
Crystals of C
10H
8N
3O
2Br undergo two reversible phase transitions between 295 and 100 K. The first, of an order–disorder nature, is a second-order transition and takes place continuously over a wide temperature range. This transition is connected with the doubling of the length of the
c axis of the unit cell and with the change of the space group from
P2
1/
m with
Z′ = 1/2 (room-temperature α-phase) to
P2
1/
c,
Z′ = 1 (β-phase, 200–120 K). During this transition the molecule loses the
Cs symmetry of the α-phase. The second transition takes place between 118 and 115 K, and is accompanied by a change of the crystal symmetry to the triclinic space group
P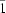
(low-temperature γ-phase). This second phase transition is accompanied by the twinning of the crystal. Neither the molecular geometry nor the crystal packing shows any dramatic changes during these phase transitions. Halogen bonds C—Br

N and dihalogen interactions Br

Br play a crucial role in determining the crystal packing and compete successfully with other kinds of weak intermolecular interactions.
Supporting information
CCDC references: 241464; 241465; 241466; 241467; 241468; 241469; 241470
For all compounds, data collection: CrysAlis CCD v.169 (Oxford Diffraction, 2003a); cell refinement: CrysAlis CCD v.169 (Oxford Diffraction, 2003a); data reduction: CrysAlis RED v.169 (Oxford Diffraction, 2003b). Program(s) used to solve structure: SHELXS97 (Sheldrick, 1990) for 295K, 250K, 200K, 170K, 150K, 120K; SIR2002 (Burla et al., 2003) for 100K. For all compounds, program(s) used to refine structure: SHELXL97 (Sheldrick, 1997).
(295K) 1-phenyl-4-nitro-5-bromo-imidazole
top
Crystal data top
| C10H8BrN3O2 | F(000) = 280 |
| Mr = 282.10 | Dx = 1.735 Mg m−3 |
| Monoclinic, P21/m | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybm | Cell parameters from 4253 reflections |
| a = 7.0281 (10) Å | θ = 3–20° |
| b = 6.9388 (9) Å | µ = 3.80 mm−1 |
| c = 11.5612 (15) Å | T = 295 K |
| β = 106.735 (12)° | Prism, colourless |
| V = 539.92 (13) Å3 | 0.3 × 0.25 × 0.1 mm |
| Z = 2 | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 1521 independent reflections |
| Radiation source: fine-focus sealed tube | 1342 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.042 |
| ω–scan | θmax = 29.7°, θmin = 3.0° |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | h = −9→9 |
| Tmin = 0.382, Tmax = 0.696 | k = −9→9 |
| 5105 measured reflections | l = −6→15 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.029 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.075 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0509P)2]
where P = (Fo2 + 2Fc2)/3 |
| 1521 reflections | (Δ/σ)max = 0.001 |
| 93 parameters | Δρmax = 0.36 e Å−3 |
| 0 restraints | Δρmin = −1.18 e Å−3 |
Crystal data top
| C10H8BrN3O2 | V = 539.92 (13) Å3 |
| Mr = 282.10 | Z = 2 |
| Monoclinic, P21/m | Mo Kα radiation |
| a = 7.0281 (10) Å | µ = 3.80 mm−1 |
| b = 6.9388 (9) Å | T = 295 K |
| c = 11.5612 (15) Å | 0.3 × 0.25 × 0.1 mm |
| β = 106.735 (12)° | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 1521 independent reflections |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | 1342 reflections with I > 2σ(I) |
| Tmin = 0.382, Tmax = 0.696 | Rint = 0.042 |
| 5105 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.029 | 0 restraints |
| wR(F2) = 0.075 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.02 | Δρmax = 0.36 e Å−3 |
| 1521 reflections | Δρmin = −1.18 e Å−3 |
| 93 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N1 | 0.8140 (3) | 0.2500 | 0.6559 (2) | 0.0348 (5) | |
| C11 | 0.9755 (4) | 0.2500 | 0.7656 (3) | 0.0404 (6) | |
| C12 | 1.0488 (4) | 0.4233 (5) | 0.8165 (3) | 0.0633 (7) | |
| H12 | 0.9998 | 0.5391 | 0.7792 | 0.101 (9)* | |
| C13 | 1.1989 (5) | 0.4205 (7) | 0.9258 (3) | 0.0859 (11) | |
| H13 | 1.2510 | 0.5361 | 0.9622 | 0.101 (9)* | |
| C14 | 1.2701 (6) | 0.2500 | 0.9801 (4) | 0.0897 (19) | |
| H14 | 1.3674 | 0.2500 | 1.0543 | 0.101 (9)* | |
| C2 | 0.6149 (4) | 0.2500 | 0.6500 (3) | 0.0388 (6) | |
| C21 | 0.5452 (5) | 0.2500 | 0.7604 (3) | 0.0580 (10) | |
| H21B | 0.4027 | 0.2500 | 0.7370 | 0.096 (10)* | |
| H21A | 0.5940 | 0.3629 | 0.8075 | 0.096 (10)* | |
| N3 | 0.5025 (3) | 0.2500 | 0.5386 (2) | 0.0384 (5) | |
| C4 | 0.6319 (4) | 0.2500 | 0.4693 (3) | 0.0346 (5) | |
| N4 | 0.5583 (4) | 0.2500 | 0.3412 (2) | 0.0477 (6) | |
| O41 | 0.3787 (4) | 0.2500 | 0.2954 (2) | 0.0762 (9) | |
| O42 | 0.6777 (4) | 0.2500 | 0.2822 (2) | 0.0709 (9) | |
| C5 | 0.8257 (4) | 0.2500 | 0.5388 (3) | 0.0317 (5) | |
| Br5 | 1.07071 (3) | 0.2500 | 0.50807 (2) | 0.03540 (12) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N1 | 0.0254 (9) | 0.0524 (13) | 0.0271 (11) | 0.000 | 0.0086 (8) | 0.000 |
| C11 | 0.0265 (11) | 0.0667 (18) | 0.0283 (14) | 0.000 | 0.0082 (10) | 0.000 |
| C12 | 0.0515 (13) | 0.0791 (19) | 0.0522 (14) | −0.0040 (12) | 0.0038 (11) | −0.0177 (14) |
| C13 | 0.0587 (15) | 0.133 (3) | 0.0548 (18) | −0.0138 (18) | −0.0012 (13) | −0.041 (2) |
| C14 | 0.047 (2) | 0.190 (6) | 0.0266 (18) | 0.000 | 0.0010 (15) | 0.000 |
| C2 | 0.0265 (11) | 0.0560 (17) | 0.0349 (15) | 0.000 | 0.0104 (10) | 0.000 |
| C21 | 0.0391 (15) | 0.102 (3) | 0.0369 (17) | 0.000 | 0.0181 (13) | 0.000 |
| N3 | 0.0255 (10) | 0.0557 (14) | 0.0352 (13) | 0.000 | 0.0104 (9) | 0.000 |
| C4 | 0.0271 (12) | 0.0456 (14) | 0.0310 (14) | 0.000 | 0.0081 (10) | 0.000 |
| N4 | 0.0378 (12) | 0.0725 (18) | 0.0299 (13) | 0.000 | 0.0054 (10) | 0.000 |
| O41 | 0.0365 (12) | 0.143 (3) | 0.0404 (14) | 0.000 | −0.0030 (10) | 0.000 |
| O42 | 0.0525 (14) | 0.130 (3) | 0.0337 (13) | 0.000 | 0.0177 (11) | 0.000 |
| C5 | 0.0254 (10) | 0.0367 (13) | 0.0339 (14) | 0.000 | 0.0096 (10) | 0.000 |
| Br5 | 0.02744 (15) | 0.04150 (17) | 0.04068 (19) | 0.000 | 0.01527 (11) | 0.000 |
Geometric parameters (Å, º) top
| N1—C5 | 1.380 (3) | C2—N3 | 1.303 (4) |
| N1—C2 | 1.381 (3) | C2—C21 | 1.493 (4) |
| N1—C11 | 1.438 (4) | C21—H21B | 0.9598 |
| C11—C12 | 1.372 (3) | C21—H21A | 0.9595 |
| C11—C12i | 1.372 (3) | N3—C4 | 1.374 (4) |
| C12—C13 | 1.394 (4) | C4—C5 | 1.367 (4) |
| C12—H12 | 0.9300 | C4—N4 | 1.421 (4) |
| C13—C14 | 1.365 (5) | N4—O41 | 1.220 (4) |
| C13—H13 | 0.9300 | N4—O42 | 1.225 (4) |
| C14—C13i | 1.365 (5) | C5—Br5 | 1.855 (2) |
| C14—H14 | 0.9300 | | |
| | | |
| C5—N1—C2 | 107.3 (2) | N3—C2—C21 | 126.2 (3) |
| C5—N1—C11 | 127.6 (2) | N1—C2—C21 | 122.3 (3) |
| C2—N1—C11 | 125.1 (2) | C2—C21—H21B | 109.4 |
| C12—C11—C12i | 122.4 (3) | C2—C21—H21A | 109.5 |
| C12—C11—N1 | 118.79 (16) | H21B—C21—H21A | 109.5 |
| C12i—C11—N1 | 118.79 (16) | C2—N3—C4 | 105.1 (2) |
| C11—C12—C13 | 118.0 (3) | C5—C4—N3 | 111.9 (3) |
| C11—C12—H12 | 121.0 | C5—C4—N4 | 127.9 (2) |
| C13—C12—H12 | 121.0 | N3—C4—N4 | 120.2 (2) |
| C14—C13—C12 | 120.7 (4) | O41—N4—O42 | 123.1 (3) |
| C14—C13—H13 | 119.7 | O41—N4—C4 | 118.3 (3) |
| C12—C13—H13 | 119.7 | O42—N4—C4 | 118.6 (2) |
| C13i—C14—C13 | 120.2 (4) | C4—C5—N1 | 104.2 (2) |
| C13i—C14—H14 | 119.9 | C4—C5—Br5 | 135.2 (2) |
| C13—C14—H14 | 119.9 | N1—C5—Br5 | 120.56 (19) |
| N3—C2—N1 | 111.5 (2) | | |
| | | |
| C5—N1—C11—C12 | 90.5 (2) | C2—N3—C4—C5 | 0.000 (1) |
| C2—N1—C11—C12 | −89.5 (2) | C2—N3—C4—N4 | 180.0 |
| C5—N1—C11—C12i | −90.5 (2) | C5—C4—N4—O41 | 180.0 |
| C2—N1—C11—C12i | 89.5 (2) | N3—C4—N4—O41 | 0.0 |
| C12i—C11—C12—C13 | −2.0 (5) | C5—C4—N4—O42 | 0.0 |
| N1—C11—C12—C13 | 177.0 (3) | N3—C4—N4—O42 | 180.0 |
| C11—C12—C13—C14 | 0.0 (5) | N3—C4—C5—N1 | 0.0 |
| C12—C13—C14—C13i | 2.0 (7) | N4—C4—C5—N1 | 180.0 |
| C5—N1—C2—N3 | 0.0 | N3—C4—C5—Br5 | 180.0 |
| C11—N1—C2—N3 | 180.000 (1) | N4—C4—C5—Br5 | 0.000 (1) |
| C5—N1—C2—C21 | 180.000 (1) | C2—N1—C5—C4 | 0.0 |
| C11—N1—C2—C21 | 0.000 (1) | C11—N1—C5—C4 | 180.000 (1) |
| N1—C2—N3—C4 | 0.0 | C2—N1—C5—Br5 | 180.0 |
| C21—C2—N3—C4 | 180.000 (1) | C11—N1—C5—Br5 | 0.000 (1) |
| Symmetry code: (i) x, −y+1/2, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14···O41ii | 0.93 | 2.77 | 3.502 (5) | 137 |
| C21—H21B···Br5iii | 0.96 | 2.98 | 3.746 (4) | 137 |
| Symmetry codes: (ii) x+1, y, z+1; (iii) x−1, y, z. |
(250K) 1-phenyl-4-nitro-5-bromo-imidazole
top
Crystal data top
| C10H8BrN3O2 | F(000) = 280 |
| Mr = 282.10 | Dx = 1.743 Mg m−3 |
| Monoclinic, P21/m | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybm | Cell parameters from 668 reflections |
| a = 7.0180 (14) Å | θ = 3–20° |
| b = 6.9160 (12) Å | µ = 3.81 mm−1 |
| c = 11.546 (2) Å | T = 250 K |
| β = 106.410 (15)° | Prism, colourless |
| V = 537.57 (17) Å3 | 0.5 × 0.3 × 0.1 mm |
| Z = 2 | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 1473 independent reflections |
| Radiation source: fine-focus sealed tube | 1097 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.054 |
| ω–scan | θmax = 29.6°, θmin = 3.0° |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | h = −6→9 |
| Tmin = 0.326, Tmax = 0.673 | k = −9→9 |
| 3360 measured reflections | l = −15→15 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.042 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.100 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.047P)2]
where P = (Fo2 + 2Fc2)/3 |
| 1473 reflections | (Δ/σ)max < 0.001 |
| 91 parameters | Δρmax = 0.74 e Å−3 |
| 0 restraints | Δρmin = −0.96 e Å−3 |
Crystal data top
| C10H8BrN3O2 | V = 537.57 (17) Å3 |
| Mr = 282.10 | Z = 2 |
| Monoclinic, P21/m | Mo Kα radiation |
| a = 7.0180 (14) Å | µ = 3.81 mm−1 |
| b = 6.9160 (12) Å | T = 250 K |
| c = 11.546 (2) Å | 0.5 × 0.3 × 0.1 mm |
| β = 106.410 (15)° | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 1473 independent reflections |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | 1097 reflections with I > 2σ(I) |
| Tmin = 0.326, Tmax = 0.673 | Rint = 0.054 |
| 3360 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.042 | 0 restraints |
| wR(F2) = 0.100 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.00 | Δρmax = 0.74 e Å−3 |
| 1473 reflections | Δρmin = −0.96 e Å−3 |
| 91 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N1 | 0.8125 (5) | 0.2500 | 0.6549 (4) | 0.0314 (9) | |
| C11 | 0.9758 (7) | 0.2500 | 0.7651 (5) | 0.0375 (12) | |
| C12 | 1.0493 (6) | 0.4233 (7) | 0.8177 (4) | 0.0562 (11) | |
| H12 | 0.9870 | 0.5380 | 0.7743 | 0.077* | |
| C13 | 1.1975 (6) | 0.4206 (10) | 0.9259 (4) | 0.0760 (16) | |
| H13 | 1.2508 | 0.5418 | 0.9592 | 0.074* | |
| C14 | 1.2719 (10) | 0.2500 | 0.9819 (6) | 0.077 (2) | |
| H14 | 1.3699 | 0.2500 | 1.0549 | 0.090* | |
| C2 | 0.6162 (6) | 0.2500 | 0.6506 (4) | 0.0330 (11) | |
| C21 | 0.5449 (8) | 0.2500 | 0.7609 (5) | 0.0512 (15) | |
| H21A | 0.4002 | 0.2500 | 0.7365 | 0.115* | |
| H21B | 0.5936 | 0.3651 | 0.8090 | 0.109* | |
| N3 | 0.5020 (6) | 0.2500 | 0.5381 (4) | 0.0327 (9) | |
| C4 | 0.6321 (6) | 0.2500 | 0.4698 (4) | 0.0294 (10) | |
| N4 | 0.5584 (6) | 0.2500 | 0.3410 (4) | 0.0422 (11) | |
| O41 | 0.6790 (6) | 0.2500 | 0.2821 (4) | 0.0647 (13) | |
| O42 | 0.3789 (6) | 0.2500 | 0.2951 (4) | 0.0669 (13) | |
| C5 | 0.8259 (6) | 0.2500 | 0.5377 (5) | 0.0291 (10) | |
| Br5 | 1.07177 (6) | 0.2500 | 0.50783 (5) | 0.03340 (19) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N1 | 0.0212 (18) | 0.042 (2) | 0.032 (2) | 0.000 | 0.0098 (16) | 0.000 |
| C11 | 0.024 (2) | 0.057 (3) | 0.033 (3) | 0.000 | 0.010 (2) | 0.000 |
| C12 | 0.042 (2) | 0.067 (3) | 0.055 (3) | −0.003 (2) | 0.0063 (19) | −0.015 (2) |
| C13 | 0.050 (3) | 0.112 (4) | 0.059 (3) | −0.009 (3) | 0.002 (2) | −0.034 (3) |
| C14 | 0.038 (3) | 0.156 (8) | 0.034 (3) | 0.000 | 0.004 (3) | 0.000 |
| C2 | 0.017 (2) | 0.050 (3) | 0.032 (3) | 0.000 | 0.0062 (18) | 0.000 |
| C21 | 0.034 (3) | 0.087 (4) | 0.039 (3) | 0.000 | 0.020 (2) | 0.000 |
| N3 | 0.0216 (17) | 0.042 (2) | 0.036 (2) | 0.000 | 0.0114 (17) | 0.000 |
| C4 | 0.020 (2) | 0.039 (3) | 0.031 (2) | 0.000 | 0.0093 (18) | 0.000 |
| N4 | 0.027 (2) | 0.060 (3) | 0.040 (3) | 0.000 | 0.0109 (19) | 0.000 |
| O41 | 0.046 (2) | 0.115 (4) | 0.040 (2) | 0.000 | 0.0229 (19) | 0.000 |
| O42 | 0.030 (2) | 0.119 (4) | 0.047 (3) | 0.000 | 0.0034 (18) | 0.000 |
| C5 | 0.023 (2) | 0.023 (2) | 0.045 (3) | 0.000 | 0.017 (2) | 0.000 |
| Br5 | 0.0239 (2) | 0.0355 (3) | 0.0453 (3) | 0.000 | 0.0170 (2) | 0.000 |
Geometric parameters (Å, º) top
| N1—C2 | 1.365 (6) | C2—N3 | 1.320 (6) |
| N1—C5 | 1.382 (6) | C2—C21 | 1.494 (7) |
| N1—C11 | 1.452 (6) | C21—H21A | 0.9745 |
| C11—C12 | 1.376 (5) | C21—H21B | 0.9750 |
| C11—C12i | 1.376 (5) | N3—C4 | 1.364 (6) |
| C12—C13 | 1.382 (6) | C4—C5 | 1.364 (6) |
| C12—H12 | 0.9735 | C4—N4 | 1.430 (6) |
| C13—C14 | 1.376 (7) | N4—O42 | 1.221 (5) |
| C13—H13 | 0.9528 | N4—O41 | 1.226 (5) |
| C14—C13i | 1.376 (7) | C5—Br5 | 1.853 (4) |
| C14—H14 | 0.9251 | | |
| | | |
| C2—N1—C5 | 108.1 (4) | N3—C2—C21 | 125.7 (4) |
| C2—N1—C11 | 124.8 (4) | N1—C2—C21 | 123.1 (4) |
| C5—N1—C11 | 127.1 (4) | C2—C21—H21A | 109.0 |
| C12—C11—C12i | 121.1 (5) | C2—C21—H21B | 109.7 |
| C12—C11—N1 | 119.4 (3) | H21A—C21—H21B | 109.5 |
| C12i—C11—N1 | 119.4 (3) | C2—N3—C4 | 104.4 (4) |
| C11—C12—C13 | 118.7 (5) | C5—C4—N3 | 112.9 (4) |
| C11—C12—H12 | 115.1 | C5—C4—N4 | 127.3 (4) |
| C13—C12—H12 | 126.2 | N3—C4—N4 | 119.7 (4) |
| C14—C13—C12 | 121.7 (6) | O42—N4—O41 | 123.3 (5) |
| C14—C13—H13 | 120.7 | O42—N4—C4 | 118.5 (4) |
| C12—C13—H13 | 117.5 | O41—N4—C4 | 118.2 (4) |
| C13i—C14—C13 | 118.2 (6) | C4—C5—N1 | 103.3 (4) |
| C13i—C14—H14 | 120.9 | C4—C5—Br5 | 136.3 (4) |
| C13—C14—H14 | 120.9 | N1—C5—Br5 | 120.4 (3) |
| N3—C2—N1 | 111.2 (4) | | |
| | | |
| C2—N1—C11—C12 | −88.7 (4) | C2—N3—C4—C5 | 0.000 (2) |
| C5—N1—C11—C12 | 91.3 (4) | C2—N3—C4—N4 | 180.000 (1) |
| C2—N1—C11—C12i | 88.7 (4) | C5—C4—N4—O42 | 180.000 (1) |
| C5—N1—C11—C12i | −91.3 (4) | N3—C4—N4—O42 | 0.000 (1) |
| C12i—C11—C12—C13 | −0.9 (8) | C5—C4—N4—O41 | 0.000 (1) |
| N1—C11—C12—C13 | 176.6 (4) | N3—C4—N4—O41 | 180.000 (1) |
| C11—C12—C13—C14 | −0.2 (8) | N3—C4—C5—N1 | 0.000 (1) |
| C12—C13—C14—C13i | 1.3 (10) | N4—C4—C5—N1 | 180.000 (1) |
| C5—N1—C2—N3 | 0.000 (1) | N3—C4—C5—Br5 | 180.000 (1) |
| C11—N1—C2—N3 | 180.000 (2) | N4—C4—C5—Br5 | 0.000 (2) |
| C5—N1—C2—C21 | 180.000 (2) | C2—N1—C5—C4 | 0.000 (2) |
| C11—N1—C2—C21 | 0.000 (2) | C11—N1—C5—C4 | 180.000 (2) |
| N1—C2—N3—C4 | 0.000 (1) | C2—N1—C5—Br5 | 180.000 (1) |
| C21—C2—N3—C4 | 180.000 (2) | C11—N1—C5—Br5 | 0.000 (2) |
| Symmetry code: (i) x, −y+1/2, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14···O41ii | 0.93 | 2.89 | 3.819 (8) | 180 |
| C21—H21A···Br5iii | 0.97 | 2.97 | 3.753 (6) | 138 |
| Symmetry codes: (ii) x+1, y, z+1; (iii) x−1, y, z. |
(200K) 1-phenyl-4-nitro-5-bromo-imidazole
top
Crystal data top
| C10H8BrN3O2 | F(000) = 560 |
| Mr = 282.10 | Dx = 1.769 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 2101 reflections |
| a = 6.9955 (9) Å | θ = 3–22° |
| b = 6.8657 (8) Å | µ = 3.87 mm−1 |
| c = 22.990 (2) Å | T = 200 K |
| β = 106.356 (10)° | Prism, colourless |
| V = 1059.5 (2) Å3 | 0.5 × 0.3 × 0.1 mm |
| Z = 4 | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 2719 independent reflections |
| Radiation source: fine-focus sealed tube | 1610 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.061 |
| ω–scan | θmax = 29.4°, θmin = 3.0° |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | h = −9→9 |
| Tmin = 0.355, Tmax = 0.725 | k = 0→9 |
| 13871 measured reflections | l = 0→30 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.049 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.137 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.13 | w = 1/[σ2(Fo2) + (0.0626P)2]
where P = (Fo2 + 2Fc2)/3 |
| 2719 reflections | (Δ/σ)max = 0.001 |
| 148 parameters | Δρmax = 1.14 e Å−3 |
| 0 restraints | Δρmin = −1.08 e Å−3 |
Crystal data top
| C10H8BrN3O2 | V = 1059.5 (2) Å3 |
| Mr = 282.10 | Z = 4 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 6.9955 (9) Å | µ = 3.87 mm−1 |
| b = 6.8657 (8) Å | T = 200 K |
| c = 22.990 (2) Å | 0.5 × 0.3 × 0.1 mm |
| β = 106.356 (10)° | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 2719 independent reflections |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | 1610 reflections with I > 2σ(I) |
| Tmin = 0.355, Tmax = 0.725 | Rint = 0.061 |
| 13871 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.049 | 0 restraints |
| wR(F2) = 0.137 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.13 | Δρmax = 1.14 e Å−3 |
| 2719 reflections | Δρmin = −1.08 e Å−3 |
| 148 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N1 | 0.6874 (5) | 0.2365 (5) | 0.42266 (16) | 0.0270 (8) | |
| C11 | 0.5246 (6) | 0.2265 (6) | 0.36738 (19) | 0.0267 (9) | |
| C12 | 0.4554 (7) | 0.0490 (8) | 0.3435 (2) | 0.0438 (14) | |
| H12 | 0.5092 | −0.0684 | 0.3634 | 0.084 (10)* | |
| C13 | 0.3039 (8) | 0.0456 (10) | 0.2894 (3) | 0.0588 (18) | |
| H13 | 0.2517 | −0.0764 | 0.2728 | 0.084 (10)* | |
| C14 | 0.2277 (8) | 0.2124 (10) | 0.2593 (2) | 0.0567 (19) | |
| H14 | 0.1277 | 0.2058 | 0.2215 | 0.084 (10)* | |
| C15 | 0.2973 (9) | 0.3899 (10) | 0.2842 (3) | 0.0618 (19) | |
| H15 | 0.2431 | 0.5066 | 0.2640 | 0.084 (10)* | |
| C16 | 0.4474 (8) | 0.3992 (8) | 0.3391 (3) | 0.0482 (15) | |
| H16 | 0.4955 | 0.5213 | 0.3566 | 0.084 (10)* | |
| C2 | 0.8855 (6) | 0.2356 (6) | 0.4247 (2) | 0.0276 (9) | |
| C21 | 0.9561 (7) | 0.2258 (7) | 0.3698 (2) | 0.0400 (13) | |
| H21A | 0.9114 | 0.3420 | 0.3449 | 0.062 (11)* | |
| H21B | 0.9016 | 0.1092 | 0.3464 | 0.062 (11)* | |
| H21C | 1.1020 | 0.2199 | 0.3817 | 0.062 (11)* | |
| N3 | 0.9993 (5) | 0.2477 (5) | 0.48111 (16) | 0.0270 (7) | |
| C4 | 0.8681 (6) | 0.2571 (6) | 0.51534 (19) | 0.0250 (8) | |
| N4 | 0.9422 (5) | 0.2716 (6) | 0.57967 (18) | 0.0330 (9) | |
| O41 | 1.1223 (5) | 0.2786 (5) | 0.60241 (16) | 0.0506 (10) | |
| O42 | 0.8201 (5) | 0.2801 (5) | 0.60897 (16) | 0.0475 (10) | |
| C5 | 0.6737 (5) | 0.2473 (6) | 0.48091 (19) | 0.0237 (8) | |
| Br5 | 0.42742 (5) | 0.25117 (6) | 0.496046 (18) | 0.02643 (16) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N1 | 0.0172 (15) | 0.036 (2) | 0.0296 (18) | −0.0028 (17) | 0.0103 (13) | 0.0021 (17) |
| C11 | 0.0172 (17) | 0.038 (3) | 0.027 (2) | −0.0019 (19) | 0.0091 (15) | 0.001 (2) |
| C12 | 0.033 (3) | 0.053 (3) | 0.042 (3) | −0.005 (2) | 0.004 (2) | −0.013 (3) |
| C13 | 0.043 (4) | 0.085 (5) | 0.043 (4) | −0.005 (3) | 0.005 (3) | −0.024 (3) |
| C14 | 0.030 (3) | 0.112 (6) | 0.027 (2) | −0.001 (3) | 0.006 (2) | 0.000 (3) |
| C15 | 0.039 (4) | 0.089 (5) | 0.050 (4) | 0.008 (3) | 0.001 (3) | 0.025 (4) |
| C16 | 0.037 (3) | 0.052 (3) | 0.053 (4) | −0.006 (3) | 0.007 (3) | 0.011 (3) |
| C2 | 0.0178 (17) | 0.037 (2) | 0.028 (2) | −0.003 (2) | 0.0058 (15) | 0.003 (2) |
| C21 | 0.028 (2) | 0.064 (4) | 0.032 (2) | −0.006 (2) | 0.0152 (19) | −0.009 (3) |
| N3 | 0.0182 (14) | 0.0350 (18) | 0.0302 (18) | −0.0025 (18) | 0.0105 (14) | −0.0024 (18) |
| C4 | 0.0174 (17) | 0.0294 (19) | 0.030 (2) | −0.001 (2) | 0.0099 (15) | −0.001 (2) |
| N4 | 0.0262 (18) | 0.042 (2) | 0.033 (2) | 0.0006 (18) | 0.0113 (15) | −0.0009 (18) |
| O41 | 0.0229 (16) | 0.090 (3) | 0.0369 (19) | 0.0004 (19) | 0.0052 (14) | −0.002 (2) |
| O42 | 0.0365 (18) | 0.078 (3) | 0.0338 (18) | −0.0026 (19) | 0.0198 (15) | −0.0088 (18) |
| C5 | 0.0178 (16) | 0.0215 (17) | 0.035 (2) | −0.0057 (19) | 0.0132 (15) | −0.001 (2) |
| Br5 | 0.0186 (2) | 0.0283 (2) | 0.0360 (3) | −0.0004 (2) | 0.01365 (16) | 0.0003 (2) |
Geometric parameters (Å, º) top
| N1—C5 | 1.371 (5) | C16—H16 | 0.9500 |
| N1—C2 | 1.373 (5) | C2—N3 | 1.319 (5) |
| N1—C11 | 1.450 (5) | C2—C21 | 1.482 (6) |
| C11—C12 | 1.368 (6) | C21—H21A | 0.9800 |
| C11—C16 | 1.387 (6) | C21—H21B | 0.9800 |
| C12—C13 | 1.389 (7) | C21—H21C | 0.9800 |
| C12—H12 | 0.9500 | N3—C4 | 1.369 (6) |
| C13—C14 | 1.367 (8) | C4—C5 | 1.369 (5) |
| C13—H13 | 0.9500 | C4—N4 | 1.426 (6) |
| C14—C15 | 1.376 (9) | N4—O41 | 1.221 (5) |
| C14—H14 | 0.9500 | N4—O42 | 1.229 (5) |
| C15—C16 | 1.397 (8) | C5—Br5 | 1.851 (4) |
| C15—H15 | 0.9500 | | |
| | | |
| C5—N1—C2 | 108.3 (3) | C15—C16—H16 | 120.7 |
| C5—N1—C11 | 127.2 (3) | N3—C2—N1 | 111.0 (4) |
| C2—N1—C11 | 124.5 (4) | N3—C2—C21 | 125.9 (4) |
| C12—C11—C16 | 121.8 (5) | N1—C2—C21 | 123.1 (4) |
| C12—C11—N1 | 119.7 (4) | C2—C21—H21A | 109.5 |
| C16—C11—N1 | 118.5 (4) | C2—C21—H21B | 109.5 |
| C11—C12—C13 | 117.9 (5) | H21A—C21—H21B | 109.5 |
| C11—C12—H12 | 121.0 | C2—C21—H21C | 109.5 |
| C13—C12—H12 | 121.0 | H21A—C21—H21C | 109.5 |
| C14—C13—C12 | 122.1 (6) | H21B—C21—H21C | 109.5 |
| C14—C13—H13 | 119.0 | C2—N3—C4 | 104.5 (3) |
| C12—C13—H13 | 119.0 | N3—C4—C5 | 112.5 (4) |
| C13—C14—C15 | 119.3 (5) | N3—C4—N4 | 119.5 (4) |
| C13—C14—H14 | 120.4 | C5—C4—N4 | 127.9 (4) |
| C15—C14—H14 | 120.4 | O41—N4—O42 | 123.8 (4) |
| C14—C15—C16 | 120.3 (6) | O41—N4—C4 | 118.4 (4) |
| C14—C15—H15 | 119.8 | O42—N4—C4 | 117.8 (4) |
| C16—C15—H15 | 119.8 | C4—C5—N1 | 103.6 (3) |
| C11—C16—C15 | 118.6 (5) | C4—C5—Br5 | 135.7 (3) |
| C11—C16—H16 | 120.7 | N1—C5—Br5 | 120.6 (3) |
| | | |
| C5—N1—C11—C12 | −92.2 (6) | N1—C2—N3—C4 | −0.2 (5) |
| C2—N1—C11—C12 | 87.6 (6) | C21—C2—N3—C4 | 178.7 (5) |
| C5—N1—C11—C16 | 89.8 (6) | C2—N3—C4—C5 | 1.1 (5) |
| C2—N1—C11—C16 | −90.4 (6) | C2—N3—C4—N4 | −180.0 (4) |
| C16—C11—C12—C13 | 0.2 (8) | N3—C4—N4—O41 | 1.2 (7) |
| N1—C11—C12—C13 | −177.8 (5) | C5—C4—N4—O41 | 180.0 (5) |
| C11—C12—C13—C14 | 1.7 (9) | N3—C4—N4—O42 | 179.9 (4) |
| C12—C13—C14—C15 | −2.5 (9) | C5—C4—N4—O42 | −1.4 (8) |
| C13—C14—C15—C16 | 1.4 (9) | N3—C4—C5—N1 | −1.6 (6) |
| C12—C11—C16—C15 | −1.2 (8) | N4—C4—C5—N1 | 179.6 (4) |
| N1—C11—C16—C15 | 176.8 (5) | N3—C4—C5—Br5 | 180.0 (3) |
| C14—C15—C16—C11 | 0.4 (9) | N4—C4—C5—Br5 | 1.2 (9) |
| C5—N1—C2—N3 | −0.8 (5) | C2—N1—C5—C4 | 1.4 (5) |
| C11—N1—C2—N3 | 179.5 (4) | C11—N1—C5—C4 | −178.9 (4) |
| C5—N1—C2—C21 | −179.7 (4) | C2—N1—C5—Br5 | −179.9 (3) |
| C11—N1—C2—C21 | 0.5 (7) | C11—N1—C5—Br5 | −0.1 (7) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14···O41i | 0.95 | 2.73 | 3.472 (6) | 136 |
| C21—H21C···Br5ii | 0.98 | 2.96 | 3.733 (5) | 137 |
| Symmetry codes: (i) x−1, −y+1/2, z−1/2; (ii) x+1, y, z. |
(170K) 1-phenyl-4-nitro-5-bromo-imidazole
top
Crystal data top
| C10H8BrN3O2 | F(000) = 560 |
| Mr = 282.10 | Dx = 1.776 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 1833 reflections |
| a = 6.9923 (12) Å | θ = 3–20° |
| b = 6.8497 (10) Å | µ = 3.88 mm−1 |
| c = 22.9702 (16) Å | T = 170 K |
| β = 106.419 (14)° | Prism, colourless |
| V = 1055.3 (2) Å3 | 0.5 × 0.3 × 0.1 mm |
| Z = 4 | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 2673 independent reflections |
| Radiation source: fine-focus sealed tube | 1387 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.060 |
| ω–scan | θmax = 29.5°, θmin = 3.0° |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | h = −6→9 |
| Tmin = 0.348, Tmax = 0.659 | k = −9→9 |
| 6354 measured reflections | l = −30→30 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.045 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.100 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.01 | w = 1/[σ2(Fo2) + (0.032P)2]
where P = (Fo2 + 2Fc2)/3 |
| 2673 reflections | (Δ/σ)max < 0.001 |
| 148 parameters | Δρmax = 0.82 e Å−3 |
| 0 restraints | Δρmin = −0.99 e Å−3 |
Crystal data top
| C10H8BrN3O2 | V = 1055.3 (2) Å3 |
| Mr = 282.10 | Z = 4 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 6.9923 (12) Å | µ = 3.88 mm−1 |
| b = 6.8497 (10) Å | T = 170 K |
| c = 22.9702 (16) Å | 0.5 × 0.3 × 0.1 mm |
| β = 106.419 (14)° | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 2673 independent reflections |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | 1387 reflections with I > 2σ(I) |
| Tmin = 0.348, Tmax = 0.659 | Rint = 0.060 |
| 6354 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.045 | 0 restraints |
| wR(F2) = 0.100 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.01 | Δρmax = 0.82 e Å−3 |
| 2673 reflections | Δρmin = −0.99 e Å−3 |
| 148 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N1 | 0.6880 (4) | 0.2287 (6) | 0.42271 (15) | 0.0230 (8) | |
| C11 | 0.5251 (6) | 0.2174 (6) | 0.36776 (19) | 0.0257 (11) | |
| C12 | 0.4562 (7) | 0.0384 (8) | 0.3440 (2) | 0.0385 (14) | |
| H12 | 0.5121 | −0.0779 | 0.3643 | 0.054 (8)* | |
| C13 | 0.3053 (8) | 0.0289 (9) | 0.2903 (3) | 0.0517 (17) | |
| H13 | 0.2546 | −0.0949 | 0.2745 | 0.054 (8)* | |
| C14 | 0.2267 (8) | 0.1959 (9) | 0.2593 (2) | 0.0507 (18) | |
| H14 | 0.1258 | 0.1881 | 0.2217 | 0.054 (8)* | |
| C15 | 0.2973 (8) | 0.3743 (9) | 0.2837 (3) | 0.0529 (17) | |
| H15 | 0.2426 | 0.4906 | 0.2631 | 0.054 (8)* | |
| C16 | 0.4469 (8) | 0.3867 (8) | 0.3381 (3) | 0.0408 (15) | |
| H16 | 0.4949 | 0.5103 | 0.3546 | 0.054 (8)* | |
| C2 | 0.8860 (5) | 0.2287 (7) | 0.42496 (19) | 0.0233 (9) | |
| C21 | 0.9558 (6) | 0.2127 (7) | 0.3697 (2) | 0.0343 (13) | |
| H21A | 0.9054 | 0.3237 | 0.3429 | 0.057 (9)* | |
| H21B | 0.9063 | 0.0908 | 0.3485 | 0.057 (9)* | |
| H21C | 1.1019 | 0.2130 | 0.3815 | 0.057 (9)* | |
| N3 | 0.9992 (5) | 0.2474 (6) | 0.48141 (14) | 0.0229 (7) | |
| C4 | 0.8686 (5) | 0.2584 (7) | 0.51525 (17) | 0.0204 (8) | |
| N4 | 0.9422 (5) | 0.2829 (5) | 0.57967 (16) | 0.0275 (9) | |
| O41 | 1.1226 (4) | 0.2910 (5) | 0.60235 (15) | 0.0422 (10) | |
| O42 | 0.8193 (5) | 0.2936 (5) | 0.60888 (15) | 0.0415 (9) | |
| C5 | 0.6742 (5) | 0.2484 (7) | 0.48074 (18) | 0.0214 (8) | |
| Br5 | 0.42698 (5) | 0.25148 (7) | 0.495917 (18) | 0.02324 (13) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N1 | 0.0159 (14) | 0.029 (2) | 0.0264 (18) | −0.0010 (17) | 0.0106 (13) | −0.0012 (18) |
| C11 | 0.0154 (18) | 0.041 (3) | 0.022 (2) | −0.001 (2) | 0.0071 (16) | −0.005 (2) |
| C12 | 0.032 (3) | 0.039 (3) | 0.042 (4) | −0.003 (2) | 0.007 (3) | −0.009 (3) |
| C13 | 0.039 (3) | 0.070 (4) | 0.042 (4) | −0.009 (3) | 0.005 (3) | −0.024 (3) |
| C14 | 0.026 (3) | 0.101 (6) | 0.023 (3) | −0.002 (3) | 0.004 (2) | −0.001 (3) |
| C15 | 0.036 (4) | 0.071 (5) | 0.046 (4) | 0.008 (3) | 0.002 (3) | 0.022 (3) |
| C16 | 0.032 (3) | 0.040 (3) | 0.048 (4) | −0.005 (2) | 0.009 (3) | 0.011 (3) |
| C2 | 0.0145 (17) | 0.032 (3) | 0.024 (2) | −0.001 (2) | 0.0063 (15) | 0.001 (2) |
| C21 | 0.023 (2) | 0.052 (4) | 0.033 (3) | −0.001 (2) | 0.0149 (19) | −0.002 (2) |
| N3 | 0.0179 (14) | 0.0276 (16) | 0.0253 (18) | 0.000 (2) | 0.0091 (13) | −0.002 (2) |
| C4 | 0.0159 (17) | 0.026 (2) | 0.0209 (19) | −0.003 (2) | 0.0077 (14) | −0.004 (2) |
| N4 | 0.0216 (17) | 0.036 (3) | 0.027 (2) | −0.0027 (17) | 0.0101 (15) | −0.0013 (17) |
| O41 | 0.0203 (15) | 0.072 (3) | 0.0339 (19) | −0.0033 (17) | 0.0062 (14) | −0.0023 (18) |
| O42 | 0.0339 (17) | 0.065 (3) | 0.0319 (19) | −0.0047 (17) | 0.0191 (15) | −0.0065 (17) |
| C5 | 0.0182 (16) | 0.0181 (17) | 0.033 (2) | −0.001 (2) | 0.0155 (16) | 0.001 (3) |
| Br5 | 0.01722 (18) | 0.0237 (2) | 0.0323 (2) | 0.0002 (2) | 0.01257 (15) | −0.0001 (3) |
Geometric parameters (Å, º) top
| N1—C5 | 1.370 (5) | C16—H16 | 0.9500 |
| N1—C2 | 1.371 (5) | C2—N3 | 1.321 (5) |
| N1—C11 | 1.443 (5) | C2—C21 | 1.486 (6) |
| C11—C12 | 1.373 (6) | C21—H21A | 0.9800 |
| C11—C16 | 1.378 (6) | C21—H21B | 0.9800 |
| C12—C13 | 1.379 (7) | C21—H21C | 0.9800 |
| C12—H12 | 0.9500 | N3—C4 | 1.359 (5) |
| C13—C14 | 1.378 (7) | C4—C5 | 1.368 (5) |
| C13—H13 | 0.9500 | C4—N4 | 1.432 (5) |
| C14—C15 | 1.377 (8) | N4—O41 | 1.223 (4) |
| C14—H14 | 0.9500 | N4—O42 | 1.233 (4) |
| C15—C16 | 1.386 (7) | C5—Br5 | 1.857 (3) |
| C15—H15 | 0.9500 | | |
| | | |
| C5—N1—C2 | 108.2 (3) | C15—C16—H16 | 120.4 |
| C5—N1—C11 | 126.9 (3) | N3—C2—N1 | 110.8 (3) |
| C2—N1—C11 | 124.9 (3) | N3—C2—C21 | 126.5 (3) |
| C12—C11—C16 | 120.6 (5) | N1—C2—C21 | 122.7 (4) |
| C12—C11—N1 | 119.8 (4) | C2—C21—H21A | 109.5 |
| C16—C11—N1 | 119.5 (4) | C2—C21—H21B | 109.5 |
| C11—C12—C13 | 119.5 (5) | H21A—C21—H21B | 109.5 |
| C11—C12—H12 | 120.3 | C2—C21—H21C | 109.5 |
| C13—C12—H12 | 120.3 | H21A—C21—H21C | 109.5 |
| C14—C13—C12 | 121.0 (6) | H21B—C21—H21C | 109.5 |
| C14—C13—H13 | 119.5 | C2—N3—C4 | 104.7 (3) |
| C12—C13—H13 | 119.5 | N3—C4—C5 | 112.6 (4) |
| C15—C14—C13 | 118.8 (5) | N3—C4—N4 | 119.6 (3) |
| C15—C14—H14 | 120.6 | C5—C4—N4 | 127.8 (4) |
| C13—C14—H14 | 120.6 | O41—N4—O42 | 124.0 (4) |
| C14—C15—C16 | 120.9 (5) | O41—N4—C4 | 118.1 (3) |
| C14—C15—H15 | 119.5 | O42—N4—C4 | 117.8 (3) |
| C16—C15—H15 | 119.5 | C4—C5—N1 | 103.7 (3) |
| C11—C16—C15 | 119.2 (5) | C4—C5—Br5 | 135.7 (3) |
| C11—C16—H16 | 120.4 | N1—C5—Br5 | 120.6 (3) |
| | | |
| C5—N1—C11—C12 | −95.2 (6) | N1—C2—N3—C4 | 0.3 (6) |
| C2—N1—C11—C12 | 87.4 (6) | C21—C2—N3—C4 | 179.3 (5) |
| C5—N1—C11—C16 | 88.0 (6) | C2—N3—C4—C5 | −0.5 (6) |
| C2—N1—C11—C16 | −89.4 (6) | C2—N3—C4—N4 | −179.0 (4) |
| C16—C11—C12—C13 | −1.0 (7) | N3—C4—N4—O41 | −0.8 (7) |
| N1—C11—C12—C13 | −177.8 (5) | C5—C4—N4—O41 | −179.1 (5) |
| C11—C12—C13—C14 | 2.0 (9) | N3—C4—N4—O42 | 179.6 (4) |
| C12—C13—C14—C15 | −2.1 (9) | C5—C4—N4—O42 | 1.2 (8) |
| C13—C14—C15—C16 | 1.1 (9) | N3—C4—C5—N1 | 0.5 (6) |
| C12—C11—C16—C15 | 0.0 (7) | N4—C4—C5—N1 | 178.9 (5) |
| N1—C11—C16—C15 | 176.8 (5) | N3—C4—C5—Br5 | 179.2 (4) |
| C14—C15—C16—C11 | −0.1 (9) | N4—C4—C5—Br5 | −2.4 (10) |
| C5—N1—C2—N3 | 0.0 (6) | C2—N1—C5—C4 | −0.3 (6) |
| C11—N1—C2—N3 | 177.8 (4) | C11—N1—C5—C4 | −178.0 (4) |
| C5—N1—C2—C21 | −179.1 (4) | C2—N1—C5—Br5 | −179.3 (3) |
| C11—N1—C2—C21 | −1.3 (7) | C11—N1—C5—Br5 | 3.0 (7) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14···O41i | 0.95 | 2.74 | 3.471 (6) | 135 |
| C21—H21C···Br5ii | 0.98 | 2.96 | 3.732 (5) | 136 |
| Symmetry codes: (i) x−1, −y+1/2, z−1/2; (ii) x+1, y, z. |
(150K) 1-phenyl-4-nitro-5-bromo-imidazole
top
Crystal data top
| C10H8BrN3O2 | F(000) = 560 |
| Mr = 282.10 | Dx = 1.783 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 2489 reflections |
| a = 6.9854 (12) Å | θ = 3–20° |
| b = 6.8362 (9) Å | µ = 3.90 mm−1 |
| c = 22.945 (2) Å | T = 150 K |
| β = 106.466 (13)° | Prism, colourless |
| V = 1050.8 (2) Å3 | 0.5 × 0.3 × 0.1 mm |
| Z = 4 | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 2676 independent reflections |
| Radiation source: fine-focus sealed tube | 1445 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.069 |
| ω–scan | θmax = 29.5°, θmin = 3.1° |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | h = −9→6 |
| Tmin = 0.321, Tmax = 0.717 | k = −9→9 |
| 6609 measured reflections | l = −30→30 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.044 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.095 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0305P)2]
where P = (Fo2 + 2Fc2)/3 |
| 2676 reflections | (Δ/σ)max = 0.001 |
| 148 parameters | Δρmax = 0.85 e Å−3 |
| 0 restraints | Δρmin = −1.00 e Å−3 |
Crystal data top
| C10H8BrN3O2 | V = 1050.8 (2) Å3 |
| Mr = 282.10 | Z = 4 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 6.9854 (12) Å | µ = 3.90 mm−1 |
| b = 6.8362 (9) Å | T = 150 K |
| c = 22.945 (2) Å | 0.5 × 0.3 × 0.1 mm |
| β = 106.466 (13)° | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 2676 independent reflections |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | 1445 reflections with I > 2σ(I) |
| Tmin = 0.321, Tmax = 0.717 | Rint = 0.069 |
| 6609 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.044 | 0 restraints |
| wR(F2) = 0.095 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.02 | Δρmax = 0.85 e Å−3 |
| 2676 reflections | Δρmin = −1.00 e Å−3 |
| 148 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N1 | 0.6877 (4) | 0.2255 (5) | 0.42295 (15) | 0.0199 (7) | |
| C11 | 0.5249 (5) | 0.2125 (6) | 0.36758 (18) | 0.0213 (10) | |
| C12 | 0.4569 (7) | 0.0327 (7) | 0.3447 (2) | 0.0345 (12) | |
| H12 | 0.5128 | −0.0829 | 0.3657 | 0.050 (7)* | |
| C13 | 0.3065 (7) | 0.0207 (8) | 0.2908 (2) | 0.0466 (15) | |
| H13 | 0.2570 | −0.1037 | 0.2752 | 0.050 (7)* | |
| C14 | 0.2270 (7) | 0.1887 (8) | 0.2594 (2) | 0.0451 (15) | |
| H14 | 0.1260 | 0.1800 | 0.2217 | 0.050 (7)* | |
| C15 | 0.2965 (8) | 0.3688 (9) | 0.2835 (3) | 0.0482 (15) | |
| H15 | 0.2408 | 0.4849 | 0.2628 | 0.050 (7)* | |
| C16 | 0.4463 (7) | 0.3813 (7) | 0.3375 (2) | 0.0375 (13) | |
| H16 | 0.4948 | 0.5054 | 0.3538 | 0.050 (7)* | |
| C2 | 0.8870 (5) | 0.2261 (6) | 0.42508 (18) | 0.0215 (9) | |
| C21 | 0.9567 (6) | 0.2079 (6) | 0.3698 (2) | 0.0307 (12) | |
| H21A | 0.9042 | 0.3170 | 0.3422 | 0.051 (9)* | |
| H21B | 0.9090 | 0.0840 | 0.3493 | 0.051 (9)* | |
| H21C | 1.1030 | 0.2104 | 0.3815 | 0.051 (9)* | |
| N3 | 1.0000 (4) | 0.2476 (5) | 0.48130 (14) | 0.0211 (6) | |
| C4 | 0.8686 (5) | 0.2594 (7) | 0.51525 (16) | 0.0182 (7) | |
| N4 | 0.9424 (5) | 0.2860 (5) | 0.57954 (16) | 0.0261 (9) | |
| O41 | 1.1228 (4) | 0.2965 (5) | 0.60233 (14) | 0.0391 (9) | |
| O42 | 0.8202 (4) | 0.2996 (5) | 0.60881 (15) | 0.0380 (9) | |
| C5 | 0.6743 (5) | 0.2476 (7) | 0.48080 (17) | 0.0199 (7) | |
| Br5 | 0.42680 (5) | 0.25172 (7) | 0.495877 (17) | 0.02096 (12) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N1 | 0.0153 (14) | 0.023 (2) | 0.0235 (17) | −0.0014 (15) | 0.0085 (12) | −0.0024 (16) |
| C11 | 0.0139 (17) | 0.032 (3) | 0.019 (2) | −0.0037 (17) | 0.0058 (15) | −0.0037 (18) |
| C12 | 0.030 (3) | 0.035 (3) | 0.037 (3) | −0.001 (2) | 0.008 (2) | −0.008 (2) |
| C13 | 0.040 (3) | 0.062 (4) | 0.032 (3) | −0.006 (3) | 0.002 (2) | −0.020 (3) |
| C14 | 0.023 (2) | 0.087 (5) | 0.025 (3) | −0.001 (3) | 0.006 (2) | −0.003 (3) |
| C15 | 0.035 (3) | 0.060 (4) | 0.044 (4) | 0.002 (3) | 0.003 (3) | 0.023 (3) |
| C16 | 0.031 (3) | 0.038 (3) | 0.042 (3) | −0.001 (2) | 0.006 (2) | 0.009 (2) |
| C2 | 0.0138 (16) | 0.028 (3) | 0.022 (2) | −0.0014 (19) | 0.0037 (14) | 0.001 (2) |
| C21 | 0.023 (2) | 0.049 (4) | 0.024 (2) | −0.003 (2) | 0.0137 (18) | −0.005 (2) |
| N3 | 0.0159 (13) | 0.0260 (16) | 0.0241 (17) | 0.002 (2) | 0.0099 (12) | −0.0015 (19) |
| C4 | 0.0158 (16) | 0.0214 (18) | 0.0193 (18) | −0.004 (2) | 0.0080 (13) | −0.001 (2) |
| N4 | 0.0205 (17) | 0.032 (3) | 0.026 (2) | 0.0000 (16) | 0.0086 (14) | −0.0016 (16) |
| O41 | 0.0176 (14) | 0.066 (3) | 0.0324 (19) | −0.0008 (15) | 0.0046 (13) | −0.0040 (16) |
| O42 | 0.0303 (16) | 0.057 (2) | 0.0315 (19) | −0.0012 (15) | 0.0169 (14) | −0.0082 (15) |
| C5 | 0.0157 (15) | 0.0153 (16) | 0.032 (2) | 0.000 (2) | 0.0125 (15) | 0.001 (2) |
| Br5 | 0.01606 (17) | 0.02094 (18) | 0.0288 (2) | −0.0002 (2) | 0.01110 (13) | −0.0004 (2) |
Geometric parameters (Å, º) top
| N1—C5 | 1.365 (5) | C16—H16 | 0.9500 |
| N1—C2 | 1.379 (5) | C2—N3 | 1.316 (5) |
| N1—C11 | 1.448 (5) | C2—C21 | 1.487 (5) |
| C11—C12 | 1.368 (5) | C21—H21A | 0.9800 |
| C11—C16 | 1.377 (6) | C21—H21B | 0.9800 |
| C12—C13 | 1.379 (6) | C21—H21C | 0.9800 |
| C12—H12 | 0.9500 | N3—C4 | 1.364 (4) |
| C13—C14 | 1.386 (7) | C4—C5 | 1.366 (5) |
| C13—H13 | 0.9500 | C4—N4 | 1.430 (5) |
| C14—C15 | 1.381 (8) | N4—O41 | 1.222 (4) |
| C14—H14 | 0.9500 | N4—O42 | 1.230 (4) |
| C15—C16 | 1.379 (7) | C5—Br5 | 1.857 (3) |
| C15—H15 | 0.9500 | | |
| | | |
| C5—N1—C2 | 108.2 (3) | C15—C16—H16 | 120.3 |
| C5—N1—C11 | 127.3 (3) | N3—C2—N1 | 110.7 (3) |
| C2—N1—C11 | 124.4 (3) | N3—C2—C21 | 126.5 (3) |
| C12—C11—C16 | 121.0 (4) | N1—C2—C21 | 122.8 (3) |
| C12—C11—N1 | 119.5 (4) | C2—C21—H21A | 109.5 |
| C16—C11—N1 | 119.4 (4) | C2—C21—H21B | 109.5 |
| C11—C12—C13 | 119.4 (5) | H21A—C21—H21B | 109.5 |
| C11—C12—H12 | 120.3 | C2—C21—H21C | 109.5 |
| C13—C12—H12 | 120.3 | H21A—C21—H21C | 109.5 |
| C12—C13—C14 | 120.6 (5) | H21B—C21—H21C | 109.5 |
| C12—C13—H13 | 119.7 | C2—N3—C4 | 104.6 (3) |
| C14—C13—H13 | 119.7 | N3—C4—C5 | 112.7 (3) |
| C15—C14—C13 | 119.1 (5) | N3—C4—N4 | 119.5 (3) |
| C15—C14—H14 | 120.4 | C5—C4—N4 | 127.8 (3) |
| C13—C14—H14 | 120.4 | O41—N4—O42 | 123.6 (4) |
| C16—C15—C14 | 120.4 (5) | O41—N4—C4 | 118.4 (3) |
| C16—C15—H15 | 119.8 | O42—N4—C4 | 118.0 (3) |
| C14—C15—H15 | 119.8 | N1—C5—C4 | 103.8 (3) |
| C11—C16—C15 | 119.5 (5) | N1—C5—Br5 | 120.5 (3) |
| C11—C16—H16 | 120.3 | C4—C5—Br5 | 135.7 (3) |
| | | |
| C5—N1—C11—C12 | −95.1 (5) | N1—C2—N3—C4 | 0.4 (5) |
| C2—N1—C11—C12 | 88.0 (5) | C21—C2—N3—C4 | 179.4 (5) |
| C5—N1—C11—C16 | 87.5 (6) | C2—N3—C4—C5 | −0.7 (6) |
| C2—N1—C11—C16 | −89.4 (5) | C2—N3—C4—N4 | −179.2 (4) |
| C16—C11—C12—C13 | −0.4 (7) | N3—C4—N4—O41 | −0.4 (7) |
| N1—C11—C12—C13 | −177.8 (4) | C5—C4—N4—O41 | −178.7 (5) |
| C11—C12—C13—C14 | 1.2 (8) | N3—C4—N4—O42 | 178.7 (4) |
| C12—C13—C14—C15 | −1.8 (8) | C5—C4—N4—O42 | 0.4 (8) |
| C13—C14—C15—C16 | 1.5 (8) | C2—N1—C5—C4 | −0.4 (5) |
| C12—C11—C16—C15 | 0.1 (7) | C11—N1—C5—C4 | −177.7 (4) |
| N1—C11—C16—C15 | 177.5 (4) | C2—N1—C5—Br5 | −179.6 (3) |
| C14—C15—C16—C11 | −0.7 (8) | C11—N1—C5—Br5 | 3.1 (6) |
| C5—N1—C2—N3 | 0.0 (5) | N3—C4—C5—N1 | 0.6 (6) |
| C11—N1—C2—N3 | 177.4 (4) | N4—C4—C5—N1 | 179.1 (4) |
| C5—N1—C2—C21 | −179.1 (4) | N3—C4—C5—Br5 | 179.7 (4) |
| C11—N1—C2—C21 | −1.7 (7) | N4—C4—C5—Br5 | −1.9 (9) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14···O41i | 0.95 | 2.74 | 3.469 (6) | 134 |
| C21—H21C···Br5ii | 0.98 | 2.95 | 3.722 (4) | 136 |
| Symmetry codes: (i) x−1, −y+1/2, z−1/2; (ii) x+1, y, z. |
(120K) 1-phenyl-4-nitro-5-bromo-imidazole
top
Crystal data top
| C10H8BrN3O2 | F(000) = 560 |
| Mr = 282.10 | Dx = 1.795 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3121 reflections |
| a = 6.9723 (5) Å | θ = 3–20° |
| b = 6.8169 (4) Å | µ = 3.93 mm−1 |
| c = 22.9000 (12) Å | T = 120 K |
| β = 106.478 (6)° | Prism, colourless |
| V = 1043.72 (11) Å3 | 0.3 × 0.25 × 0.1 mm |
| Z = 4 | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 2788 independent reflections |
| Radiation source: fine-focus sealed tube | 2349 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.045 |
| ω–scan | θmax = 29.8°, θmin = 3.1° |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | h = −7→9 |
| Tmin = 0.378, Tmax = 0.573 | k = −7→9 |
| 13861 measured reflections | l = −31→30 |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.033 | All H-atom parameters refined |
| wR(F2) = 0.068 | w = 1/[σ2(Fo2) + (0.P)2 + 2.850P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.07 | (Δ/σ)max = 0.001 |
| 2788 reflections | Δρmax = 0.71 e Å−3 |
| 178 parameters | Δρmin = −0.52 e Å−3 |
| 0 restraints | Extinction correction: SHELXL, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0095 (9) |
Crystal data top
| C10H8BrN3O2 | V = 1043.72 (11) Å3 |
| Mr = 282.10 | Z = 4 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 6.9723 (5) Å | µ = 3.93 mm−1 |
| b = 6.8169 (4) Å | T = 120 K |
| c = 22.9000 (12) Å | 0.3 × 0.25 × 0.1 mm |
| β = 106.478 (6)° | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 2788 independent reflections |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | 2349 reflections with I > 2σ(I) |
| Tmin = 0.378, Tmax = 0.573 | Rint = 0.045 |
| 13861 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.033 | 0 restraints |
| wR(F2) = 0.068 | All H-atom parameters refined |
| S = 1.07 | Δρmax = 0.71 e Å−3 |
| 2788 reflections | Δρmin = −0.52 e Å−3 |
| 178 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N1 | 0.6875 (3) | 0.2210 (3) | 0.42263 (9) | 0.0140 (4) | |
| C11 | 0.5253 (3) | 0.2058 (4) | 0.36720 (11) | 0.0151 (5) | |
| C12 | 0.4574 (4) | 0.0223 (5) | 0.34512 (14) | 0.0256 (6) | |
| H12 | 0.518 (5) | −0.087 (5) | 0.3681 (16) | 0.030 (9)* | |
| C13 | 0.3061 (5) | 0.0104 (6) | 0.29057 (15) | 0.0338 (7) | |
| H13 | 0.259 (7) | −0.107 (8) | 0.275 (2) | 0.069 (15)* | |
| C14 | 0.2258 (4) | 0.1786 (6) | 0.25930 (14) | 0.0326 (7) | |
| H14 | 0.128 (6) | 0.169 (6) | 0.2209 (19) | 0.044 (11)* | |
| C15 | 0.2940 (5) | 0.3607 (6) | 0.28268 (15) | 0.0356 (8) | |
| H15 | 0.235 (6) | 0.472 (6) | 0.264 (2) | 0.052 (12)* | |
| C16 | 0.4448 (4) | 0.3763 (5) | 0.33738 (14) | 0.0273 (6) | |
| H16 | 0.497 (6) | 0.497 (6) | 0.3532 (18) | 0.040 (11)* | |
| C2 | 0.8879 (3) | 0.2209 (4) | 0.42527 (11) | 0.0149 (5) | |
| C21 | 0.9579 (4) | 0.1999 (5) | 0.37037 (13) | 0.0220 (6) | |
| H21A | 0.905 (6) | 0.302 (6) | 0.3432 (18) | 0.043 (11)* | |
| H21B | 0.908 (6) | 0.077 (6) | 0.3511 (17) | 0.040 (10)* | |
| H21C | 1.088 (6) | 0.197 (5) | 0.3815 (16) | 0.033 (10)* | |
| N3 | 1.0005 (3) | 0.2457 (4) | 0.48137 (9) | 0.0153 (4) | |
| C4 | 0.8695 (3) | 0.2636 (4) | 0.51549 (10) | 0.0132 (4) | |
| N4 | 0.9426 (3) | 0.2931 (3) | 0.57956 (10) | 0.0175 (4) | |
| O41 | 1.1249 (3) | 0.3044 (3) | 0.60261 (9) | 0.0275 (5) | |
| O42 | 0.8205 (3) | 0.3077 (3) | 0.60895 (9) | 0.0268 (5) | |
| C5 | 0.6734 (3) | 0.2467 (4) | 0.48058 (10) | 0.0138 (4) | |
| Br5 | 0.42674 (3) | 0.25220 (4) | 0.495852 (10) | 0.01471 (9) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N1 | 0.0112 (8) | 0.0174 (11) | 0.0127 (9) | −0.0010 (8) | 0.0025 (7) | −0.0015 (7) |
| C11 | 0.0107 (10) | 0.0218 (13) | 0.0125 (11) | −0.0011 (9) | 0.0026 (8) | −0.0004 (9) |
| C12 | 0.0252 (14) | 0.0258 (15) | 0.0217 (14) | −0.0008 (12) | −0.0001 (11) | −0.0041 (11) |
| C13 | 0.0278 (15) | 0.0435 (19) | 0.0258 (16) | −0.0046 (14) | 0.0005 (12) | −0.0141 (14) |
| C14 | 0.0197 (13) | 0.062 (2) | 0.0136 (13) | −0.0005 (14) | 0.0009 (10) | −0.0004 (13) |
| C15 | 0.0272 (16) | 0.047 (2) | 0.0271 (17) | 0.0027 (14) | −0.0020 (13) | 0.0157 (14) |
| C16 | 0.0240 (14) | 0.0271 (15) | 0.0271 (15) | −0.0010 (12) | 0.0013 (11) | 0.0081 (12) |
| C2 | 0.0116 (10) | 0.0155 (12) | 0.0176 (11) | −0.0011 (9) | 0.0040 (8) | 0.0001 (9) |
| C21 | 0.0165 (12) | 0.0340 (16) | 0.0176 (13) | −0.0016 (11) | 0.0083 (10) | −0.0033 (11) |
| N3 | 0.0127 (8) | 0.0198 (10) | 0.0138 (9) | 0.0009 (9) | 0.0045 (7) | −0.0011 (9) |
| C4 | 0.0118 (9) | 0.0143 (10) | 0.0134 (10) | 0.0009 (9) | 0.0033 (8) | −0.0017 (9) |
| N4 | 0.0170 (9) | 0.0181 (11) | 0.0163 (10) | −0.0005 (8) | 0.0030 (8) | −0.0010 (8) |
| O41 | 0.0152 (9) | 0.0457 (13) | 0.0184 (9) | −0.0016 (8) | −0.0006 (7) | −0.0027 (8) |
| O42 | 0.0235 (9) | 0.0434 (13) | 0.0158 (9) | −0.0015 (9) | 0.0092 (7) | −0.0053 (8) |
| C5 | 0.0124 (9) | 0.0128 (10) | 0.0169 (11) | −0.0013 (9) | 0.0052 (8) | −0.0023 (10) |
| Br5 | 0.01168 (12) | 0.01573 (13) | 0.01764 (13) | 0.00000 (10) | 0.00566 (8) | −0.00018 (10) |
Geometric parameters (Å, º) top
| N1—C5 | 1.369 (3) | C16—H16 | 0.93 (4) |
| N1—C2 | 1.382 (3) | C2—N3 | 1.313 (3) |
| N1—C11 | 1.444 (3) | C2—C21 | 1.479 (3) |
| C11—C12 | 1.382 (4) | C21—H21A | 0.93 (4) |
| C11—C16 | 1.384 (4) | C21—H21B | 0.97 (4) |
| C12—C13 | 1.390 (4) | C21—H21C | 0.87 (4) |
| C12—H12 | 0.94 (4) | N3—C4 | 1.366 (3) |
| C13—C14 | 1.383 (5) | C4—C5 | 1.379 (3) |
| C13—H13 | 0.90 (5) | C4—N4 | 1.424 (3) |
| C14—C15 | 1.382 (5) | N4—O42 | 1.230 (3) |
| C14—H14 | 0.95 (4) | N4—O41 | 1.233 (3) |
| C15—C16 | 1.392 (4) | C5—Br5 | 1.850 (2) |
| C15—H15 | 0.91 (4) | | |
| | | |
| C5—N1—C2 | 107.88 (19) | C15—C16—H16 | 122 (2) |
| C5—N1—C11 | 127.41 (19) | N3—C2—N1 | 111.1 (2) |
| C2—N1—C11 | 124.6 (2) | N3—C2—C21 | 126.5 (2) |
| C12—C11—C16 | 122.1 (3) | N1—C2—C21 | 122.4 (2) |
| C12—C11—N1 | 119.2 (2) | C2—C21—H21A | 109 (2) |
| C16—C11—N1 | 118.7 (2) | C2—C21—H21B | 108 (2) |
| C11—C12—C13 | 118.4 (3) | H21A—C21—H21B | 109 (3) |
| C11—C12—H12 | 117 (2) | C2—C21—H21C | 109 (2) |
| C13—C12—H12 | 124 (2) | H21A—C21—H21C | 113 (3) |
| C14—C13—C12 | 120.6 (3) | H21B—C21—H21C | 109 (3) |
| C14—C13—H13 | 119 (3) | C2—N3—C4 | 105.07 (19) |
| C12—C13—H13 | 120 (3) | N3—C4—C5 | 112.0 (2) |
| C15—C14—C13 | 120.0 (3) | N3—C4—N4 | 120.03 (19) |
| C15—C14—H14 | 120 (2) | C5—C4—N4 | 127.9 (2) |
| C13—C14—H14 | 120 (3) | O42—N4—O41 | 123.4 (2) |
| C14—C15—C16 | 120.4 (3) | O42—N4—C4 | 118.3 (2) |
| C14—C15—H15 | 120 (3) | O41—N4—C4 | 118.3 (2) |
| C16—C15—H15 | 119 (3) | N1—C5—C4 | 103.94 (19) |
| C11—C16—C15 | 118.5 (3) | N1—C5—Br5 | 120.76 (16) |
| C11—C16—H16 | 120 (2) | C4—C5—Br5 | 135.30 (18) |
| | | |
| C5—N1—C11—C12 | −95.2 (3) | N1—C2—N3—C4 | −0.5 (3) |
| C2—N1—C11—C12 | 88.1 (3) | C21—C2—N3—C4 | 178.1 (3) |
| C5—N1—C11—C16 | 85.8 (3) | C2—N3—C4—C5 | 1.0 (3) |
| C2—N1—C11—C16 | −91.0 (3) | C2—N3—C4—N4 | −179.8 (2) |
| C16—C11—C12—C13 | 1.3 (4) | N3—C4—N4—O42 | −179.7 (2) |
| N1—C11—C12—C13 | −177.7 (3) | C5—C4—N4—O42 | −0.6 (4) |
| C11—C12—C13—C14 | −0.1 (5) | N3—C4—N4—O41 | 0.9 (4) |
| C12—C13—C14—C15 | −0.9 (5) | C5—C4—N4—O41 | 179.9 (3) |
| C13—C14—C15—C16 | 0.6 (5) | C2—N1—C5—C4 | 0.8 (3) |
| C12—C11—C16—C15 | −1.6 (4) | C11—N1—C5—C4 | −176.4 (2) |
| N1—C11—C16—C15 | 177.4 (3) | C2—N1—C5—Br5 | −179.33 (18) |
| C14—C15—C16—C11 | 0.6 (5) | C11—N1—C5—Br5 | 3.5 (4) |
| C5—N1—C2—N3 | −0.2 (3) | N3—C4—C5—N1 | −1.1 (3) |
| C11—N1—C2—N3 | 177.1 (2) | N4—C4—C5—N1 | 179.8 (2) |
| C5—N1—C2—C21 | −178.9 (2) | N3—C4—C5—Br5 | 179.0 (2) |
| C11—N1—C2—C21 | −1.6 (4) | N4—C4—C5—Br5 | −0.1 (5) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14···O41i | 0.95 (4) | 2.71 (4) | 3.457 (4) | 136 (3) |
| C21—H21C···Br5ii | 0.87 (4) | 3.01 (4) | 3.707 (3) | 138 (3) |
| Symmetry codes: (i) x−1, −y+1/2, z−1/2; (ii) x+1, y, z. |
(100K) 1-phenyl-4-nitro-5-bromo-imidazole
top
Crystal data top
| C10H8BrN3O2 | Z = 2 |
| Mr = 282.10 | F(000) = 280 |
| Triclinic, P1 | Dx = 1.806 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 6.9654 (10) Å | Cell parameters from 1221 reflections |
| b = 6.8215 (10) Å | θ = 4–22° |
| c = 11.3983 (17) Å | µ = 3.95 mm−1 |
| α = 92.683 (13)° | T = 100 K |
| β = 106.506 (12)° | Prism, colourless |
| γ = 90.277 (12)° | 0.5 × 0.3 × 0.1 mm |
| V = 518.60 (14) Å3 | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 1720 independent reflections |
| Radiation source: fine-focus sealed tube | 1586 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.072 |
| ω–scan | θmax = 29.9°, θmin = 3.7° |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | h = −9→9 |
| Tmin = 0.305, Tmax = 0.594 | k = −9→9 |
| 6344 measured reflections | l = −15→15 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.058 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.161 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.11 | w = 1/[σ2(Fo2) + (0.0209P)2 + 5.8358P]
where P = (Fo2 + 2Fc2)/3 |
| 1720 reflections | (Δ/σ)max = 0.027 |
| 146 parameters | Δρmax = 1.82 e Å−3 |
| 0 restraints | Δρmin = −1.85 e Å−3 |
Crystal data top
| C10H8BrN3O2 | γ = 90.277 (12)° |
| Mr = 282.10 | V = 518.60 (14) Å3 |
| Triclinic, P1 | Z = 2 |
| a = 6.9654 (10) Å | Mo Kα radiation |
| b = 6.8215 (10) Å | µ = 3.95 mm−1 |
| c = 11.3983 (17) Å | T = 100 K |
| α = 92.683 (13)° | 0.5 × 0.3 × 0.1 mm |
| β = 106.506 (12)° | |
Data collection top
KUMA KM4CCD four-circle
diffractometer | 1720 independent reflections |
Absorption correction: multi-scan
SORTAV (Blessing, 1989) | 1586 reflections with I > 2σ(I) |
| Tmin = 0.305, Tmax = 0.594 | Rint = 0.072 |
| 6344 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.058 | 0 restraints |
| wR(F2) = 0.161 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.11 | Δρmax = 1.82 e Å−3 |
| 1720 reflections | Δρmin = −1.85 e Å−3 |
| 146 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N1 | 0.8092 (9) | 0.7313 (11) | 0.1534 (6) | 0.0217 (14) | |
| C11 | 0.9712 (10) | 0.7195 (14) | 0.2643 (6) | 0.0228 (17) | |
| C12 | 1.0368 (12) | 0.5357 (14) | 0.3087 (8) | 0.0232 (16) | |
| H12 | 0.9795 | 0.4172 | 0.2655 | 0.028* | |
| C13 | 1.1894 (13) | 0.5338 (14) | 0.4187 (8) | 0.0251 (17) | |
| H13 | 1.2374 | 0.4118 | 0.4512 | 0.030* | |
| C14 | 1.2728 (12) | 0.7090 (15) | 0.4819 (7) | 0.0278 (18) | |
| H14 | 1.3757 | 0.7057 | 0.5572 | 0.033* | |
| C15 | 1.2063 (14) | 0.8830 (17) | 0.4354 (9) | 0.035 (2) | |
| H15 | 1.2647 | 1.0015 | 0.4781 | 0.042* | |
| C16 | 1.0547 (14) | 0.8918 (17) | 0.3266 (9) | 0.034 (2) | |
| H16 | 1.0088 | 1.0151 | 0.2953 | 0.041* | |
| C2 | 0.6094 (11) | 0.7311 (14) | 0.1477 (7) | 0.0232 (17) | |
| C21 | 0.5387 (11) | 0.7104 (15) | 0.2581 (7) | 0.0256 (18) | |
| H21A | 0.6152 | 0.6084 | 0.3081 | 0.033* | |
| H21B | 0.3962 | 0.6735 | 0.2327 | 0.033* | |
| H21C | 0.5582 | 0.8355 | 0.3062 | 0.033* | |
| N3 | 0.4962 (9) | 0.7493 (9) | 0.0351 (5) | 0.0127 (11) | |
| C4 | 0.6271 (10) | 0.7560 (11) | −0.0337 (6) | 0.0161 (14) | |
| N4 | 0.5557 (10) | 0.7883 (12) | −0.1618 (6) | 0.0253 (15) | |
| O41 | 0.6784 (8) | 0.7969 (10) | −0.2206 (5) | 0.0271 (13) | |
| O42 | 0.3742 (8) | 0.8015 (10) | −0.2077 (5) | 0.0267 (13) | |
| C5 | 0.8233 (10) | 0.7505 (10) | 0.0378 (6) | 0.0147 (13) | |
| Br5 | 1.07127 (10) | 0.75271 (10) | 0.00703 (6) | 0.0131 (2) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N1 | 0.007 (3) | 0.049 (4) | 0.009 (3) | −0.004 (3) | 0.004 (2) | 0.002 (3) |
| C11 | 0.006 (3) | 0.056 (5) | 0.005 (3) | −0.003 (3) | 0.001 (2) | −0.003 (3) |
| C12 | 0.015 (4) | 0.038 (5) | 0.014 (4) | 0.004 (3) | 0.001 (3) | 0.007 (3) |
| C13 | 0.022 (4) | 0.035 (5) | 0.017 (4) | 0.002 (3) | 0.001 (3) | 0.005 (3) |
| C14 | 0.016 (4) | 0.057 (6) | 0.008 (3) | 0.002 (4) | 0.000 (3) | 0.005 (3) |
| C15 | 0.019 (4) | 0.053 (6) | 0.026 (5) | −0.003 (4) | −0.002 (3) | −0.014 (4) |
| C16 | 0.018 (4) | 0.060 (7) | 0.020 (4) | 0.005 (4) | −0.002 (3) | −0.002 (4) |
| C2 | 0.007 (3) | 0.052 (5) | 0.012 (4) | −0.001 (3) | 0.005 (3) | 0.001 (3) |
| C21 | 0.010 (3) | 0.056 (6) | 0.012 (3) | 0.000 (3) | 0.004 (3) | 0.003 (3) |
| N3 | 0.010 (3) | 0.020 (3) | 0.010 (3) | −0.002 (2) | 0.004 (2) | −0.001 (2) |
| C4 | 0.006 (3) | 0.030 (4) | 0.012 (3) | −0.005 (3) | 0.003 (2) | 0.000 (3) |
| N4 | 0.009 (3) | 0.056 (5) | 0.011 (3) | −0.003 (3) | 0.004 (2) | 0.001 (3) |
| O41 | 0.016 (3) | 0.054 (4) | 0.014 (3) | 0.000 (3) | 0.009 (2) | 0.003 (2) |
| O42 | 0.011 (3) | 0.054 (4) | 0.013 (3) | 0.002 (2) | 0.000 (2) | 0.002 (2) |
| C5 | 0.012 (3) | 0.020 (3) | 0.013 (3) | −0.005 (3) | 0.006 (3) | −0.005 (2) |
| Br5 | 0.0081 (3) | 0.0181 (4) | 0.0141 (4) | −0.0012 (2) | 0.0049 (2) | −0.0002 (2) |
Geometric parameters (Å, º) top
| N1—C5 | 1.360 (9) | C2—N3 | 1.313 (10) |
| N1—C2 | 1.375 (9) | C2—C21 | 1.488 (10) |
| N1—C11 | 1.442 (9) | N3—C4 | 1.363 (8) |
| C11—C16 | 1.380 (14) | C4—C5 | 1.380 (9) |
| C11—C12 | 1.404 (13) | C4—N4 | 1.430 (10) |
| C12—C13 | 1.395 (11) | N4—O42 | 1.229 (8) |
| C13—C14 | 1.400 (13) | N4—O41 | 1.230 (8) |
| C14—C15 | 1.352 (15) | C5—Br5 | 1.858 (7) |
| C15—C16 | 1.386 (13) | | |
| | | |
| C5—N1—C2 | 107.7 (6) | N3—C2—C21 | 126.3 (6) |
| C5—N1—C11 | 127.4 (6) | N1—C2—C21 | 122.2 (7) |
| C2—N1—C11 | 124.9 (6) | C2—N3—C4 | 104.8 (6) |
| C16—C11—C12 | 121.4 (8) | N3—C4—C5 | 111.7 (6) |
| C16—C11—N1 | 118.5 (8) | N3—C4—N4 | 120.1 (6) |
| C12—C11—N1 | 120.0 (8) | C5—C4—N4 | 127.8 (6) |
| C13—C12—C11 | 117.4 (8) | O42—N4—O41 | 123.5 (7) |
| C12—C13—C14 | 121.0 (8) | O42—N4—C4 | 118.1 (6) |
| C15—C14—C13 | 119.7 (8) | O41—N4—C4 | 118.4 (6) |
| C14—C15—C16 | 121.2 (10) | N1—C5—C4 | 104.3 (6) |
| C11—C16—C15 | 119.3 (10) | N1—C5—Br5 | 120.7 (5) |
| N3—C2—N1 | 111.5 (6) | C4—C5—Br5 | 134.9 (5) |
| | | |
| C5—N1—C11—C16 | 86.3 (11) | N1—C2—N3—C4 | 1.7 (9) |
| C2—N1—C11—C16 | −91.5 (11) | C21—C2—N3—C4 | −177.8 (9) |
| C5—N1—C11—C12 | −95.6 (10) | C2—N3—C4—C5 | −2.7 (9) |
| C2—N1—C11—C12 | 86.6 (11) | C2—N3—C4—N4 | −176.0 (8) |
| C16—C11—C12—C13 | 0.2 (11) | N3—C4—N4—O42 | −2.9 (12) |
| N1—C11—C12—C13 | −177.8 (7) | C5—C4—N4—O42 | −175.1 (8) |
| C11—C12—C13—C14 | 0.1 (13) | N3—C4—N4—O41 | 178.8 (7) |
| C12—C13—C14—C15 | −0.6 (12) | C5—C4—N4—O41 | 6.6 (13) |
| C13—C14—C15—C16 | 0.8 (14) | C2—N1—C5—C4 | −1.5 (9) |
| C12—C11—C16—C15 | −0.1 (13) | C11—N1—C5—C4 | −179.6 (8) |
| N1—C11—C16—C15 | 178.0 (8) | C2—N1—C5—Br5 | −179.0 (6) |
| C14—C15—C16—C11 | −0.5 (15) | C11—N1—C5—Br5 | 2.9 (12) |
| C5—N1—C2—N3 | −0.1 (10) | N3—C4—C5—N1 | 2.6 (9) |
| C11—N1—C2—N3 | 178.1 (8) | N4—C4—C5—N1 | 175.3 (8) |
| C5—N1—C2—C21 | 179.4 (8) | N3—C4—C5—Br5 | 179.5 (6) |
| C11—N1—C2—C21 | −2.5 (14) | N4—C4—C5—Br5 | −7.8 (13) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C12—H12···O42i | 0.95 | 2.77 | 3.553 (11) | 140 |
| C21—H21B···Br5ii | 0.98 | 2.98 | 3.697 (8) | 131 |
| Symmetry codes: (i) −x+1, −y+1, −z; (ii) x−1, y, z. |
Experimental details
| | (295K) | (250K) | (200K) | (170K) |
| Crystal data |
| Chemical formula | C10H8BrN3O2 | C10H8BrN3O2 | C10H8BrN3O2 | C10H8BrN3O2 |
| Mr | 282.10 | 282.10 | 282.10 | 282.10 |
| Crystal system, space group | Monoclinic, P21/m | Monoclinic, P21/m | Monoclinic, P21/c | Monoclinic, P21/c |
| Temperature (K) | 295 | 250 | 200 | 170 |
| a, b, c (Å) | 7.0281 (10), 6.9388 (9), 11.5612 (15) | 7.0180 (14), 6.9160 (12), 11.546 (2) | 6.9955 (9), 6.8657 (8), 22.990 (2) | 6.9923 (12), 6.8497 (10), 22.9702 (16) |
| α, β, γ (°) | 90, 106.735 (12), 90 | 90, 106.410 (15), 90 | 90, 106.356 (10), 90 | 90, 106.419 (14), 90 |
| V (Å3) | 539.92 (13) | 537.57 (17) | 1059.5 (2) | 1055.3 (2) |
| Z | 2 | 2 | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα | Mo Kα |
| µ (mm−1) | 3.80 | 3.81 | 3.87 | 3.88 |
| Crystal size (mm) | 0.3 × 0.25 × 0.1 | 0.5 × 0.3 × 0.1 | 0.5 × 0.3 × 0.1 | 0.5 × 0.3 × 0.1 |
| |
| Data collection |
| Diffractometer | KUMA KM4CCD four-circle
diffractometer | KUMA KM4CCD four-circle
diffractometer | KUMA KM4CCD four-circle
diffractometer | KUMA KM4CCD four-circle
diffractometer |
| Absorption correction | Multi-scan
SORTAV (Blessing, 1989) | Multi-scan
SORTAV (Blessing, 1989) | Multi-scan
SORTAV (Blessing, 1989) | Multi-scan
SORTAV (Blessing, 1989) |
| Tmin, Tmax | 0.382, 0.696 | 0.326, 0.673 | 0.355, 0.725 | 0.348, 0.659 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 5105, 1521, 1342 | 3360, 1473, 1097 | 13871, 2719, 1610 | 6354, 2673, 1387 |
| Rint | 0.042 | 0.054 | 0.061 | 0.060 |
| (sin θ/λ)max (Å−1) | 0.697 | 0.694 | 0.691 | 0.693 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.029, 0.075, 1.02 | 0.042, 0.100, 1.00 | 0.049, 0.137, 1.13 | 0.045, 0.100, 1.01 |
| No. of reflections | 1521 | 1473 | 2719 | 2673 |
| No. of parameters | 93 | 91 | 148 | 148 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.36, −1.18 | 0.74, −0.96 | 1.14, −1.08 | 0.82, −0.99 |
| | (150K) | (120K) | (100K) |
| Crystal data |
| Chemical formula | C10H8BrN3O2 | C10H8BrN3O2 | C10H8BrN3O2 |
| Mr | 282.10 | 282.10 | 282.10 |
| Crystal system, space group | Monoclinic, P21/c | Monoclinic, P21/c | Triclinic, P1 |
| Temperature (K) | 150 | 120 | 100 |
| a, b, c (Å) | 6.9854 (12), 6.8362 (9), 22.945 (2) | 6.9723 (5), 6.8169 (4), 22.9000 (12) | 6.9654 (10), 6.8215 (10), 11.3983 (17) |
| α, β, γ (°) | 90, 106.466 (13), 90 | 90, 106.478 (6), 90 | 92.683 (13), 106.506 (12), 90.277 (12) |
| V (Å3) | 1050.8 (2) | 1043.72 (11) | 518.60 (14) |
| Z | 4 | 4 | 2 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα |
| µ (mm−1) | 3.90 | 3.93 | 3.95 |
| Crystal size (mm) | 0.5 × 0.3 × 0.1 | 0.3 × 0.25 × 0.1 | 0.5 × 0.3 × 0.1 |
| |
| Data collection |
| Diffractometer | KUMA KM4CCD four-circle
diffractometer | KUMA KM4CCD four-circle
diffractometer | KUMA KM4CCD four-circle
diffractometer |
| Absorption correction | Multi-scan
SORTAV (Blessing, 1989) | Multi-scan
SORTAV (Blessing, 1989) | Multi-scan
SORTAV (Blessing, 1989) |
| Tmin, Tmax | 0.321, 0.717 | 0.378, 0.573 | 0.305, 0.594 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 6609, 2676, 1445 | 13861, 2788, 2349 | 6344, 1720, 1586 |
| Rint | 0.069 | 0.045 | 0.072 |
| (sin θ/λ)max (Å−1) | 0.693 | 0.699 | 0.702 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.044, 0.095, 1.02 | 0.033, 0.068, 1.07 | 0.058, 0.161, 1.11 |
| No. of reflections | 2676 | 2788 | 1720 |
| No. of parameters | 148 | 178 | 146 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | All H-atom parameters refined | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.85, −1.00 | 0.71, −0.52 | 1.82, −1.85 |
Hydrogen-bond geometry (Å, º) for (295K) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14···O41i | 0.93 | 2.77 | 3.502 (5) | 136.8 |
| C21—H21B···Br5ii | 0.96 | 2.98 | 3.746 (4) | 137.4 |
| Symmetry codes: (i) x+1, y, z+1; (ii) x−1, y, z. |
Hydrogen-bond geometry (Å, º) for (250K) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14···O41i | 0.93 | 2.89 | 3.819 (8) | 179.5 |
| C21—H21A···Br5ii | 0.97 | 2.97 | 3.753 (6) | 137.7 |
| Symmetry codes: (i) x+1, y, z+1; (ii) x−1, y, z. |
Hydrogen-bond geometry (Å, º) for (200K) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14···O41i | 0.95 | 2.73 | 3.472 (6) | 135.5 |
| C21—H21C···Br5ii | 0.98 | 2.96 | 3.733 (5) | 136.5 |
| Symmetry codes: (i) x−1, −y+1/2, z−1/2; (ii) x+1, y, z. |
Hydrogen-bond geometry (Å, º) for (170K) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14···O41i | 0.95 | 2.74 | 3.471 (6) | 134.5 |
| C21—H21C···Br5ii | 0.98 | 2.96 | 3.732 (5) | 136.3 |
| Symmetry codes: (i) x−1, −y+1/2, z−1/2; (ii) x+1, y, z. |
Hydrogen-bond geometry (Å, º) for (150K) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14···O41i | 0.95 | 2.74 | 3.469 (6) | 134.4 |
| C21—H21C···Br5ii | 0.98 | 2.95 | 3.722 (4) | 136.2 |
| Symmetry codes: (i) x−1, −y+1/2, z−1/2; (ii) x+1, y, z. |
Hydrogen-bond geometry (Å, º) for (120K) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C14—H14···O41i | 0.95 (4) | 2.71 (4) | 3.457 (4) | 136 (3) |
| C21—H21C···Br5ii | 0.87 (4) | 3.01 (4) | 3.707 (3) | 138 (3) |
| Symmetry codes: (i) x−1, −y+1/2, z−1/2; (ii) x+1, y, z. |
Hydrogen-bond geometry (Å, º) for (100K) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C12—H12···O42i | 0.95 | 2.77 | 3.553 (11) | 140.2 |
| C21—H21B···Br5ii | 0.98 | 2.98 | 3.697 (8) | 131.2 |
| Symmetry codes: (i) −x+1, −y+1, −z; (ii) x−1, y, z. |
 N and dihalogen interactions Br
N and dihalogen interactions Br Br play a crucial role in determining the crystal packing and compete successfully with other kinds of weak intermolecular interactions.
Br play a crucial role in determining the crystal packing and compete successfully with other kinds of weak intermolecular interactions.
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/b/issues/2004/03/00/bk5005/bk5005fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/b/issues/2004/03/00/bk5005/bk5005fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/b/issues/2004/03/00/bk5005/bk5005fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/b/issues/2004/03/00/bk5005/bk5005fig4thm.gif)
![[Figure 5]](http://journals.iucr.org/b/issues/2004/03/00/bk5005/bk5005fig5thm.gif)
![[Figure 6]](http://journals.iucr.org/b/issues/2004/03/00/bk5005/bk5005fig6thm.gif)
![[Figure 7]](http://journals.iucr.org/b/issues/2004/03/00/bk5005/bk5005fig7thm.gif)
![[Figure 8]](http://journals.iucr.org/b/issues/2004/03/00/bk5005/bk5005fig8thm.gif)
![[Figure 9]](http://journals.iucr.org/b/issues/2004/03/00/bk5005/bk5005fig9thm.gif)
![[Figure 10]](http://journals.iucr.org/b/issues/2004/03/00/bk5005/bk5005fig10thm.gif)
![[Figure 11]](http://journals.iucr.org/b/issues/2004/03/00/bk5005/bk5005fig11thm.gif)
![[Figure 12]](http://journals.iucr.org/b/issues/2004/03/00/bk5005/bk5005fig12thm.gif)



