Mg
2Ru
2Cl
10O·16H
2O {dimagnesium μ-oxo-bis[pentachlororuthenate(IV)] hexadecahydrate} crystallizes in the monoclinic system (space group
P2
1/
c). The structure consists of layers of [Ru
2Cl
10O]
4− anions, [Mg(H
2O)
6]
2+ cations and water molecules stacked along the
a axis. Only the O atom bonded to Ru occupies the 2
a site with
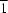
symmetry. All the other atoms occupy general 4
e sites. The crystal structure is stabilized by O—H

O and O—H

Cl interactions.
Supporting information
The title compound was crystallized from a supersaturated hydrochloric acid solution (50%, 5 ml) prepared using doubly distilled water and a mixture of ruthenium(III) chloride trihydrate (2.61 g) and magnesium chloride (0.95 g) in a 1:1 ratio. Supersaturation was obtained by gentle warming of the solution. Thin, brown needle-shaped single crystals of Mg2Ru2Cl10O.16H2O were obtained at ambient temperature by slow evaporation of the solution.
H atoms were fixed by geometric constraints using HFIX and allowed to ride on the attached O atom (O—H = 0.92–0.99 Å). CIF suggests that these atoms were refined with distance restraints applied, not as riding. Please check all values in Table 2; many of the O—H distances do not match the geom_bond_distance values in CIF.
Data collection: COLLECT (Nonius, 1997); cell refinement: SCALEPACK (Otwinowski & Minor, 1997); data reduction: SCALEPACK and DENZO (Otwinowski & Minor, 1997); program(s) used to solve structure: SIR92 (Altomare et al., 1993); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: ORTEP-3 (Farrugia, 1997) and ATOMS (Dowty, 1995); software used to prepare material for publication: WinGX (Farrugia, 1999) and PARST (Nardelli, 1995).
dimagnesium µ-oxo-bis(pentachlororuthenate(IV) hexadecahydrate
top
Crystal data top
| Mg2Ru2Cl10O·16H2O | F(000) = 900 |
| Mr = 906.88 | Dx = 2.112 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 4491 reflections |
| a = 8.256 (2) Å | θ = 2.3–26.4° |
| b = 16.343 (3) Å | µ = 2.09 mm−1 |
| c = 10.647 (5) Å | T = 173 K |
| β = 95.499 (3)° | Needle, brown |
| V = 1430.0 (8) Å3 | 0.45 × 0.06 × 0.03 mm |
| Z = 2 | |
Data collection top
Nonius KappaCCD
diffractometer | 2918 independent reflections |
| Radiation source: fine-focus sealed tube | 2295 reflections with I > 2σ(I) |
| Detector resolution: 9 pixels mm-1 | Rint = 0.038 |
| ω or ϕ scans? | θmax = 26.4°, θmin = 2.3° |
Absorption correction: part of the refinement model (ΔF)
(Sheldrick, 1990) | h = −9→10 |
| Tmin = 0.760, Tmax = 0.939 | k = −20→18 |
| 8087 measured reflections | l = −11→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.028 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.063 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0098P)2 + 0.1594P]
where P = (Fo2 + 2Fc2)/3 |
| 2918 reflections | (Δ/σ)max = 0.002 |
| 190 parameters | Δρmax = 0.58 e Å−3 |
| 16 restraints | Δρmin = −0.66 e Å−3 |
Crystal data top
| Mg2Ru2Cl10O·16H2O | V = 1430.0 (8) Å3 |
| Mr = 906.88 | Z = 2 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 8.256 (2) Å | µ = 2.09 mm−1 |
| b = 16.343 (3) Å | T = 173 K |
| c = 10.647 (5) Å | 0.45 × 0.06 × 0.03 mm |
| β = 95.499 (3)° | |
Data collection top
Nonius KappaCCD
diffractometer | 2918 independent reflections |
Absorption correction: part of the refinement model (ΔF)
(Sheldrick, 1990) | 2295 reflections with I > 2σ(I) |
| Tmin = 0.760, Tmax = 0.939 | Rint = 0.038 |
| 8087 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.028 | 16 restraints |
| wR(F2) = 0.063 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | Δρmax = 0.58 e Å−3 |
| 2918 reflections | Δρmin = −0.66 e Å−3 |
| 190 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Ru | 0.46786 (3) | 0.439501 (13) | 0.36057 (2) | 0.01211 (9) | |
| Cl1 | 0.44264 (11) | 0.55535 (4) | 0.22846 (8) | 0.0223 (2) | |
| Cl2 | 0.74948 (10) | 0.43557 (4) | 0.33572 (8) | 0.01764 (19) | |
| Cl3 | 0.18124 (10) | 0.43860 (4) | 0.36637 (8) | 0.01985 (19) | |
| Cl4 | 0.42362 (10) | 0.35620 (5) | 0.18105 (8) | 0.0246 (2) | |
| Cl5 | 0.49101 (10) | 0.31640 (4) | 0.47910 (8) | 0.0210 (2) | |
| Mg | 0.01007 (12) | 0.18896 (5) | 0.25193 (10) | 0.0173 (3) | |
| O1W | 0.0345 (3) | 0.30061 (12) | 0.1593 (2) | 0.0222 (6) | |
| H11W | 0.087 (4) | 0.3393 (16) | 0.215 (3) | 0.033* | |
| H21W | −0.067 (3) | 0.3217 (17) | 0.120 (3) | 0.033* | |
| O2W | 0.1570 (3) | 0.22248 (14) | 0.4098 (2) | 0.0308 (6) | |
| H12W | 0.264 (3) | 0.2452 (19) | 0.416 (4) | 0.046* | |
| H22W | 0.122 (4) | 0.227 (2) | 0.493 (2) | 0.046* | |
| O3W | 0.2037 (3) | 0.14318 (14) | 0.1661 (3) | 0.0326 (6) | |
| H13W | 0.204 (5) | 0.0980 (16) | 0.110 (3) | 0.049* | |
| H23W | 0.304 (3) | 0.1677 (19) | 0.204 (3) | 0.049* | |
| O4W | −0.0214 (3) | 0.07874 (13) | 0.3412 (2) | 0.0247 (6) | |
| H14W | 0.074 (3) | 0.0545 (17) | 0.386 (3) | 0.037* | |
| H24W | −0.084 (4) | 0.0402 (17) | 0.289 (3) | 0.037* | |
| O5W | −0.1394 (3) | 0.15077 (13) | 0.0987 (2) | 0.0280 (6) | |
| H15W | −0.117 (4) | 0.1083 (16) | 0.041 (3) | 0.042* | |
| H25W | −0.254 (2) | 0.151 (2) | 0.105 (4) | 0.042* | |
| O6W | −0.1955 (3) | 0.23270 (13) | 0.3223 (2) | 0.0259 (6) | |
| H16W | −0.212 (4) | 0.2876 (12) | 0.336 (3) | 0.039* | |
| H26W | −0.234 (4) | 0.2079 (18) | 0.394 (2) | 0.039* | |
| O7W | −0.2495 (3) | 0.35047 (13) | 0.0389 (2) | 0.0262 (6) | |
| H17W | −0.250 (4) | 0.4089 (11) | 0.030 (3) | 0.039* | |
| H27W | −0.339 (3) | 0.3416 (19) | 0.088 (3) | 0.039* | |
| O8W | −0.2682 (3) | 0.52003 (14) | 0.0458 (2) | 0.0288 (6) | |
| H18W | −0.305 (4) | 0.5504 (17) | −0.031 (2) | 0.043* | |
| H28W | −0.349 (4) | 0.532 (2) | 0.102 (3) | 0.043* | |
| O1 | 0.5 | 0.5 | 0.5 | 0.0114 (6) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Ru | 0.01097 (16) | 0.01256 (15) | 0.01243 (15) | 0.00045 (10) | −0.00084 (11) | −0.00163 (10) |
| Cl1 | 0.0258 (5) | 0.0221 (4) | 0.0186 (5) | 0.0045 (3) | −0.0011 (4) | 0.0052 (3) |
| Cl2 | 0.0122 (4) | 0.0229 (4) | 0.0179 (5) | 0.0013 (3) | 0.0016 (3) | −0.0001 (3) |
| Cl3 | 0.0115 (4) | 0.0212 (4) | 0.0265 (5) | 0.0007 (3) | 0.0000 (4) | −0.0031 (3) |
| Cl4 | 0.0216 (5) | 0.0306 (4) | 0.0212 (5) | −0.0010 (4) | 0.0004 (4) | −0.0095 (4) |
| Cl5 | 0.0202 (5) | 0.0153 (4) | 0.0272 (5) | 0.0002 (3) | 0.0004 (4) | 0.0036 (3) |
| Mg | 0.0157 (6) | 0.0175 (6) | 0.0185 (7) | −0.0006 (5) | 0.0002 (5) | −0.0001 (4) |
| O1W | 0.0229 (15) | 0.0178 (12) | 0.0251 (15) | −0.0042 (10) | −0.0024 (11) | 0.0007 (10) |
| O2W | 0.0336 (17) | 0.0353 (13) | 0.0220 (15) | −0.0166 (12) | −0.0054 (13) | −0.0003 (12) |
| O3W | 0.0257 (15) | 0.0356 (15) | 0.0377 (18) | 0.0058 (12) | 0.0094 (13) | −0.0045 (12) |
| O4W | 0.0214 (15) | 0.0203 (12) | 0.0315 (16) | −0.0034 (10) | −0.0022 (12) | 0.0038 (10) |
| O5W | 0.0295 (15) | 0.0283 (13) | 0.0246 (15) | 0.0005 (11) | −0.0055 (12) | −0.0052 (11) |
| O6W | 0.0276 (15) | 0.0206 (12) | 0.0320 (16) | 0.0053 (11) | 0.0150 (12) | 0.0033 (11) |
| O7W | 0.0262 (15) | 0.0251 (12) | 0.0278 (15) | 0.0016 (11) | 0.0056 (12) | 0.0014 (11) |
| O8W | 0.0271 (16) | 0.0366 (14) | 0.0221 (16) | −0.0015 (12) | −0.0016 (12) | −0.0002 (11) |
| O1 | 0.0093 (16) | 0.0116 (14) | 0.0131 (17) | 0.0017 (12) | −0.0007 (13) | 0.0006 (12) |
Geometric parameters (Å, º) top
| Ru—O1 | 1.7822 (6) | O1W—H21W | 0.960 (18) |
| Ru—Cl4 | 2.3464 (11) | O7W—H17W | 0.959 (18) |
| Ru—Cl1 | 2.3559 (9) | O7W—H27W | 0.959 (18) |
| Ru—Cl2 | 2.3660 (10) | O6W—H16W | 0.921 (18) |
| Ru—Cl5 | 2.3728 (9) | O6W—H26W | 0.950 (18) |
| Ru—Cl3 | 2.3731 (10) | O2W—H12W | 0.957 (18) |
| Mg—O5W | 2.047 (3) | O2W—H22W | 0.964 (18) |
| Mg—O6W | 2.048 (2) | O3W—H13W | 0.949 (18) |
| Mg—O2W | 2.051 (3) | O3W—H23W | 0.974 (19) |
| Mg—O3W | 2.056 (3) | O5W—H15W | 0.954 (18) |
| Mg—O4W | 2.065 (2) | O5W—H25W | 0.954 (18) |
| Mg—O1W | 2.093 (2) | O1—Rui | 1.7822 (6) |
| O4W—H14W | 0.968 (18) | O8W—H18W | 0.983 (18) |
| O4W—H24W | 0.959 (18) | O8W—H28W | 0.958 (18) |
| O1W—H11W | 0.946 (18) | | |
| | | |
| O1—Ru—Cl4 | 178.14 (2) | O6W—Mg—O4W | 89.48 (9) |
| O1—Ru—Cl1 | 92.81 (3) | O2W—Mg—O4W | 86.93 (10) |
| O1—Ru—Cl5 | 91.68 (3) | O3W—Mg—O4W | 91.41 (10) |
| O1—Ru—Cl3 | 92.92 (2) | O5W—Mg—O6W | 86.60 (11) |
| O1—Ru—Cl2 | 92.22 (2) | Mg—O1W—H11W | 110 (2) |
| Cl1—Ru—Cl2 | 89.24 (3) | Mg—O1W—H21W | 113.5 (19) |
| Cl1—Ru—Cl5 | 175.50 (3) | H11W—O1W—H21W | 111 (3) |
| Cl1—Ru—Cl3 | 89.38 (3) | H17W—O7W—H27W | 102 (3) |
| Cl2—Ru—Cl3 | 174.73 (3) | Mg—O2W—H12W | 129 (2) |
| Cl2—Ru—Cl5 | 90.37 (3) | Mg—O2W—H22W | 125 (2) |
| Cl4—Ru—Cl5 | 86.54 (4) | H12W—O2W—H22W | 105 (3) |
| Cl4—Ru—Cl1 | 88.97 (4) | Mg—O3W—H13W | 128 (2) |
| Cl4—Ru—Cl2 | 88.34 (3) | Mg—O3W—H23W | 110 (2) |
| Cl4—Ru—Cl3 | 86.55 (3) | H13W—O3W—H23W | 122 (3) |
| Cl5—Ru—Cl3 | 90.61 (3) | Mg—O4W—H14W | 116.8 (19) |
| O5W—Mg—O1W | 88.18 (10) | Mg—O4W—H24W | 113 (2) |
| O6W—Mg—O1W | 89.34 (9) | H14W—O4W—H24W | 113 (3) |
| O2W—Mg—O1W | 94.43 (10) | Mg—O5W—H15W | 126 (2) |
| O3W—Mg—O1W | 89.65 (10) | Mg—O5W—H25W | 118 (2) |
| O4W—Mg—O1W | 178.25 (11) | H15W—O5W—H25W | 108 (3) |
| O5W—Mg—O2W | 177.37 (10) | Mg—O6W—H16W | 123 (2) |
| O6W—Mg—O2W | 93.02 (11) | Mg—O6W—H26W | 120 (2) |
| O5W—Mg—O3W | 88.27 (11) | H16W—O6W—H26W | 103 (3) |
| O6W—Mg—O3W | 174.80 (12) | H18W—O8W—H28W | 104 (3) |
| O2W—Mg—O3W | 92.14 (12) | Rui—O1—Ru | 180 |
| O5W—Mg—O4W | 90.46 (10) | | |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1W—H11W···Cl3 | 0.94 (3) | 2.37 (3) | 3.302 (3) | 172 (3) |
| O1W—H21W···O7W | 0.96 (3) | 1.73 (3) | 2.688 (4) | 174 (3) |
| O2W—H12W···Cl5 | 0.95 (3) | 2.25 (3) | 3.180 (3) | 164 (3) |
| O2W—H22W···O1Wii | 0.96 (2) | 2.03 (2) | 2.956 (3) | 162 (3) |
| O3W—H13W···Cl2iii | 0.95 (3) | 2.74 (3) | 3.415 (3) | 129 (2) |
| O3W—H13W···Cl3iv | 0.95 (3) | 2.65 (3) | 3.448 (4) | 142 (2) |
| O3W—H23W···Cl1iii | 0.97 (3) | 2.82 (3) | 3.349 (3) | 115 (2) |
| O4W—H14W···O8Wv | 0.97 (3) | 1.79 (3) | 2.745 (4) | 171 (3) |
| O4W—H24W···Cl3v | 0.96 (3) | 2.43 (3) | 3.363 (3) | 166 (3) |
| O5W—H15W···Cl2vi | 0.96 (3) | 2.46 (3) | 3.191 (3) | 133 (2) |
| O5W—H25W···Cl5vi | 0.96 (2) | 2.44 (3) | 3.237 (3) | 140 (3) |
| O6W—H16W···Cl2vii | 0.92 (2) | 2.44 (2) | 3.351 (3) | 171 (3) |
| O6W—H26W···O7Wii | 0.95 (2) | 1.83 (2) | 2.750 (3) | 164 (3) |
| O7W—H17W···O8W | 0.96 (2) | 1.83 (2) | 2.777 (3) | 168 (3) |
| O7W—H27W···Cl4vii | 0.96 (3) | 2.29 (3) | 3.220 (3) | 163 (3) |
| O8W—H28W···Cl1vii | 0.96 (3) | 2.32 (3) | 3.272 (3) | 176 (3) |
| O8W—H18W···Cl4viii | 0.98 (2) | 2.35 (3) | 3.313 (3) | 166 (2) |
| Symmetry codes: (ii) x, −y+1/2, z+1/2; (iii) −x+1, y−1/2, −z+1/2; (iv) x, −y+1/2, z−1/2; (v) −x, y−1/2, −z+1/2; (vi) x−1, −y+1/2, z−1/2; (vii) x−1, y, z; (viii) −x, −y+1, −z. |
Experimental details
| Crystal data |
| Chemical formula | Mg2Ru2Cl10O·16H2O |
| Mr | 906.88 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 173 |
| a, b, c (Å) | 8.256 (2), 16.343 (3), 10.647 (5) |
| β (°) | 95.499 (3) |
| V (Å3) | 1430.0 (8) |
| Z | 2 |
| Radiation type | Mo Kα |
| µ (mm−1) | 2.09 |
| Crystal size (mm) | 0.45 × 0.06 × 0.03 |
| |
| Data collection |
| Diffractometer | Nonius KappaCCD
diffractometer |
| Absorption correction | Part of the refinement model (ΔF)
(Sheldrick, 1990) |
| Tmin, Tmax | 0.760, 0.939 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 8087, 2918, 2295 |
| Rint | 0.038 |
| (sin θ/λ)max (Å−1) | 0.625 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.028, 0.063, 1.04 |
| No. of reflections | 2918 |
| No. of parameters | 190 |
| No. of restraints | 16 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.58, −0.66 |
Selected geometric parameters (Å, º) top| Ru—O1 | 1.7822 (6) | Mg—O5W | 2.047 (3) |
| Ru—Cl4 | 2.3464 (11) | Mg—O6W | 2.048 (2) |
| Ru—Cl1 | 2.3559 (9) | Mg—O2W | 2.051 (3) |
| Ru—Cl2 | 2.3660 (10) | Mg—O3W | 2.056 (3) |
| Ru—Cl5 | 2.3728 (9) | Mg—O4W | 2.065 (2) |
| Ru—Cl3 | 2.3731 (10) | Mg—O1W | 2.093 (2) |
| | | |
| O1—Ru—Cl4 | 178.14 (2) | O5W—Mg—O3W | 88.27 (11) |
| O1—Ru—Cl1 | 92.81 (3) | O2W—Mg—O3W | 92.14 (12) |
| O1—Ru—Cl5 | 91.68 (3) | O2W—Mg—O4W | 86.93 (10) |
| O1—Ru—Cl3 | 92.92 (2) | O5W—Mg—O6W | 86.60 (11) |
| O1—Ru—Cl2 | 92.22 (2) | Rui—O1—Ru | 180 |
| O4W—Mg—O1W | 178.25 (11) | | |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1W—H11W···Cl3 | 0.94 (3) | 2.37 (3) | 3.302 (3) | 172 (3) |
| O1W—H21W···O7W | 0.96 (3) | 1.73 (3) | 2.688 (4) | 174 (3) |
| O2W—H12W···Cl5 | 0.95 (3) | 2.25 (3) | 3.180 (3) | 164 (3) |
| O2W—H22W···O1Wii | 0.96 (2) | 2.03 (2) | 2.956 (3) | 162 (3) |
| O3W—H13W···Cl2iii | 0.95 (3) | 2.74 (3) | 3.415 (3) | 129 (2) |
| O3W—H13W···Cl3iv | 0.95 (3) | 2.65 (3) | 3.448 (4) | 142 (2) |
| O3W—H23W···Cl1iii | 0.97 (3) | 2.82 (3) | 3.349 (3) | 115 (2) |
| O4W—H14W···O8Wv | 0.97 (3) | 1.79 (3) | 2.745 (4) | 171 (3) |
| O4W—H24W···Cl3v | 0.96 (3) | 2.43 (3) | 3.363 (3) | 166 (3) |
| O5W—H15W···Cl2vi | 0.96 (3) | 2.46 (3) | 3.191 (3) | 133 (2) |
| O5W—H25W···Cl5vi | 0.955 (18) | 2.44 (3) | 3.237 (3) | 140 (3) |
| O6W—H16W···Cl2vii | 0.92 (2) | 2.44 (2) | 3.351 (3) | 171 (3) |
| O6W—H26W···O7Wii | 0.95 (2) | 1.83 (2) | 2.750 (3) | 164 (3) |
| O7W—H17W···O8W | 0.960 (18) | 1.831 (18) | 2.777 (3) | 168 (3) |
| O7W—H27W···Cl4vii | 0.96 (3) | 2.29 (3) | 3.220 (3) | 163 (3) |
| O8W—H28W···Cl1vii | 0.96 (3) | 2.32 (3) | 3.272 (3) | 176 (3) |
| O8W—H18W···Cl4viii | 0.98 (2) | 2.35 (3) | 3.313 (3) | 166 (2) |
| Symmetry codes: (ii) x, −y+1/2, z+1/2; (iii) −x+1, y−1/2, −z+1/2; (iv) x, −y+1/2, z−1/2; (v) −x, y−1/2, −z+1/2; (vi) x−1, −y+1/2, z−1/2; (vii) x−1, y, z; (viii) −x, −y+1, −z. |
 O and O—H
O and O—H Cl interactions.
Cl interactions.
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2007/01/00/bc3021/bc3021fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2007/01/00/bc3021/bc3021fig2thm.gif)




As part of an ongoing study of ruthenium materials in high oxidation states, the chemistry of ruthenium chlorides, fluorides and oxide fluorides is of interest. These materials are also of some value to the nuclear industry (Bourgeois & Cochet-Muchy, 1971).
The structure of Mg2Ru2Cl10O.16H2O presented here is the first example of a salt containing the RuIV oxychloride anion together with magnesium. Interestingly, few other derivatives of [Ru2Cl10O]4− are known. The only previous structure reports are for K4Ru2Cl10O (Deloume et al., 1979) and Cs4Ru2Cl10O (Santana Da Silva et al., 1999) where RuIV is present with anhydrous alkali ions. One goal of the present work was therefore to examine the nature of the interactions between the [Ru2Cl10O]4− anion and hydrated cations capable of forming hydrogen bonds.
The asymmetric unit contains one [Ru2Cl10O]4− anion, one [Mg(H2O)6]2+ cation and two water molecules (Fig. 1). The overall structure consists of layers stacked along the a direction, with [Ru2Cl10O]4− dimeric units bridging adjacent layers (Fig. 2). The Ru atom resides in a distorted octahedron of one O and five Cl atoms, with an average Ru—Cl distance of 2.362 Å, similar to that in K4Ru2Cl10O (2.362 Å), Cs4Ru2Cl10O (2.357 Å) and K4Ru2Cl10O·H2O (2.360 Å). As expected, the Ru—O distance compares well with those reported for Cs4Ru2Cl10O (1.791 Å), K4Ru2Cl10O (1.800 Å) and K4Ru2Cl10O·H2O (1.797 Å). In the latter case, the structure was first determined (Mathieson et al., 1952) from a zero-level Weissenberg photograph for the formula K4Ru2Cl10O. H2O. A subsequent fully anisotropic refinement (R = 0.034 for 245 measured reflections) has shown that there is, in fact, no water molecule in the structure (Deloume et al., 1979). The correct formula is therefore K4Ru2Cl10O.
The Mg2+ ion is surrounded by six water molecules via Mg2+—OH2 ion-dipole interactions. The Mg—O distances (average Mg—O = 2.057 Å) are longer than the values reported for magnesium bis(triazide) hexahydrate (average Mg—O = 2.024 Å; Mautner & Krischner, 1986), and similar to those in [Mg(H2O)6](C14H10Cl2NO2).22H2O (average Mg—O = 2.058 Å), where the Mg2+ ion is also hexahydrated (Castellari et al., 1999).
Two types of intermolecular interactions are present in the structure, including O—H···O hydrogen bonds and O—H···Cl dipole–dipole interactions that contribute to hold cations and anions together. The presence of water molecules in the structure results in the presence of additional hydrogen bonds (Table 2 and Fig. 3). Five O—H···O hydrogen bonds are formed by the two water molecules that ensure the cohesion between the [Mg(H2O)6]2+ ions located in the same layer. The environment of the [Ru2Cl10O]4− anion contains twelve O—H···Cl dipole–dipole interactions between the anion and the water molecules. Overall, the various interactions bridge the rutenate anions within and between layers and form cavities occupied by the [Mg(H2O)6]2+ cations.