The double salt [CuCl
2(H
2O)
2{CdCl
2}]·2H
2O crystallizes in the triclinic rather than the monoclinic system as reported previously. The structure consists of sheets in the
ac plane with slightly distorted octahedral CdCl
6 [Cd—Cl 2.5813 (8)–2.6943 (8) Å] connected by Cd—Cl—Cd bridges in the Cd equatorial plane along
a, and by Cd—Cl—Cu bridges to layers of square-planar CuCl
2(H
2O)
2 along
c. There are long axial Cu—Cl interactions of 2.8623 (7) Å and additional water of hydration is hydrogen bonded to coordinated water and chloride ligands. The additional water connects the
ac sheets into a three-dimensional network. Both Cd and Cu occupy different
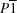
sites. The Cu

Cu and Cd

Cd distances are 3.8274 (6) Å.
Supporting information
The CdCl2–CuCl2–H2O system has been explored previously (Bassett &
Strain, 1952). Crystals were obtained after about four weeks by evaporation
from a solution containing CuCl2.2H2O (18.75 g, 0.11 mol) and CdCl2
(9.17 g, 0.05 mol) dissolved in H2O (20 ml).
Crystal decay was monitored by recollection of the first 50 frames periodically
throughout the data collection. H atoms were refined isotropically in observed
positions.
Data collection: SMART (Bruker, 1998); cell refinement: SMART; data reduction: SHELXTL (Sheldrick, 1997a); program(s) used to solve structure: SHELXS97 (Sheldrick, 1990); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997b); molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Crystal data top
| [CuCl2(H2O)2{CdCl2}]·2H2O | Z = 1 |
| Mr = 389.80 | F(000) = 185 |
| Triclinic, P1 | Dx = 2.801 Mg m−3 |
| a = 3.8274 (6) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 7.1591 (11) Å | Cell parameters from 2232 reflections |
| c = 8.7756 (13) Å | θ = 5.9–56.6° |
| α = 87.595 (2)° | µ = 5.72 mm−1 |
| β = 82.735 (2)° | T = 299 K |
| γ = 75.639 (2)° | Column, green |
| V = 231.06 (6) Å3 | 0.38 × 0.10 × 0.05 mm |
Data collection top
CCD area-detector
diffractometer | 1094 independent reflections |
| Radiation source: normal-focus sealed tube | 1033 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.028 |
| ω scans | θmax = 28.3°, θmin = 2.3° |
Absorption correction: multi-scan
(Blessing, 1995) | h = −5→5 |
| Tmin = 0.475, Tmax = 0.810 | k = −9→9 |
| 2771 measured reflections | l = −11→11 |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.027 | All H-atom parameters refined |
| wR(F2) = 0.071 | w = 1/[σ2(Fo2) + (0.0468P)2 + 0.0268P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.07 | (Δ/σ)max < 0.001 |
| 1094 reflections | Δρmax = 0.65 e Å−3 |
| 66 parameters | Δρmin = −0.84 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.417 (14) |
Crystal data top
| [CuCl2(H2O)2{CdCl2}]·2H2O | γ = 75.639 (2)° |
| Mr = 389.80 | V = 231.06 (6) Å3 |
| Triclinic, P1 | Z = 1 |
| a = 3.8274 (6) Å | Mo Kα radiation |
| b = 7.1591 (11) Å | µ = 5.72 mm−1 |
| c = 8.7756 (13) Å | T = 299 K |
| α = 87.595 (2)° | 0.38 × 0.10 × 0.05 mm |
| β = 82.735 (2)° | |
Data collection top
CCD area-detector
diffractometer | 1094 independent reflections |
Absorption correction: multi-scan
(Blessing, 1995) | 1033 reflections with I > 2σ(I) |
| Tmin = 0.475, Tmax = 0.810 | Rint = 0.028 |
| 2771 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.027 | 0 restraints |
| wR(F2) = 0.071 | All H-atom parameters refined |
| S = 1.07 | Δρmax = 0.65 e Å−3 |
| 1094 reflections | Δρmin = −0.84 e Å−3 |
| 66 parameters | |
Special details top
Experimental. Crystal decay was monitored by re-collection of the first fifty frames
periodically throughout the data collection. |
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Cd | 0 | 0 | 0.5000 | 0.02412 (17) | |
| Cl2 | 0.38166 (18) | 0.25311 (10) | 0.47309 (8) | 0.0263 (2) | |
| Cl1 | 0.06418 (18) | −0.05094 (11) | 0.19434 (8) | 0.0288 (2) | |
| Cu | 0.5000 | 0 | 0 | 0.02544 (18) | |
| O1 | 0.3722 (7) | 0.2746 (3) | 0.0369 (3) | 0.0314 (5) | |
| H1A | 0.342 (11) | 0.344 (6) | −0.044 (5) | 0.043 (11)* | |
| H1B | 0.197 (14) | 0.323 (8) | 0.099 (6) | 0.061 (15)* | |
| O2 | 0.2219 (9) | 0.5051 (4) | −0.2125 (4) | 0.0374 (6) | |
| H2B | 0.196 (19) | 0.446 (11) | −0.291 (9) | 0.09 (2)* | |
| H2A | 0.372 (18) | 0.556 (9) | −0.233 (7) | 0.065 (19)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cd | 0.0220 (2) | 0.0295 (2) | 0.0218 (2) | −0.00814 (13) | −0.00270 (12) | 0.00072 (12) |
| Cl2 | 0.0249 (3) | 0.0253 (4) | 0.0285 (4) | −0.0069 (3) | −0.0018 (3) | 0.0016 (3) |
| Cl1 | 0.0270 (4) | 0.0423 (4) | 0.0178 (3) | −0.0111 (3) | −0.0003 (3) | −0.0001 (3) |
| Cu | 0.0270 (3) | 0.0238 (3) | 0.0228 (3) | −0.0045 (2) | 0.0040 (2) | −0.0003 (2) |
| O1 | 0.0373 (12) | 0.0267 (12) | 0.0273 (11) | −0.0045 (10) | 0.0009 (10) | −0.0016 (9) |
| O2 | 0.0498 (16) | 0.0310 (13) | 0.0304 (12) | −0.0104 (12) | −0.0007 (10) | 0.0015 (9) |
Geometric parameters (Å, º) top
| Cd—Cl2i | 2.5813 (8) | Cu—O1 | 1.935 (2) |
| Cd—Cl2 | 2.5813 (8) | Cu—O1v | 1.935 (2) |
| Cd—Cl2ii | 2.6303 (7) | Cu—Cl1v | 2.3139 (7) |
| Cd—Cl2iii | 2.6303 (7) | Cu—Cl1iv | 2.8623 (7) |
| Cd—Cl1i | 2.6943 (8) | O1—H1A | 0.85 (5) |
| Cd—Cl1 | 2.6943 (8) | O1—H1B | 0.82 (5) |
| Cl2—Cdiv | 2.6303 (7) | O2—H2B | 0.85 (8) |
| Cl1—Cu | 2.3139 (7) | O2—H2A | 0.75 (7) |
| | | |
| Cl2i—Cd—Cl2 | 180.00 (3) | Cl1i—Cd—Cl1 | 180.0 |
| Cl2i—Cd—Cl2ii | 85.49 (3) | Cd—Cl2—Cdiv | 94.51 (3) |
| Cl2—Cd—Cl2ii | 94.51 (3) | Cu—Cl1—Cd | 130.99 (3) |
| Cl2i—Cd—Cl2iii | 94.51 (3) | Cuiv—Cl1—Cdiv | 128.44 (3) |
| Cl2—Cd—Cl2iii | 85.49 (3) | O1—Cu—O1v | 180.0 |
| Cl2ii—Cd—Cl2iii | 180.00 (3) | O1—Cu—Cl1 | 92.00 (8) |
| Cl2i—Cd—Cl1i | 93.24 (2) | O1v—Cu—Cl1 | 88.00 (8) |
| Cl2—Cd—Cl1i | 86.76 (2) | O1—Cu—Cl1v | 88.00 (8) |
| Cl2ii—Cd—Cl1i | 92.51 (2) | O1v—Cu—Cl1v | 92.00 (8) |
| Cl2iii—Cd—Cl1i | 87.49 (2) | Cl1—Cu—Cl1v | 180.00 (5) |
| Cl2i—Cd—Cl1 | 86.76 (2) | Cu—O1—H1A | 114 (3) |
| Cl2—Cd—Cl1 | 93.24 (2) | Cu—O1—H1B | 120 (4) |
| Cl2ii—Cd—Cl1 | 87.49 (2) | H1A—O1—H1B | 105 (4) |
| Cl2iii—Cd—Cl1 | 92.51 (2) | H2B—O2—H2A | 109 (7) |
| Symmetry codes: (i) −x, −y, −z+1; (ii) x−1, y, z; (iii) −x+1, −y, −z+1; (iv) x+1, y, z; (v) −x+1, −y, −z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···O2 | 0.85 (5) | 1.87 (5) | 2.718 (4) | 174 (4) |
| O1—H1B···O2vi | 0.82 (5) | 1.95 (5) | 2.752 (4) | 165 (5) |
| O2—H2A···O1vii | 0.76 (7) | 2.57 (6) | 3.050 (4) | 124 (6) |
| O2—H2A···Cl2vii | 0.76 (7) | 2.66 (7) | 3.259 (3) | 138 (6) |
| O2—H2B···Cl2viii | 0.85 (8) | 2.46 (8) | 3.269 (3) | 158 (6) |
| Symmetry codes: (vi) −x, −y+1, −z; (vii) −x+1, −y+1, −z; (viii) x, y, z−1. |
Experimental details
| Crystal data |
| Chemical formula | [CuCl2(H2O)2{CdCl2}]·2H2O |
| Mr | 389.80 |
| Crystal system, space group | Triclinic, P1 |
| Temperature (K) | 299 |
| a, b, c (Å) | 3.8274 (6), 7.1591 (11), 8.7756 (13) |
| α, β, γ (°) | 87.595 (2), 82.735 (2), 75.639 (2) |
| V (Å3) | 231.06 (6) |
| Z | 1 |
| Radiation type | Mo Kα |
| µ (mm−1) | 5.72 |
| Crystal size (mm) | 0.38 × 0.10 × 0.05 |
| |
| Data collection |
| Diffractometer | CCD area-detector
diffractometer |
| Absorption correction | Multi-scan
(Blessing, 1995) |
| Tmin, Tmax | 0.475, 0.810 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 2771, 1094, 1033 |
| Rint | 0.028 |
| (sin θ/λ)max (Å−1) | 0.667 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.027, 0.071, 1.07 |
| No. of reflections | 1094 |
| No. of parameters | 66 |
| H-atom treatment | All H-atom parameters refined |
| Δρmax, Δρmin (e Å−3) | 0.65, −0.84 |
Selected geometric parameters (Å, º) top| Cd—Cl2 | 2.5813 (8) | Cl1—Cu | 2.3139 (7) |
| Cd—Cl2i | 2.6303 (7) | Cu—O1 | 1.935 (2) |
| Cd—Cl1 | 2.6943 (8) | Cu—Cl1ii | 2.8623 (7) |
| | | |
| Cu—Cl1—Cd | 130.99 (3) | Cuii—Cl1—Cdii | 128.44 (3) |
| Symmetry codes: (i) x−1, y, z; (ii) x+1, y, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···O2 | 0.85 (5) | 1.87 (5) | 2.718 (4) | 174 (4) |
| O1—H1B···O2iii | 0.82 (5) | 1.95 (5) | 2.752 (4) | 165 (5) |
| O2—H2A···O1iv | 0.76 (7) | 2.57 (6) | 3.050 (4) | 124 (6) |
| O2—H2A···Cl2iv | 0.76 (7) | 2.66 (7) | 3.259 (3) | 138 (6) |
| O2—H2B···Cl2v | 0.85 (8) | 2.46 (8) | 3.269 (3) | 158 (6) |
| Symmetry codes: (iii) −x, −y+1, −z; (iv) −x+1, −y+1, −z; (v) x, y, z−1. |
 Cu and Cd
Cu and Cd Cd distances are 3.8274 (6) Å.
Cd distances are 3.8274 (6) Å.![[Figure 1]](http://journals.iucr.org/c/issues/2000/03/00/br1267/br1267fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2000/03/00/br1267/br1267fig2thm.gif)

 journal menu
journal menu












The structural analysis of the title salt was performed in support of an examination of its magnetic behavior. A previous Weissenberg camera diffraction study using Cu radiation (Thrierr-Sorel et al., 1969) indicated a C-centered monoclinic cell. In contrast, we find the structure to be triclinic with a cell one-quarter the volume of that reported earlier. The green columnar crystals tend to twin and only parallelepiped-shaped crystals were indexed successfully.
The title structure (Fig. 1) consists of approximately octahedral CdCl6 linked into a linear polymer by double Cl bridges running along a. The Cl—Cd—Cl angles are within 8° of 90 or 180°. The axial Cl ligand bridges to Cu, which forms square-planar CuCl2(H2O)2, and these Cl bridges connect the Cd and Cu atoms into sheets in the ac plane (Fig. 2). Cl1 also forms a long interaction with a neighboring Cu atom with an axial Cu—Cl distance of 2.8623 (7) Å. Angles around copper are within 6° of 90 or 180°. The water coordinated to Cu is hydrogen bonded to non-coordinated lattice water, which is in turn hydrogen bonded to Cl2. These hydrogen bonds connect the sheets along the b direction to form a three-dimensional network. The Cu and Cd atoms occupy different 1 sites such that the closest Cd···Cd and Cu···Cu distances are both one-unit translations along the a axis, i.e. 3.8274 (6) Å.