 | 0.017* | |
| N1B | 0.13842 (11) | 1.1464 (4) | 0.45883 (8) | 0.0148 (4) | |
| H1BC | 0.121974 | 1.050526 | 0.484829 | 0.018* | |
| H1BB | 0.101125 | 1.221370 | 0.441821 | 0.018* | |
| H1BA | 0.168759 | 1.251415 | 0.474764 | 0.018* | |
| C3B | 0.18124 (14) | 1.1420 (5) | 0.36488 (10) | 0.0163 (5) | |
| C4B | 0.22151 (15) | 1.3414 (5) | 0.36345 (10) | 0.0216 (6) | |
| H4B | 0.248452 | 1.389678 | 0.395110 | 0.026* | |
| C5B | 0.22272 (16) | 1.4717 (5) | 0.31577 (11) | 0.0254 (6) | |
| H5B | 0.250175 | 1.609230 | 0.315000 | 0.030* | |
| C6B | 0.18404 (16) | 1.4008 (5) | 0.26972 (11) | 0.0275 (6) | |
| H6B | 0.185179 | 1.489075 | 0.237117 | 0.033* | |
| C7B | 0.14350 (16) | 1.2013 (6) | 0.27080 (11) | 0.0286 (7) | |
| H7B | 0.116787 | 1.153213 | 0.239005 | 0.034* | |
| C8B | 0.14190 (15) | 1.0707 (5) | 0.31863 (10) | 0.0237 (6) | |
| H8B | 0.114061 | 0.934021 | 0.319512 | 0.028* | |
| O1W | 0.40477 (10) | 0.6858 (4) | 0.46303 (7) | 0.0176 (4) | |
| H1W | 0.420 (2) | 0.813 (8) | 0.4692 (15) | 0.045 (12)* | |
| H2W | 0.437 (3) | 0.599 (8) | 0.4450 (17) | 0.069 (14)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| S1A | 0.0104 (3) | 0.0164 (3) | 0.0177 (3) | 0.0006 (2) | −0.0001 (2) | −0.0003 (2) |
| O1A | 0.0169 (10) | 0.0214 (10) | 0.0334 (10) | 0.0068 (8) | −0.0046 (8) | −0.0016 (8) |
| O2A | 0.0228 (11) | 0.0204 (10) | 0.0263 (10) | −0.0049 (8) | 0.0088 (8) | −0.0023 (7) |
| O3A | 0.0130 (9) | 0.0178 (10) | 0.0251 (9) | −0.0031 (7) | 0.0011 (7) | 0.0012 (7) |
| C1A | 0.0200 (14) | 0.0309 (16) | 0.0232 (13) | −0.0044 (12) | −0.0030 (10) | 0.0027 (11) |
| F1A | 0.0406 (11) | 0.0309 (10) | 0.0310 (9) | −0.0151 (8) | −0.0031 (7) | −0.0077 (7) |
| F2A | 0.0337 (11) | 0.0524 (14) | 0.0515 (11) | 0.0037 (9) | −0.0268 (9) | 0.0046 (9) |
| F3A | 0.0516 (14) | 0.0612 (15) | 0.0208 (9) | −0.0245 (11) | 0.0077 (8) | −0.0041 (8) |
| O1B | 0.0086 (9) | 0.0249 (11) | 0.0241 (9) | 0.0053 (7) | −0.0003 (7) | −0.0022 (7) |
| O2B | 0.0127 (9) | 0.0162 (10) | 0.0218 (9) | 0.0006 (7) | −0.0022 (6) | −0.0007 (7) |
| C1B | 0.0086 (11) | 0.0110 (12) | 0.0217 (12) | −0.0011 (9) | 0.0022 (8) | 0.0010 (9) |
| C2B | 0.0094 (12) | 0.0157 (13) | 0.0182 (12) | 0.0002 (9) | −0.0007 (8) | −0.0017 (9) |
| N1B | 0.0078 (10) | 0.0173 (11) | 0.0193 (10) | −0.0001 (8) | 0.0002 (7) | −0.0011 (8) |
| C3B | 0.0104 (11) | 0.0188 (13) | 0.0199 (12) | 0.0025 (10) | 0.0017 (9) | −0.0008 (9) |
| C4B | 0.0191 (14) | 0.0247 (15) | 0.0211 (12) | −0.0013 (11) | 0.0001 (10) | 0.0007 (10) |
| C5B | 0.0283 (16) | 0.0220 (15) | 0.0260 (14) | 0.0000 (12) | 0.0032 (11) | 0.0023 (11) |
| C6B | 0.0278 (16) | 0.0315 (17) | 0.0234 (14) | 0.0088 (13) | 0.0029 (11) | 0.0078 (11) |
| C7B | 0.0255 (15) | 0.0392 (18) | 0.0205 (13) | 0.0005 (13) | −0.0041 (11) | 0.0001 (12) |
| C8B | 0.0177 (14) | 0.0301 (17) | 0.0231 (13) | −0.0038 (12) | −0.0019 (10) | −0.0005 (11) |
| O1W | 0.0110 (9) | 0.0171 (10) | 0.0250 (9) | 0.0027 (8) | 0.0023 (7) | −0.0005 (7) |
Geometric parameters (Å, º) top
| S1A—O1A | 1.440 (2) | N1B—H1BB | 0.9100 |
| S1A—O2A | 1.4439 (19) | N1B—H1BA | 0.9100 |
| S1A—O3A | 1.455 (2) | C3B—C4B | 1.382 (4) |
| S1A—C1A | 1.825 (3) | C3B—C8B | 1.389 (4) |
| C1A—F1A | 1.335 (3) | C4B—H4B | 0.9500 |
| C1A—F2A | 1.327 (4) | C4B—C5B | 1.393 (4) |
| C1A—F3A | 1.326 (3) | C5B—H5B | 0.9500 |
| O1B—H1B | 0.8400 | C5B—C6B | 1.378 (4) |
| O1B—C1B | 1.312 (3) | C6B—H6B | 0.9500 |
| O2B—C1B | 1.221 (3) | C6B—C7B | 1.385 (5) |
| C1B—C2B | 1.523 (3) | C7B—H7B | 0.9500 |
| C2B—H2B | 1.0000 | C7B—C8B | 1.397 (4) |
| C2B—N1B | 1.503 (3) | C8B—H8B | 0.9500 |
| C2B—C3B | 1.516 (3) | O1W—H1W | 0.80 (5) |
| N1B—H1BC | 0.9100 | O1W—H2W | 0.91 (5) |
| | | |
| O1A—S1A—O2A | 115.19 (12) | C2B—N1B—H1BA | 109.5 |
| O1A—S1A—O3A | 114.63 (11) | H1BC—N1B—H1BB | 109.5 |
| O1A—S1A—C1A | 104.11 (13) | H1BC—N1B—H1BA | 109.5 |
| O2A—S1A—O3A | 113.46 (11) | H1BB—N1B—H1BA | 109.5 |
| O2A—S1A—C1A | 104.13 (12) | C4B—C3B—C2B | 120.0 (2) |
| O3A—S1A—C1A | 103.42 (12) | C4B—C3B—C8B | 120.0 (2) |
| F1A—C1A—S1A | 110.56 (18) | C8B—C3B—C2B | 119.9 (2) |
| F2A—C1A—S1A | 110.4 (2) | C3B—C4B—H4B | 119.9 |
| F2A—C1A—F1A | 108.3 (2) | C3B—C4B—C5B | 120.2 (2) |
| F3A—C1A—S1A | 111.21 (19) | C5B—C4B—H4B | 119.9 |
| F3A—C1A—F1A | 108.2 (2) | C4B—C5B—H5B | 120.0 |
| F3A—C1A—F2A | 108.1 (2) | C6B—C5B—C4B | 119.9 (3) |
| C1B—O1B—H1B | 109.5 | C6B—C5B—H5B | 120.0 |
| O1B—C1B—C2B | 111.6 (2) | C5B—C6B—H6B | 119.9 |
| O2B—C1B—O1B | 125.6 (2) | C5B—C6B—C7B | 120.2 (3) |
| O2B—C1B—C2B | 122.8 (2) | C7B—C6B—H6B | 119.9 |
| C1B—C2B—H2B | 108.5 | C6B—C7B—H7B | 120.0 |
| N1B—C2B—C1B | 107.57 (18) | C6B—C7B—C8B | 120.0 (3) |
| N1B—C2B—H2B | 108.5 | C8B—C7B—H7B | 120.0 |
| N1B—C2B—C3B | 110.1 (2) | C3B—C8B—C7B | 119.6 (3) |
| C3B—C2B—C1B | 113.4 (2) | C3B—C8B—H8B | 120.2 |
| C3B—C2B—H2B | 108.5 | C7B—C8B—H8B | 120.2 |
| C2B—N1B—H1BC | 109.5 | H1W—O1W—H2W | 110 (4) |
| C2B—N1B—H1BB | 109.5 | | |
| | | |
| O1A—S1A—C1A—F1A | 61.1 (2) | C1B—C2B—C3B—C4B | −53.8 (3) |
| O1A—S1A—C1A—F2A | −179.15 (19) | C1B—C2B—C3B—C8B | 129.3 (3) |
| O1A—S1A—C1A—F3A | −59.1 (2) | C2B—C3B—C4B—C5B | −176.8 (2) |
| O2A—S1A—C1A—F1A | −60.0 (2) | C2B—C3B—C8B—C7B | 177.1 (3) |
| O2A—S1A—C1A—F2A | 59.8 (2) | N1B—C2B—C3B—C4B | 66.8 (3) |
| O2A—S1A—C1A—F3A | 179.8 (2) | N1B—C2B—C3B—C8B | −110.1 (3) |
| O3A—S1A—C1A—F1A | −178.83 (19) | C3B—C4B—C5B—C6B | −0.5 (4) |
| O3A—S1A—C1A—F2A | −59.0 (2) | C4B—C3B—C8B—C7B | 0.2 (4) |
| O3A—S1A—C1A—F3A | 61.0 (2) | C4B—C5B—C6B—C7B | 0.5 (4) |
| O1B—C1B—C2B—N1B | 179.1 (2) | C5B—C6B—C7B—C8B | −0.2 (5) |
| O1B—C1B—C2B—C3B | −58.9 (3) | C6B—C7B—C8B—C3B | −0.1 (5) |
| O2B—C1B—C2B—N1B | −0.1 (3) | C8B—C3B—C4B—C5B | 0.1 (4) |
| O2B—C1B—C2B—C3B | 121.9 (2) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1B—H1B···O1W | 0.84 | 1.79 | 2.632 (3) | 175 |
| C2B—H2B···O1Ai | 1.00 | 2.53 | 3.481 (3) | 158 |
| N1B—H1BC···O1Wii | 0.91 | 1.95 | 2.859 (3) | 172 |
| N1B—H1BB···O1Aiii | 0.91 | 2.16 | 2.954 (3) | 145 |
| N1B—H1BA···O2Biv | 0.91 | 2.02 | 2.925 (3) | 177 |
| O1W—H1W···O2Av | 0.80 (5) | 2.03 (5) | 2.781 (3) | 156 (4) |
| O1W—H2W···O3A | 0.91 (5) | 1.86 (5) | 2.771 (3) | 174 (4) |
| Symmetry codes: (i) x−1/2, y+1/2, z; (ii) −x+1/2, −y+3/2, −z+1; (iii) x−1/2, y+3/2, z; (iv) −x+1/2, −y+5/2, −z+1; (v) x, y+1, z. |
DL-Phenylglycinium trifluoromethanesulfonate (ptf_1)
top
Crystal data top
| C8H10NO2+·CF3O3S− | Z = 4 |
| Mr = 301.24 | F(000) = 616 |
| Triclinic, P1 | Dx = 1.608 Mg m−3 |
| a = 10.485 (4) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 10.635 (4) Å | Cell parameters from 2218 reflections |
| c = 13.734 (5) Å | θ = 3.1–25.9° |
| α = 104.79 (3)° | µ = 0.31 mm−1 |
| β = 98.04 (3)° | T = 373 K |
| γ = 118.07 (3)° | Irregular |
| V = 1244.4 (9) Å3 | |
Data collection top
Rigaku Xcalibur, Sapphire1, long nozzle
diffractometer | 6208 independent reflections |
| Radiation source: fine-focus sealed X-ray tube, Enhance (Mo) X-ray Source | 2625 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.086 |
| Detector resolution: 8.4350 pixels mm-1 | θmax = 29.6°, θmin = 3.2° |
| ω scans | h = −14→14 |
Absorption correction: multi-scan
(CrysAlis PRO; Rigaku OD, 2018) | k = −14→12 |
| Tmin = 0.729, Tmax = 1.000 | l = −17→18 |
| 14080 measured reflections | |
Refinement top
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.092 | H-atom parameters constrained |
| wR(F2) = 0.274 | w = 1/[σ2(Fo2) + (0.0827P)2 + 1.8829P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max < 0.001 |
| 6208 reflections | Δρmax = 0.36 e Å−3 |
| 372 parameters | Δρmin = −0.43 e Å−3 |
| 0 restraints | |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. The single-crystal XRD measurements for hydrated form PGTFH and anhydrous
form PGTF were performed on an Xcalibur CCD diffractometer (Mo Kα
radiation, λ = 0.71073 nm). The crystallographic data for PGTFH were
collected at 100 K. CrysAlis CCD and CrysAlis RED were used for data
collection and reduction (Rigaku OD, 2015). The crystal structure was solved
and refined using the SHELXT (Sheldrick, 2015a) and SHELXL (Sheldrick, 2015b)
software packages. Non-hydrogen bonded atoms were refined with anisotropic
displacement parameters. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| S1A1 | 0.10042 (17) | 0.21499 (17) | 0.87773 (12) | 0.0458 (4) | |
| O1A1 | 0.0000 (5) | 0.0878 (5) | 0.9018 (4) | 0.0687 (13) | |
| O2A1 | 0.1137 (6) | 0.3569 (5) | 0.9338 (3) | 0.0644 (12) | |
| O3A1 | 0.2388 (5) | 0.2235 (6) | 0.8783 (4) | 0.0782 (15) | |
| C1A1 | 0.0143 (9) | 0.1714 (9) | 0.7418 (5) | 0.0668 (19) | |
| F1A1 | 0.0891 (7) | 0.2817 (7) | 0.7109 (4) | 0.130 (2) | |
| F2A1 | −0.1203 (6) | 0.1570 (7) | 0.7276 (4) | 0.1218 (19) | |
| F3A1 | −0.0049 (8) | 0.0489 (7) | 0.6779 (4) | 0.134 (2) | |
| S1A2 | 0.55384 (18) | 0.65876 (17) | 0.84295 (12) | 0.0486 (4) | |
| O1A2 | 0.6600 (5) | 0.6116 (5) | 0.8498 (4) | 0.0627 (14) | 0.948 (5) |
| O2A2 | 0.6213 (7) | 0.8183 (5) | 0.8697 (4) | 0.088 (2) | 0.948 (5) |
| O3A2 | 0.4361 (6) | 0.5851 (7) | 0.8858 (4) | 0.0863 (19) | 0.948 (5) |
| O1X2 | 0.487 (9) | 0.756 (9) | 0.878 (6) | 0.05 (2)* | 0.052 (5) |
| O2X2 | 0.539 (7) | 0.568 (7) | 0.890 (5) | 0.029 (16)* | 0.052 (5) |
| O3X2 | 0.727 (7) | 0.773 (7) | 0.880 (4) | 0.027 (15)* | 0.052 (5) |
| C1A2 | 0.4592 (8) | 0.5810 (8) | 0.7032 (5) | 0.0604 (17) | |
| F1A2 | 0.3883 (8) | 0.4312 (6) | 0.6644 (4) | 0.112 (2) | 0.948 (5) |
| F2A2 | 0.5500 (6) | 0.6274 (9) | 0.6476 (4) | 0.125 (3) | 0.948 (5) |
| F3A2 | 0.3567 (6) | 0.6142 (7) | 0.6811 (4) | 0.112 (2) | 0.948 (5) |
| F1X2 | 0.493 (6) | 0.699 (6) | 0.669 (4) | 0.030 (14)* | 0.052 (5) |
| F2X2 | 0.322 (5) | 0.470 (6) | 0.663 (3) | 0.019 (12)* | 0.052 (5) |
| F3X2 | 0.527 (7) | 0.519 (7) | 0.667 (5) | 0.039 (15)* | 0.052 (5) |
| O1B1 | −0.0017 (5) | 0.5153 (5) | 0.8687 (4) | 0.0659 (13) | |
| H1B1 | 0.041384 | 0.469028 | 0.874748 | 0.099* | |
| O2B1 | 0.2195 (5) | 0.7206 (5) | 0.9728 (4) | 0.0672 (13) | |
| C1B1 | 0.0977 (7) | 0.6632 (7) | 0.9103 (5) | 0.0443 (13) | |
| C2B1 | 0.0403 (6) | 0.7529 (6) | 0.8722 (4) | 0.0416 (13) | |
| H2B1 | −0.058393 | 0.722894 | 0.883974 | 0.050* | |
| N1B1 | 0.1497 (5) | 0.9189 (5) | 0.9386 (3) | 0.0451 (11) | |
| H1BA | 0.149324 | 0.935045 | 1.005373 | 0.054* | |
| H1BB | 0.122208 | 0.975498 | 0.914537 | 0.054* | |
| H1BC | 0.242549 | 0.944897 | 0.935250 | 0.054* | |
| C3B1 | 0.0209 (6) | 0.7258 (6) | 0.7572 (4) | 0.0433 (13) | |
| C4B1 | −0.1023 (8) | 0.7115 (9) | 0.6943 (5) | 0.0688 (19) | |
| H4B1 | −0.175166 | 0.716222 | 0.723965 | 0.083* | |
| C5B1 | −0.1223 (10) | 0.6905 (10) | 0.5902 (6) | 0.090 (3) | |
| H5B1 | −0.207807 | 0.680218 | 0.549464 | 0.108* | |
| C6B1 | −0.0166 (11) | 0.6849 (9) | 0.5468 (6) | 0.084 (2) | |
| H6B1 | −0.028819 | 0.671165 | 0.475753 | 0.101* | |
| C7B1 | 0.1093 (11) | 0.6994 (10) | 0.6074 (6) | 0.088 (3) | |
| H7B1 | 0.181030 | 0.693676 | 0.576751 | 0.106* | |
| C8B1 | 0.1295 (8) | 0.7223 (9) | 0.7131 (5) | 0.069 (2) | |
| H8B1 | 0.216270 | 0.735391 | 0.754396 | 0.083* | |
| O1B2 | 0.6019 (6) | 0.0230 (6) | 0.8074 (4) | 0.0758 (14) | |
| H1B2 | 0.582690 | −0.047899 | 0.828059 | 0.114* | |
| O2B2 | 0.4404 (5) | 0.0472 (5) | 0.8888 (4) | 0.0602 (12) | |
| C1B2 | 0.5346 (7) | 0.0933 (7) | 0.8460 (5) | 0.0471 (14) | |
| C2B2 | 0.5930 (7) | 0.2413 (6) | 0.8265 (4) | 0.0435 (13) | |
| H2B2 | 0.703706 | 0.293941 | 0.843800 | 0.052* | |
| N1B2 | 0.5548 (6) | 0.3384 (5) | 0.8983 (4) | 0.0485 (12) | |
| H1BD | 0.596853 | 0.355609 | 0.964950 | 0.058* | |
| H1BE | 0.454440 | 0.290863 | 0.884235 | 0.058* | |
| H1BF | 0.589893 | 0.427185 | 0.888913 | 0.058* | |
| C3B2 | 0.5274 (7) | 0.2092 (7) | 0.7120 (4) | 0.0457 (14) | |
| C4B2 | 0.6236 (9) | 0.2745 (9) | 0.6572 (5) | 0.070 (2) | |
| H4B2 | 0.727454 | 0.339771 | 0.690657 | 0.084* | |
| C5B2 | 0.5626 (12) | 0.2411 (11) | 0.5505 (7) | 0.093 (3) | |
| H5B2 | 0.626868 | 0.286125 | 0.513142 | 0.112* | |
| C6B2 | 0.4133 (11) | 0.1453 (10) | 0.5003 (6) | 0.084 (2) | |
| H6B2 | 0.374751 | 0.122497 | 0.428673 | 0.101* | |
| C7B2 | 0.3197 (10) | 0.0825 (9) | 0.5554 (6) | 0.087 (3) | |
| H7B2 | 0.216023 | 0.016921 | 0.521258 | 0.105* | |
| C8B2 | 0.3753 (8) | 0.1140 (8) | 0.6602 (5) | 0.070 (2) | |
| H8B2 | 0.309090 | 0.070416 | 0.696895 | 0.084* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| S1A1 | 0.0514 (9) | 0.0479 (9) | 0.0497 (9) | 0.0294 (8) | 0.0193 (7) | 0.0262 (7) |
| O1A1 | 0.087 (3) | 0.051 (3) | 0.077 (3) | 0.031 (3) | 0.041 (3) | 0.041 (2) |
| O2A1 | 0.097 (4) | 0.052 (3) | 0.059 (3) | 0.052 (3) | 0.019 (3) | 0.021 (2) |
| O3A1 | 0.065 (3) | 0.118 (4) | 0.082 (3) | 0.064 (3) | 0.028 (3) | 0.048 (3) |
| C1A1 | 0.072 (5) | 0.080 (5) | 0.054 (4) | 0.040 (4) | 0.021 (4) | 0.031 (4) |
| F1A1 | 0.149 (5) | 0.144 (5) | 0.083 (3) | 0.048 (4) | 0.030 (3) | 0.083 (4) |
| F2A1 | 0.081 (3) | 0.185 (6) | 0.092 (4) | 0.078 (4) | 0.000 (3) | 0.037 (4) |
| F3A1 | 0.210 (7) | 0.133 (5) | 0.058 (3) | 0.115 (5) | 0.008 (3) | 0.002 (3) |
| S1A2 | 0.0564 (10) | 0.0419 (9) | 0.0478 (9) | 0.0250 (8) | 0.0125 (7) | 0.0210 (7) |
| O1A2 | 0.073 (3) | 0.073 (3) | 0.062 (3) | 0.049 (3) | 0.015 (3) | 0.034 (3) |
| O2A2 | 0.102 (5) | 0.036 (3) | 0.098 (4) | 0.031 (3) | −0.016 (3) | 0.019 (3) |
| O3A2 | 0.076 (4) | 0.104 (5) | 0.062 (3) | 0.027 (3) | 0.037 (3) | 0.038 (3) |
| C1A2 | 0.070 (5) | 0.063 (5) | 0.063 (4) | 0.045 (4) | 0.022 (4) | 0.021 (4) |
| F1A2 | 0.144 (6) | 0.078 (4) | 0.076 (3) | 0.059 (4) | −0.010 (3) | −0.006 (3) |
| F2A2 | 0.103 (4) | 0.228 (7) | 0.073 (3) | 0.085 (5) | 0.044 (3) | 0.093 (4) |
| F3A2 | 0.114 (4) | 0.169 (6) | 0.078 (3) | 0.113 (4) | −0.002 (3) | 0.022 (3) |
| O1B1 | 0.063 (3) | 0.036 (2) | 0.085 (3) | 0.023 (2) | 0.000 (3) | 0.019 (2) |
| O2B1 | 0.061 (3) | 0.057 (3) | 0.076 (3) | 0.032 (2) | −0.005 (3) | 0.024 (2) |
| C1B1 | 0.047 (4) | 0.040 (3) | 0.050 (3) | 0.022 (3) | 0.015 (3) | 0.024 (3) |
| C2B1 | 0.042 (3) | 0.037 (3) | 0.044 (3) | 0.020 (3) | 0.013 (3) | 0.015 (3) |
| N1B1 | 0.055 (3) | 0.037 (3) | 0.046 (3) | 0.026 (2) | 0.015 (2) | 0.016 (2) |
| C3B1 | 0.041 (3) | 0.035 (3) | 0.050 (3) | 0.016 (3) | 0.012 (3) | 0.019 (3) |
| C4B1 | 0.060 (4) | 0.096 (6) | 0.059 (4) | 0.048 (4) | 0.014 (4) | 0.029 (4) |
| C5B1 | 0.098 (6) | 0.125 (8) | 0.052 (5) | 0.067 (6) | 0.009 (5) | 0.030 (5) |
| C6B1 | 0.123 (8) | 0.074 (5) | 0.043 (4) | 0.047 (5) | 0.017 (5) | 0.020 (4) |
| C7B1 | 0.103 (7) | 0.121 (7) | 0.070 (5) | 0.071 (6) | 0.051 (5) | 0.041 (5) |
| C8B1 | 0.066 (4) | 0.097 (6) | 0.057 (4) | 0.050 (4) | 0.025 (4) | 0.030 (4) |
| O1B2 | 0.108 (4) | 0.063 (3) | 0.090 (4) | 0.059 (3) | 0.046 (3) | 0.041 (3) |
| O2B2 | 0.055 (3) | 0.056 (3) | 0.076 (3) | 0.024 (2) | 0.025 (2) | 0.041 (2) |
| C1B2 | 0.052 (4) | 0.045 (3) | 0.049 (3) | 0.028 (3) | 0.011 (3) | 0.023 (3) |
| C2B2 | 0.053 (3) | 0.040 (3) | 0.047 (3) | 0.027 (3) | 0.019 (3) | 0.022 (3) |
| N1B2 | 0.058 (3) | 0.043 (3) | 0.053 (3) | 0.030 (3) | 0.018 (3) | 0.024 (2) |
| C3B2 | 0.057 (4) | 0.045 (3) | 0.044 (3) | 0.029 (3) | 0.017 (3) | 0.025 (3) |
| C4B2 | 0.074 (5) | 0.090 (5) | 0.065 (5) | 0.045 (4) | 0.036 (4) | 0.047 (4) |
| C5B2 | 0.107 (7) | 0.123 (8) | 0.068 (5) | 0.060 (7) | 0.047 (5) | 0.053 (5) |
| C6B2 | 0.108 (7) | 0.079 (6) | 0.046 (4) | 0.038 (5) | 0.009 (5) | 0.024 (4) |
| C7B2 | 0.086 (6) | 0.082 (6) | 0.056 (5) | 0.024 (5) | −0.009 (4) | 0.025 (4) |
| C8B2 | 0.062 (5) | 0.078 (5) | 0.052 (4) | 0.019 (4) | 0.014 (4) | 0.035 (4) |
Geometric parameters (Å, º) top
| S1A1—O1A1 | 1.426 (4) | C3B1—C8B1 | 1.374 (8) |
| S1A1—O2A1 | 1.439 (4) | C4B1—H4B1 | 0.9300 |
| S1A1—O3A1 | 1.409 (5) | C4B1—C5B1 | 1.359 (10) |
| S1A1—C1A1 | 1.791 (7) | C5B1—H5B1 | 0.9300 |
| C1A1—F1A1 | 1.285 (8) | C5B1—C6B1 | 1.347 (11) |
| C1A1—F2A1 | 1.326 (8) | C6B1—H6B1 | 0.9300 |
| C1A1—F3A1 | 1.276 (8) | C6B1—C7B1 | 1.376 (11) |
| S1A2—O1A2 | 1.421 (4) | C7B1—H7B1 | 0.9300 |
| S1A2—O2A2 | 1.412 (5) | C7B1—C8B1 | 1.377 (10) |
| S1A2—O3A2 | 1.419 (5) | C8B1—H8B1 | 0.9300 |
| S1A2—O1X2 | 1.52 (7) | O1B2—H1B2 | 0.8200 |
| S1A2—O2X2 | 1.26 (6) | O1B2—C1B2 | 1.318 (7) |
| S1A2—O3X2 | 1.54 (6) | O2B2—C1B2 | 1.191 (7) |
| S1A2—C1A2 | 1.800 (7) | C1B2—C2B2 | 1.509 (7) |
| C1A2—F1A2 | 1.310 (8) | C2B2—H2B2 | 0.9800 |
| C1A2—F2A2 | 1.297 (8) | C2B2—N1B2 | 1.476 (7) |
| C1A2—F3A2 | 1.301 (7) | C2B2—C3B2 | 1.505 (7) |
| C1A2—F1X2 | 1.36 (5) | N1B2—H1BD | 0.8900 |
| C1A2—F2X2 | 1.27 (5) | N1B2—H1BE | 0.8900 |
| C1A2—F3X2 | 1.25 (6) | N1B2—H1BF | 0.8900 |
| O1B1—H1B1 | 0.8200 | C3B2—C4B2 | 1.373 (8) |
| O1B1—C1B1 | 1.312 (7) | C3B2—C8B2 | 1.367 (9) |
| O2B1—C1B1 | 1.197 (7) | C4B2—H4B2 | 0.9300 |
| C1B1—C2B1 | 1.501 (7) | C4B2—C5B2 | 1.393 (10) |
| C2B1—H2B1 | 0.9800 | C5B2—H5B2 | 0.9300 |
| C2B1—N1B1 | 1.498 (7) | C5B2—C6B2 | 1.343 (12) |
| C2B1—C3B1 | 1.497 (7) | C6B2—H6B2 | 0.9300 |
| N1B1—H1BA | 0.8900 | C6B2—C7B2 | 1.353 (11) |
| N1B1—H1BB | 0.8900 | C7B2—H7B2 | 0.9300 |
| N1B1—H1BC | 0.8900 | C7B2—C8B2 | 1.364 (9) |
| C3B1—C4B1 | 1.369 (8) | C8B2—H8B2 | 0.9300 |
| | | |
| O1A1—S1A1—O2A1 | 114.5 (3) | C4B1—C3B1—C2B1 | 120.7 (5) |
| O1A1—S1A1—C1A1 | 105.5 (3) | C4B1—C3B1—C8B1 | 118.1 (6) |
| O2A1—S1A1—C1A1 | 104.5 (3) | C8B1—C3B1—C2B1 | 121.1 (5) |
| O3A1—S1A1—O1A1 | 111.9 (3) | C3B1—C4B1—H4B1 | 118.7 |
| O3A1—S1A1—O2A1 | 115.4 (3) | C5B1—C4B1—C3B1 | 122.6 (7) |
| O3A1—S1A1—C1A1 | 103.5 (3) | C5B1—C4B1—H4B1 | 118.7 |
| F1A1—C1A1—S1A1 | 112.2 (6) | C4B1—C5B1—H5B1 | 120.4 |
| F1A1—C1A1—F2A1 | 103.2 (7) | C6B1—C5B1—C4B1 | 119.1 (7) |
| F2A1—C1A1—S1A1 | 110.9 (5) | C6B1—C5B1—H5B1 | 120.4 |
| F3A1—C1A1—S1A1 | 114.3 (5) | C5B1—C6B1—H6B1 | 119.9 |
| F3A1—C1A1—F1A1 | 108.3 (7) | C5B1—C6B1—C7B1 | 120.2 (7) |
| F3A1—C1A1—F2A1 | 107.3 (7) | C7B1—C6B1—H6B1 | 119.9 |
| O1A2—S1A2—C1A2 | 103.2 (3) | C6B1—C7B1—H7B1 | 119.8 |
| O2A2—S1A2—O1A2 | 113.8 (3) | C6B1—C7B1—C8B1 | 120.3 (7) |
| O2A2—S1A2—O3A2 | 117.2 (4) | C8B1—C7B1—H7B1 | 119.8 |
| O2A2—S1A2—C1A2 | 103.5 (3) | C3B1—C8B1—C7B1 | 119.7 (7) |
| O3A2—S1A2—O1A2 | 113.0 (3) | C3B1—C8B1—H8B1 | 120.2 |
| O3A2—S1A2—C1A2 | 103.9 (3) | C7B1—C8B1—H8B1 | 120.2 |
| O1X2—S1A2—O3X2 | 105 (4) | C1B2—O1B2—H1B2 | 109.5 |
| O1X2—S1A2—C1A2 | 98 (3) | O1B2—C1B2—C2B2 | 109.9 (5) |
| O2X2—S1A2—O1X2 | 114 (4) | O2B2—C1B2—O1B2 | 125.5 (5) |
| O2X2—S1A2—O3X2 | 103 (4) | O2B2—C1B2—C2B2 | 124.5 (5) |
| O2X2—S1A2—C1A2 | 119 (3) | C1B2—C2B2—H2B2 | 108.6 |
| O3X2—S1A2—C1A2 | 117 (2) | N1B2—C2B2—C1B2 | 108.1 (4) |
| F1A2—C1A2—S1A2 | 111.9 (5) | N1B2—C2B2—H2B2 | 108.6 |
| F2A2—C1A2—S1A2 | 113.8 (5) | N1B2—C2B2—C3B2 | 112.5 (4) |
| F2A2—C1A2—F1A2 | 105.5 (6) | C3B2—C2B2—C1B2 | 110.3 (5) |
| F2A2—C1A2—F3A2 | 107.0 (6) | C3B2—C2B2—H2B2 | 108.6 |
| F3A2—C1A2—S1A2 | 112.1 (5) | C2B2—N1B2—H1BD | 109.5 |
| F3A2—C1A2—F1A2 | 106.1 (7) | C2B2—N1B2—H1BE | 109.5 |
| F1X2—C1A2—S1A2 | 109 (2) | C2B2—N1B2—H1BF | 109.5 |
| F2X2—C1A2—S1A2 | 121 (2) | H1BD—N1B2—H1BE | 109.5 |
| F2X2—C1A2—F1X2 | 115 (3) | H1BD—N1B2—H1BF | 109.5 |
| F3X2—C1A2—S1A2 | 101 (3) | H1BE—N1B2—H1BF | 109.5 |
| F3X2—C1A2—F1X2 | 107 (4) | C4B2—C3B2—C2B2 | 119.0 (6) |
| F3X2—C1A2—F2X2 | 102 (4) | C8B2—C3B2—C2B2 | 121.9 (5) |
| C1B1—O1B1—H1B1 | 109.5 | C8B2—C3B2—C4B2 | 119.0 (6) |
| O1B1—C1B1—C2B1 | 112.3 (5) | C3B2—C4B2—H4B2 | 120.6 |
| O2B1—C1B1—O1B1 | 123.9 (5) | C3B2—C4B2—C5B2 | 118.7 (7) |
| O2B1—C1B1—C2B1 | 123.7 (5) | C5B2—C4B2—H4B2 | 120.6 |
| C1B1—C2B1—H2B1 | 108.4 | C4B2—C5B2—H5B2 | 119.2 |
| N1B1—C2B1—C1B1 | 107.5 (5) | C6B2—C5B2—C4B2 | 121.5 (7) |
| N1B1—C2B1—H2B1 | 108.4 | C6B2—C5B2—H5B2 | 119.2 |
| C3B1—C2B1—C1B1 | 113.1 (5) | C5B2—C6B2—H6B2 | 120.4 |
| C3B1—C2B1—H2B1 | 108.4 | C5B2—C6B2—C7B2 | 119.1 (7) |
| C3B1—C2B1—N1B1 | 110.8 (4) | C7B2—C6B2—H6B2 | 120.4 |
| C2B1—N1B1—H1BA | 109.5 | C6B2—C7B2—H7B2 | 119.5 |
| C2B1—N1B1—H1BB | 109.5 | C6B2—C7B2—C8B2 | 120.9 (8) |
| C2B1—N1B1—H1BC | 109.5 | C8B2—C7B2—H7B2 | 119.5 |
| H1BA—N1B1—H1BB | 109.5 | C3B2—C8B2—H8B2 | 119.7 |
| H1BA—N1B1—H1BC | 109.5 | C7B2—C8B2—C3B2 | 120.7 (7) |
| H1BB—N1B1—H1BC | 109.5 | C7B2—C8B2—H8B2 | 119.7 |
| | | |
| O1A1—S1A1—C1A1—F1A1 | 176.1 (6) | O2B1—C1B1—C2B1—C3B1 | 113.8 (7) |
| O1A1—S1A1—C1A1—F2A1 | 61.3 (6) | C1B1—C2B1—C3B1—C4B1 | 139.3 (6) |
| O1A1—S1A1—C1A1—F3A1 | −60.1 (7) | C1B1—C2B1—C3B1—C8B1 | −44.0 (8) |
| O2A1—S1A1—C1A1—F1A1 | 54.9 (6) | C2B1—C3B1—C4B1—C5B1 | 178.4 (7) |
| O2A1—S1A1—C1A1—F2A1 | −59.8 (6) | C2B1—C3B1—C8B1—C7B1 | −179.1 (7) |
| O2A1—S1A1—C1A1—F3A1 | 178.8 (6) | N1B1—C2B1—C3B1—C4B1 | −99.9 (6) |
| O3A1—S1A1—C1A1—F1A1 | −66.2 (7) | N1B1—C2B1—C3B1—C8B1 | 76.9 (7) |
| O3A1—S1A1—C1A1—F2A1 | 179.0 (6) | C3B1—C4B1—C5B1—C6B1 | −0.6 (13) |
| O3A1—S1A1—C1A1—F3A1 | 57.6 (7) | C4B1—C3B1—C8B1—C7B1 | −2.2 (10) |
| O1A2—S1A2—C1A2—F1A2 | 61.5 (6) | C4B1—C5B1—C6B1—C7B1 | 0.3 (13) |
| O1A2—S1A2—C1A2—F2A2 | −57.9 (6) | C5B1—C6B1—C7B1—C8B1 | −1.1 (13) |
| O1A2—S1A2—C1A2—F3A2 | −179.5 (6) | C6B1—C7B1—C8B1—C3B1 | 2.1 (12) |
| O2A2—S1A2—C1A2—F1A2 | −179.6 (6) | C8B1—C3B1—C4B1—C5B1 | 1.5 (11) |
| O2A2—S1A2—C1A2—F2A2 | 61.0 (7) | O1B2—C1B2—C2B2—N1B2 | −162.1 (5) |
| O2A2—S1A2—C1A2—F3A2 | −60.6 (7) | O1B2—C1B2—C2B2—C3B2 | 74.5 (6) |
| O3A2—S1A2—C1A2—F1A2 | −56.6 (6) | O2B2—C1B2—C2B2—N1B2 | 18.7 (8) |
| O3A2—S1A2—C1A2—F2A2 | −176.0 (6) | O2B2—C1B2—C2B2—C3B2 | −104.7 (7) |
| O3A2—S1A2—C1A2—F3A2 | 62.4 (6) | C1B2—C2B2—C3B2—C4B2 | −128.9 (6) |
| O1X2—S1A2—C1A2—F1X2 | 57 (4) | C1B2—C2B2—C3B2—C8B2 | 49.5 (8) |
| O1X2—S1A2—C1A2—F2X2 | −79 (4) | C2B2—C3B2—C4B2—C5B2 | 178.3 (6) |
| O1X2—S1A2—C1A2—F3X2 | 170 (4) | C2B2—C3B2—C8B2—C7B2 | −177.6 (6) |
| O2X2—S1A2—C1A2—F1X2 | −179 (4) | N1B2—C2B2—C3B2—C4B2 | 110.3 (6) |
| O2X2—S1A2—C1A2—F2X2 | 45 (5) | N1B2—C2B2—C3B2—C8B2 | −71.2 (7) |
| O2X2—S1A2—C1A2—F3X2 | −66 (5) | C3B2—C4B2—C5B2—C6B2 | −1.0 (13) |
| O3X2—S1A2—C1A2—F1X2 | −54 (4) | C4B2—C3B2—C8B2—C7B2 | 0.9 (11) |
| O3X2—S1A2—C1A2—F2X2 | 170 (4) | C4B2—C5B2—C6B2—C7B2 | 1.4 (14) |
| O3X2—S1A2—C1A2—F3X2 | 58 (4) | C5B2—C6B2—C7B2—C8B2 | −0.7 (14) |
| O1B1—C1B1—C2B1—N1B1 | 170.0 (5) | C6B2—C7B2—C8B2—C3B2 | −0.5 (12) |
| O1B1—C1B1—C2B1—C3B1 | −67.3 (6) | C8B2—C3B2—C4B2—C5B2 | −0.2 (10) |
| O2B1—C1B1—C2B1—N1B1 | −8.9 (8) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1B1—H1B1···O2A1 | 0.82 | 1.96 | 2.735 (6) | 159 |
| N1B1—H1BA···O1A1i | 0.89 | 2.12 | 2.865 (6) | 141 |
| N1B1—H1BB···O1A1ii | 0.89 | 2.14 | 2.973 (6) | 155 |
| N1B1—H1BC···O2B2ii | 0.89 | 2.12 | 2.960 (6) | 157 |
| O1B2—H1B2···O2A2iii | 0.82 | 1.85 | 2.614 (6) | 155 |
| N1B2—H1BD···O3A2iv | 0.89 | 2.11 | 2.842 (7) | 139 |
| N1B2—H1BE···O3A1 | 0.89 | 2.01 | 2.883 (7) | 168 |
| N1B2—H1BF···O1A2 | 0.89 | 1.99 | 2.878 (6) | 173 |
| Symmetry codes: (i) −x, −y+1, −z+2; (ii) x, y+1, z; (iii) x, y−1, z; (iv) −x+1, −y+1, −z+2. |
Table 1 Thermodynamic parameters (Tonset, Tmax,
ΔHonset and ΔSonset) for dehydration and melting processes
estimated from DSC measurements top| Process | Tonseta (K) | Tmaxa (K) | ΔHonseta (kJ mol-1) | ΔSonseta (J mol-1 K-1) |
| Dehydration | /353.4 | /379.2 | /21.8 | /61.7 |
| Solid→liquid (1st cycle) | 456.3/461.3 | 448.7/493.5 | -20.6/56.2 | -45.2/121.8 |
| Solid→liquid (2nd cycle) | 450.6/497.1 | 444.0/503.6 | -20.1/26.2 | -44.5/52.7 |
| Note: (a) the values given before and after `/' are related to the
cooling and heating process, respectively. |
Table 3 The wavenumbers (cm-1), IR and Raman intensities of the bands
observed in the vibrational spectra of polycrystalline samples of the PGTFH
and PGTF (after annealing) at room temperature top| PGTFH | PGTF | Proposed assignmentsb,c | | |
| IR (cm-1) | Ramana (cm-1) | IR (cm-1) | Ramana (cm-1) | |
| 3418 s sh | | | | νasH2O(cryst) |
| 3361 s b | | | | νasH2O(cryst) |
| 3228 s | 3214 (2) | | 3214 (2) | νN—H···O |
| 3164 s | 3165 (2) | 3191 s sh | | νN—H···O |
| | 3160 vs b | | νOH (O—H···O) |
| 3147 s sh | | 3153 s b | 3150 (3) | νN—H···O/ν8a(νCC) + ν19a(νCC) |
| 3111 w | | | | ν8b(νCC) + ν19a(νCC) |
| 3103 w | | 3098 s sh | | |
| | 3084 s sh | 3084 (10) | |
| 3079 (15) | | 3074 (8) | |
| 3061 (8) | 3067 s sh | 3066 (13) | ν2(νCH) |
| 3051 (4) | 3054 s sh | 3055 (7) | ν20b(νCH) |
| 3046 vs b | | | | νOH (A) (O—H···Ow) |
| 3042 s | 3042 (3) | 3041 s sh | | ν7a(νCH) |
| 3024 (3) | | | ν20a(νCH) |
| 3019 (3) | | 3020 (3) | ν7b(νCH) |
| | | 3002 (5) | |
| 2991 (2) | | 2996 (4) | |
| 2968 s | 2970 (6) | 2975 s sh | 2980 (6) | νCH/2 × ν19a(CC) |
| | | 2974 (5) sh | |
| 2933 w sh | | | | |
| 2869 m | | 2852 w sh | | |
| 2776 w | | | | |
| 2728 vw | | 2723 w | | |
| 2697 vw sh | | | | |
| 2672 w | | | | |
| 2623 m b | | 2624 w b | | νOH (B) (O—H···Ow) |
| 2602 m | | 2607 w b | | |
| 2528 vw | | | | |
| | 1764 sh | 1763 (10) sh | νC═O |
| | 1742 s | 1748 (19) | νC═O |
| 1722 s | 1726 (7) | | | νC═O |
| 1652 w b | | | | δH2O |
| 1614 m | 1606 (22) | 1603 m | 1605 (33) | ν8a(νCC) |
| 1591 | 1591 (16) | | 1590 (27) | ν8b(νCC) |
| 1582 w | | | | δasNH3+ |
| | 1508 s sh | | ν19a(νCC) |
| 1501 s | 1502 (2) | 1503 s | 1502 (14) | ν19a(νCC) |
| 1460 s | 1462 (2) | 1460 m | 1462 (15) | ν19b(νCC) + ν14(νCC) |
| 1456 w sh | | | | δOH (O—H···Ow) |
| | 1420 w | 1423 (15) | δsNH3+ |
| 1399 vw | | | | νC-O(H) |
| 1358 w | 1357 (3) | 1356 m | 1357 (17) | δCH |
| 1347 s | 1344 (2) | 1341 w | | γCH + νNH3+ |
| 1307 vw sh | 1308 (2) | | 1305 (17) | ν3(νCH) |
| | 1284 vs | 1284 (18) | |
| 1277 vs | | 1278 vs sh | 1270 (19) | νasSO3 |
| 1266 vs | 1268 (9) | 1249 s | 1251 (19) | νasSO3 |
| 1237 vs | | | | νC—O(H) |
| 1227 vs | 1228 (14) | 1227 vs b | 1229 (30) | νsCF3/νC—O(H) |
| | 1220 vs sh | 1213 (22) | |
| 1194 w | 1196 (19) | 1200 w sh | 1192 (29) | ν(C—Cring) + νCH + νCN |
| 1182 s | 1184 (6) sh | 1176 vs | | ν9a(δCH) |
| 1175 s | 1174 (6) | 1176 vs sh | 1172 (19) | νasCF3 |
| 1159 w | 1160 (10) | 1158 s | 1158 (23) | ν15(νCH) |
| 1128 w | 1127 (2) | | | δOH (O—H···Ow) |
| 1111 vw | 1111 (3) | 1111 m | 1111 (18) | νasSO3 |
| 1100 w | 1099 (3) | 1097 m | 1097 (17) | νCN + ν18b(δCH)/γCH |
| 1074 w | 1073 (2) | 1071 m | 1071 (17) | ν18b(δCH) + νCN |
| 1031 vs | 1037 (56) | 1034 vs | 1039 (85) | νsSO3 |
| 1021 (18) | 1027 | 1017 (24) | ν18a(δCH) |
| 1003 (100) | 1008 vw | 1003 (100) | ν12(δCC) + ν1(νCC) |
| 991 vw | 992 (5) | 988 vw | 992 (21) sh | ν5(νCH) |
| 970 (2) | 963 vw | | ν17a(νCH) |
| 952 m b | | | | γOH (O—H···Ow) |
| 922 w | 920 (3) | 921 m | 923 (17) | ν17b(νCH) |
| 888 m | 889 (4) | 882 m | 882 (19) | ν1(νCC) |
| | 849 vw | | |
| 833 w | 833 (11) | 827 m | 825 (26) | 2 × ν16a(δCC) |
| | 795 w b | | γOH (O—H···O) |
| 766 s | 766 (38) | 766 w sh | 767 (52) | δsCF3 |
| | 761 m | | δsCF3 |
| | 744 vw | | |
| 737 m | 741 (9) | | 727 (29) | ν11(νCH) + νCOO + ν4(νCC) |
| 697 s | | 695 m | | νCCN + νCOO |
| 672 vw | | | | τH2O, ρH2O |
| | | 657 (17) | |
| 643 s | 652 (3) | 643 s | 646 (17) | δsSO3 |
| 635 (3) | 637 s sh | | δCOO |
| 615 m | 616 (14) | 616 vw | 616 (30) | ν(Cring—CN) + ν6b(νCC)/ν6a(νCC) |
| 597 vw | | | | ωH2O |
| 583 sh | 585 (9) | 585 m | 584 (23) | δsSO3 |
| 573 s | 577 (9) | 574 m sh | 575 (20) | δasCF3 |
| 522 m | | 525 w | 527 (16) | ν16b(νCC) +ν(CCC)ringC |
| 514 s | 513 (5) b | 512 m | 514 (17) | δasSO3 |
| | 498 m | 497 (21) | tNH3+(dehydrated) |
| 485 w | 484 (4) | 477 w | 479 (19) | δCC═O + νCC/skeletal vibrations |
| 352 (25) sh | | 352 (36) sh | ρSO3 |
| 349 (27) | | 348 (39) | ρSO3 |
| 318 (22) | | 322 (40) | νCS |
| 284 (4) | | 270 (16) | νr(NH3+) |
| | | 222 (12) | |
| Notes: (a) relative intensities are given in parentheses with respect
to the most intense Raman band in the PGTFH and in the PGTF crystals spectra
at 1003 cm-1, respectively. Their intensities were taken as 100.
(b) Used symbols: vs = very strong, s = strong,
m = medium, w = weak, vw = very weak, b = broad,
sh = shoulder; ν = stretching (s = symmetric and as = asymmetric),
δ = bending in plane, γ = bending out of plane, ρ = rocking, τ = torsion,
ω = wagging and t = twisting.
(c) The phenyl-ring internal vibrations are marked by symbols taken
from Varsányi (1969) and Wojtkowiak & Chabanel (1977). |
Table 3 Analysis of the fundamental (k = 0) modes of the PGTF crystal top| Cia | Nb | A | T | L | C6H5CHNH3COOH | CF3SO3- | Selection rules | | |
| | | | | νPG | νH | νac | IR | Raman |
| Ag | 174 | 0 | 12 | 12 | 108 | 6 | 36 | ia | xx,yy,zz,xy,xz,yz |
| Au | 174 | 3 | 9 | 12 | 108 | 6 | 36 | x,y,z | ia |
| Notes: (a) unit-cell symmetry; (b)
abbreviations: N is the total number of modes,
A are the acoustic modes,
T are the translation modes,
L are the libration modes;
νPG are the internal vibrations of phenylglycine,
νH are the modes of the proton in O—H···O hydrogen bonds and
νac are the internal modes of the trifluoromethanesulfonate anion. |
Table 2 Analysis of the fundamental (k = 0) modes of the PGTFH crystal top| C2ha | Nb | A | T | L | C6H5CHNH3COOH | CF3SO3- | H2O | Selection rules | | |
| | | | | νPG | νH | νac | νw | IR | Raman |
| Ag | 96 | 0 | 9 | 9 | 54 | 3 | 18 | 3 | ia | xx,yy,zz,xy |
| Bg | 96 | 0 | 9 | 9 | 54 | 3 | 18 | 3 | ia | xz,yz |
| Au | 96 | 1 | 8 | 9 | 54 | 3 | 18 | 3 | z | ia |
| Bu | 96 | 2 | 7 | 9 | 54 | 3 | 18 | 3 | x,y | ia |
| Notes: (a) unit-cell symmetry; (b)
abbreviations: N is the total number of modes,
A are the acoustic modes,
T are the translation modes,
L are the libration modes;
νPG are the internal vibrations of phenylglycine,
νH are the modes of the proton in O—H···O hydrogen bonds,
νac are the internal modes of the trifluoromethanesulfonate anion and
νw are the internal vibrations of water molecules. |
 O hydrogen bonds between ions inside layers and a reorganization of the ions within the layers. This reorganization results in the formation of two different ion pairs (DL-phenylglycinium trifluoromethanesulfonate) and the formation of a new hydrogen-bond network. The dehydration process does not destroy the nature of the crystal structure. Both crystals, i.e. hydrated and anhydrous, have a layered structure, although the layers of each crystal are arranged somewhat differently.
O hydrogen bonds between ions inside layers and a reorganization of the ions within the layers. This reorganization results in the formation of two different ion pairs (DL-phenylglycinium trifluoromethanesulfonate) and the formation of a new hydrogen-bond network. The dehydration process does not destroy the nature of the crystal structure. Both crystals, i.e. hydrated and anhydrous, have a layered structure, although the layers of each crystal are arranged somewhat differently.
 journal menu
journal menu











