The molecular structures of two salicylaldehyde thiosemicarbazone derivatives, namely salicylaldehyde 4-phenylthiosemicarbazone, C
14H
13N
3OS, (I), and 4-methoxysalicylaldehyde 4-phenylthiosemicarbazone, C
15H
15N
3O
2S, (II), both of potential pharmacological interest, are found in the keto (thione) tautomeric form. The first compound represents a second triclinic polymorph of composition β-C
14H
13N
3OS. Although both polymorphs crystallize in the same space group (
P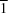
), the α-polymorph [Seena, Kurup & Suresh (2008).
J. Chem. Crystallogr. 38, 93–96] differs from the β form in its unit-cell volume at 293 K. The molecules in the crystal structures of (I) and (II) are linked into centrosymmetric
R22(8) dimers by hydrogen bonds of the N—H

S=C type. These dimers are connected through π–π stacking and T-shaped C—H

π interactions into three-dimensional networks.
Supporting information
CCDC references: 707220; 707221
Compound (I) was prepared by slight modification of the procedure described by
Purohit et al. (1989), using methanol as the solvent instead of
ethanol. White needle-like crystals were obtained directly from the reaction
mixture as the first form of compound (I). The structure of this α-polymorph,
obtained from ethanol, was recently published by Seena et al.
(2008).
The remaining mother liquor was kept at room temperature and after 2–3 weeks
yielded the β-polymorph of compound (I), in the form of transparent plates
[prismatic in CIF tables - please clarify]. The crystals were filtered off and
dried over KOH. From this batch were picked single crystals suitable for X-ray
diffraction.
Compound (II) was prepared by adopting a similar procedure as for (I).
Equimolar amounts of 4-phenylthiosemicarbazide and 4-methoxysalicylaldehyde
were dissolved in methanol and stirred until white needle-like crystals
started to precipitate (4–5 h). After standing overnight, the compound was
filtered off and dried over KOH [identification of this product?]. The mother
liquor was kept at room temperature and after 6 d yielded single crystals of
(II) suitable for X-ray diffraction. Elemental analysis, calculated for
C15H15N3O2S: C 59.78, H 5.02, N 13.94, S 10.64%; found: C 59.67, H
4.96, N 13.87, S 10.60%.
H atoms were constrained to ideal geometry using an appropriate riding model,
with C—H = 0.93–0.96 Å, N—H = 0.86 Å and O—H = 0.82 Å, and with
Uiso(H) = 1.2Ueq(C, N) or 1.5Ueq(O).
For both compounds, data collection: CrysAlis CCD (Oxford Diffraction, 2003); cell refinement: CrysAlis RED (Oxford Diffraction, 2003); data reduction: CrysAlis RED (Oxford Diffraction, 2003); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008); molecular graphics: ORTEP-3 (Farrugia, 1997) and Mercury (Macrae et al., 2006); software used to prepare material for publication: PLATON (Spek, 2003).
(I) salicylaldehyde 4-phenylthiosemicarbazone
top
Crystal data top
| C14H13N3OS | Z = 4 |
| Mr = 271.34 | F(000) = 568 |
| Triclinic, P1 | Dx = 1.345 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.6328 (3) Å | Cell parameters from 13724 reflections |
| b = 11.1114 (3) Å | θ = 3.8–34.8° |
| c = 12.8839 (10) Å | µ = 0.24 mm−1 |
| α = 75.251 (5)° | T = 293 K |
| β = 65.652 (2)° | Prismatic, colourless |
| γ = 81.539 (2)° | 0.79 × 0.59 × 0.20 mm |
| V = 1339.56 (12) Å3 | |
Data collection top
Oxford Diffraction Xcalibur CCD
diffractometer | 4676 independent reflections |
| Radiation source: fine-focus sealed tube | 4184 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.012 |
| ω scans | θmax = 25.0°, θmin = 3.8° |
Absorption correction: analytical
(CrysAlis RED; Oxford Diffraction, 2003) | h = −12→12 |
| Tmin = 0.856, Tmax = 0.936 | k = −13→13 |
| 14306 measured reflections | l = −13→15 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.031 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.079 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.038P)2 + 0.3482P]
where P = (Fo2 + 2Fc2)/3 |
| 4676 reflections | (Δ/σ)max = 0.001 |
| 345 parameters | Δρmax = 0.15 e Å−3 |
| 0 restraints | Δρmin = −0.20 e Å−3 |
Crystal data top
| C14H13N3OS | γ = 81.539 (2)° |
| Mr = 271.34 | V = 1339.56 (12) Å3 |
| Triclinic, P1 | Z = 4 |
| a = 10.6328 (3) Å | Mo Kα radiation |
| b = 11.1114 (3) Å | µ = 0.24 mm−1 |
| c = 12.8839 (10) Å | T = 293 K |
| α = 75.251 (5)° | 0.79 × 0.59 × 0.20 mm |
| β = 65.652 (2)° | |
Data collection top
Oxford Diffraction Xcalibur CCD
diffractometer | 4676 independent reflections |
Absorption correction: analytical
(CrysAlis RED; Oxford Diffraction, 2003) | 4184 reflections with I > 2σ(I) |
| Tmin = 0.856, Tmax = 0.936 | Rint = 0.012 |
| 14306 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.031 | 0 restraints |
| wR(F2) = 0.079 | H-atom parameters constrained |
| S = 1.03 | Δρmax = 0.15 e Å−3 |
| 4676 reflections | Δρmin = −0.20 e Å−3 |
| 345 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C11 | 0.57661 (13) | 0.79953 (12) | 0.39556 (11) | 0.0371 (3) | |
| C12 | 0.38367 (14) | 0.79827 (13) | 0.69151 (12) | 0.0396 (3) | |
| H12 | 0.4426 | 0.8482 | 0.6971 | 0.048* | |
| C13 | 0.26358 (14) | 0.75380 (13) | 0.79479 (11) | 0.0390 (3) | |
| C14 | 0.15581 (15) | 0.69642 (14) | 0.79333 (12) | 0.0448 (3) | |
| C15 | 0.04261 (17) | 0.65928 (16) | 0.89493 (14) | 0.0541 (4) | |
| H15 | −0.0289 | 0.6219 | 0.8929 | 0.065* | |
| C16 | 0.03510 (18) | 0.67723 (16) | 0.99887 (14) | 0.0579 (4) | |
| H16 | −0.0416 | 0.6522 | 1.0670 | 0.070* | |
| C17 | 0.14072 (19) | 0.73219 (17) | 1.00292 (13) | 0.0597 (4) | |
| H17 | 0.1360 | 0.7432 | 1.0736 | 0.072* | |
| C18 | 0.25273 (17) | 0.77046 (15) | 0.90198 (13) | 0.0501 (4) | |
| H18 | 0.3231 | 0.8084 | 0.9050 | 0.060* | |
| C19 | 0.52114 (14) | 0.70249 (13) | 0.26887 (12) | 0.0412 (3) | |
| C110 | 0.62569 (16) | 0.61911 (16) | 0.22067 (14) | 0.0546 (4) | |
| H110 | 0.6883 | 0.5852 | 0.2550 | 0.066* | |
| C111 | 0.6362 (2) | 0.58653 (17) | 0.12048 (16) | 0.0662 (5) | |
| H111 | 0.7069 | 0.5311 | 0.0867 | 0.079* | |
| C112 | 0.5424 (2) | 0.63580 (19) | 0.07068 (15) | 0.0674 (5) | |
| H112 | 0.5487 | 0.6123 | 0.0043 | 0.081* | |
| C113 | 0.43998 (19) | 0.71922 (18) | 0.11876 (14) | 0.0628 (5) | |
| H113 | 0.3772 | 0.7528 | 0.0845 | 0.075* | |
| C114 | 0.42906 (16) | 0.75399 (15) | 0.21795 (13) | 0.0487 (4) | |
| H114 | 0.3601 | 0.8116 | 0.2499 | 0.058* | |
| N11 | 0.49815 (12) | 0.72982 (12) | 0.37811 (10) | 0.0443 (3) | |
| H11N | 0.4276 | 0.6986 | 0.4378 | 0.053* | |
| N12 | 0.53128 (11) | 0.82126 (11) | 0.50405 (10) | 0.0406 (3) | |
| H12N | 0.5781 | 0.8673 | 0.5182 | 0.049* | |
| N13 | 0.41248 (11) | 0.77191 (11) | 0.59270 (9) | 0.0386 (3) | |
| O1 | 0.15616 (12) | 0.67825 (14) | 0.69303 (10) | 0.0675 (4) | |
| H1O | 0.2316 | 0.6950 | 0.6400 | 0.101* | |
| S1 | 0.72434 (4) | 0.86032 (4) | 0.29361 (3) | 0.04948 (12) | |
| C21 | −0.15678 (13) | 0.89188 (12) | 0.56879 (11) | 0.0363 (3) | |
| C22 | 0.00350 (13) | 0.67580 (12) | 0.39304 (11) | 0.0368 (3) | |
| H22 | −0.0584 | 0.6939 | 0.3565 | 0.044* | |
| C23 | 0.10949 (13) | 0.57819 (12) | 0.36235 (11) | 0.0342 (3) | |
| C24 | 0.21183 (13) | 0.54659 (12) | 0.40808 (11) | 0.0370 (3) | |
| C25 | 0.31164 (14) | 0.45392 (13) | 0.37349 (13) | 0.0460 (3) | |
| H25 | 0.3789 | 0.4328 | 0.4045 | 0.055* | |
| C26 | 0.31156 (16) | 0.39288 (14) | 0.29318 (14) | 0.0509 (4) | |
| H26 | 0.3801 | 0.3317 | 0.2691 | 0.061* | |
| C27 | 0.21119 (16) | 0.42124 (14) | 0.24788 (13) | 0.0496 (4) | |
| H27 | 0.2114 | 0.3790 | 0.1941 | 0.059* | |
| C28 | 0.11097 (15) | 0.51243 (13) | 0.28288 (12) | 0.0420 (3) | |
| H28 | 0.0426 | 0.5308 | 0.2530 | 0.050* | |
| C29 | −0.12202 (14) | 0.91015 (12) | 0.74222 (11) | 0.0371 (3) | |
| C210 | −0.25224 (16) | 0.89519 (15) | 0.82987 (14) | 0.0511 (4) | |
| H210 | −0.3211 | 0.8638 | 0.8188 | 0.061* | |
| C211 | −0.27949 (18) | 0.92724 (16) | 0.93415 (14) | 0.0595 (4) | |
| H211 | −0.3673 | 0.9178 | 0.9934 | 0.071* | |
| C212 | −0.1778 (2) | 0.97295 (16) | 0.95089 (14) | 0.0603 (4) | |
| H212 | −0.1965 | 0.9936 | 1.0215 | 0.072* | |
| C213 | −0.04887 (19) | 0.98809 (16) | 0.86358 (14) | 0.0576 (4) | |
| H213 | 0.0196 | 1.0199 | 0.8748 | 0.069* | |
| C214 | −0.01993 (16) | 0.95638 (13) | 0.75878 (13) | 0.0446 (3) | |
| H214 | 0.0679 | 0.9662 | 0.6998 | 0.054* | |
| N21 | −0.08594 (12) | 0.86846 (11) | 0.63721 (10) | 0.0408 (3) | |
| H21N | −0.0105 | 0.8234 | 0.6156 | 0.049* | |
| N22 | −0.11577 (11) | 0.82821 (10) | 0.48379 (10) | 0.0389 (3) | |
| H22N | −0.1579 | 0.8442 | 0.4370 | 0.047* | |
| N23 | −0.00950 (11) | 0.73849 (10) | 0.46782 (9) | 0.0351 (2) | |
| O2 | 0.21752 (10) | 0.60625 (11) | 0.48570 (9) | 0.0519 (3) | |
| H2O | 0.1509 | 0.6558 | 0.5029 | 0.078* | |
| S2 | −0.29168 (4) | 0.99553 (4) | 0.58034 (4) | 0.05216 (12) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C11 | 0.0352 (7) | 0.0416 (7) | 0.0377 (7) | 0.0042 (5) | −0.0193 (6) | −0.0084 (6) |
| C12 | 0.0394 (7) | 0.0455 (7) | 0.0396 (7) | 0.0028 (6) | −0.0211 (6) | −0.0118 (6) |
| C13 | 0.0423 (7) | 0.0418 (7) | 0.0363 (7) | 0.0027 (6) | −0.0190 (6) | −0.0109 (6) |
| C14 | 0.0456 (8) | 0.0543 (8) | 0.0408 (8) | 0.0003 (6) | −0.0205 (6) | −0.0165 (6) |
| C15 | 0.0502 (9) | 0.0627 (10) | 0.0512 (9) | −0.0130 (7) | −0.0178 (7) | −0.0128 (7) |
| C16 | 0.0625 (10) | 0.0623 (10) | 0.0401 (8) | −0.0157 (8) | −0.0114 (7) | −0.0036 (7) |
| C17 | 0.0745 (11) | 0.0728 (11) | 0.0341 (8) | −0.0144 (9) | −0.0196 (8) | −0.0119 (7) |
| C18 | 0.0578 (9) | 0.0592 (9) | 0.0406 (8) | −0.0110 (7) | −0.0228 (7) | −0.0124 (7) |
| C19 | 0.0390 (7) | 0.0468 (8) | 0.0372 (7) | −0.0094 (6) | −0.0097 (6) | −0.0129 (6) |
| C110 | 0.0465 (9) | 0.0595 (10) | 0.0556 (9) | 0.0001 (7) | −0.0144 (7) | −0.0195 (8) |
| C111 | 0.0679 (11) | 0.0625 (11) | 0.0577 (10) | −0.0073 (9) | −0.0030 (9) | −0.0301 (9) |
| C112 | 0.0834 (13) | 0.0781 (12) | 0.0431 (9) | −0.0297 (11) | −0.0133 (9) | −0.0223 (9) |
| C113 | 0.0672 (11) | 0.0811 (12) | 0.0471 (9) | −0.0179 (9) | −0.0267 (8) | −0.0098 (9) |
| C114 | 0.0471 (8) | 0.0547 (9) | 0.0451 (8) | −0.0045 (7) | −0.0175 (7) | −0.0118 (7) |
| N11 | 0.0367 (6) | 0.0599 (8) | 0.0368 (6) | −0.0097 (5) | −0.0094 (5) | −0.0159 (5) |
| N12 | 0.0352 (6) | 0.0530 (7) | 0.0374 (6) | −0.0032 (5) | −0.0160 (5) | −0.0125 (5) |
| N13 | 0.0345 (6) | 0.0473 (6) | 0.0354 (6) | 0.0035 (5) | −0.0160 (5) | −0.0103 (5) |
| O1 | 0.0509 (7) | 0.1168 (11) | 0.0488 (6) | −0.0156 (7) | −0.0181 (5) | −0.0371 (7) |
| S1 | 0.0430 (2) | 0.0681 (3) | 0.0388 (2) | −0.01540 (17) | −0.01489 (16) | −0.00862 (17) |
| C21 | 0.0359 (7) | 0.0354 (7) | 0.0393 (7) | −0.0017 (5) | −0.0146 (6) | −0.0110 (6) |
| C22 | 0.0367 (7) | 0.0406 (7) | 0.0353 (7) | 0.0003 (5) | −0.0161 (6) | −0.0098 (6) |
| C23 | 0.0336 (7) | 0.0355 (7) | 0.0306 (6) | −0.0030 (5) | −0.0094 (5) | −0.0071 (5) |
| C24 | 0.0341 (7) | 0.0416 (7) | 0.0345 (7) | −0.0051 (5) | −0.0107 (5) | −0.0097 (5) |
| C25 | 0.0369 (7) | 0.0462 (8) | 0.0543 (9) | 0.0028 (6) | −0.0192 (7) | −0.0103 (7) |
| C26 | 0.0451 (8) | 0.0424 (8) | 0.0606 (10) | 0.0066 (6) | −0.0140 (7) | −0.0198 (7) |
| C27 | 0.0550 (9) | 0.0468 (8) | 0.0494 (8) | 0.0019 (7) | −0.0166 (7) | −0.0235 (7) |
| C28 | 0.0453 (8) | 0.0451 (8) | 0.0399 (7) | −0.0010 (6) | −0.0189 (6) | −0.0131 (6) |
| C29 | 0.0416 (7) | 0.0354 (7) | 0.0367 (7) | 0.0003 (5) | −0.0170 (6) | −0.0100 (5) |
| C210 | 0.0453 (8) | 0.0578 (9) | 0.0524 (9) | −0.0088 (7) | −0.0131 (7) | −0.0227 (7) |
| C211 | 0.0588 (10) | 0.0633 (10) | 0.0464 (9) | −0.0089 (8) | −0.0037 (8) | −0.0205 (8) |
| C212 | 0.0838 (12) | 0.0606 (10) | 0.0424 (8) | −0.0094 (9) | −0.0242 (9) | −0.0183 (7) |
| C213 | 0.0693 (11) | 0.0644 (10) | 0.0520 (9) | −0.0166 (8) | −0.0310 (9) | −0.0144 (8) |
| C214 | 0.0475 (8) | 0.0465 (8) | 0.0411 (8) | −0.0095 (6) | −0.0188 (6) | −0.0050 (6) |
| N21 | 0.0361 (6) | 0.0501 (7) | 0.0426 (6) | 0.0081 (5) | −0.0187 (5) | −0.0209 (5) |
| N22 | 0.0399 (6) | 0.0418 (6) | 0.0402 (6) | 0.0079 (5) | −0.0207 (5) | −0.0147 (5) |
| N23 | 0.0331 (6) | 0.0366 (6) | 0.0342 (6) | 0.0010 (4) | −0.0115 (5) | −0.0098 (5) |
| O2 | 0.0424 (6) | 0.0719 (7) | 0.0555 (6) | 0.0082 (5) | −0.0258 (5) | −0.0326 (6) |
| S2 | 0.0526 (2) | 0.0511 (2) | 0.0677 (3) | 0.01951 (17) | −0.0352 (2) | −0.03039 (19) |
Geometric parameters (Å, º) top
| C11—N11 | 1.3341 (17) | C21—N21 | 1.3361 (17) |
| C11—N12 | 1.3515 (17) | C21—N22 | 1.3416 (17) |
| C11—S1 | 1.6758 (14) | C21—S2 | 1.6827 (13) |
| C12—N13 | 1.2816 (17) | C22—N23 | 1.2790 (17) |
| C12—C13 | 1.447 (2) | C22—C23 | 1.4470 (18) |
| C12—H12 | 0.9300 | C22—H22 | 0.9300 |
| C13—C18 | 1.3955 (19) | C23—C28 | 1.3953 (18) |
| C13—C14 | 1.400 (2) | C23—C24 | 1.4019 (18) |
| C14—O1 | 1.3563 (17) | C24—O2 | 1.3575 (16) |
| C14—C15 | 1.380 (2) | C24—C25 | 1.3810 (19) |
| C15—C16 | 1.372 (2) | C25—C26 | 1.374 (2) |
| C15—H15 | 0.9300 | C25—H25 | 0.9300 |
| C16—C17 | 1.380 (2) | C26—C27 | 1.378 (2) |
| C16—H16 | 0.9300 | C26—H26 | 0.9300 |
| C17—C18 | 1.371 (2) | C27—C28 | 1.373 (2) |
| C17—H17 | 0.9300 | C27—H27 | 0.9300 |
| C18—H18 | 0.9300 | C28—H28 | 0.9300 |
| C19—C114 | 1.375 (2) | C29—C214 | 1.3779 (19) |
| C19—C110 | 1.379 (2) | C29—C210 | 1.379 (2) |
| C19—N11 | 1.4300 (17) | C29—N21 | 1.4251 (17) |
| C110—C111 | 1.385 (2) | C210—C211 | 1.381 (2) |
| C110—H110 | 0.9300 | C210—H210 | 0.9300 |
| C111—C112 | 1.376 (3) | C211—C212 | 1.372 (2) |
| C111—H111 | 0.9300 | C211—H211 | 0.9300 |
| C112—C113 | 1.367 (3) | C212—C213 | 1.369 (2) |
| C112—H112 | 0.9300 | C212—H212 | 0.9300 |
| C113—C114 | 1.383 (2) | C213—C214 | 1.382 (2) |
| C113—H113 | 0.9300 | C213—H213 | 0.9300 |
| C114—H114 | 0.9300 | C214—H214 | 0.9300 |
| N11—H11N | 0.8600 | N21—H21N | 0.8600 |
| N12—N13 | 1.3770 (16) | N22—N23 | 1.3764 (15) |
| N12—H12N | 0.8600 | N22—H22N | 0.8600 |
| O1—H1O | 0.8200 | O2—H2O | 0.8200 |
| | | |
| N11—C11—N12 | 116.32 (12) | N21—C21—N22 | 117.08 (12) |
| N11—C11—S1 | 124.73 (10) | N21—C21—S2 | 125.10 (10) |
| N12—C11—S1 | 118.95 (10) | N22—C21—S2 | 117.82 (10) |
| N13—C12—C13 | 122.93 (13) | N23—C22—C23 | 123.47 (12) |
| N13—C12—H12 | 118.5 | N23—C22—H22 | 118.3 |
| C13—C12—H12 | 118.5 | C23—C22—H22 | 118.3 |
| C18—C13—C14 | 117.73 (13) | C28—C23—C24 | 118.09 (12) |
| C18—C13—C12 | 118.56 (13) | C28—C23—C22 | 118.70 (12) |
| C14—C13—C12 | 123.70 (12) | C24—C23—C22 | 123.22 (12) |
| O1—C14—C15 | 117.54 (13) | O2—C24—C25 | 118.02 (12) |
| O1—C14—C13 | 121.92 (13) | O2—C24—C23 | 121.65 (12) |
| C15—C14—C13 | 120.51 (13) | C25—C24—C23 | 120.32 (12) |
| C16—C15—C14 | 120.26 (15) | C26—C25—C24 | 119.99 (13) |
| C16—C15—H15 | 119.9 | C26—C25—H25 | 120.0 |
| C14—C15—H15 | 119.9 | C24—C25—H25 | 120.0 |
| C15—C16—C17 | 120.37 (15) | C25—C26—C27 | 120.82 (14) |
| C15—C16—H16 | 119.8 | C25—C26—H26 | 119.6 |
| C17—C16—H16 | 119.8 | C27—C26—H26 | 119.6 |
| C18—C17—C16 | 119.56 (14) | C28—C27—C26 | 119.39 (14) |
| C18—C17—H17 | 120.2 | C28—C27—H27 | 120.3 |
| C16—C17—H17 | 120.2 | C26—C27—H27 | 120.3 |
| C17—C18—C13 | 121.54 (14) | C27—C28—C23 | 121.38 (13) |
| C17—C18—H18 | 119.2 | C27—C28—H28 | 119.3 |
| C13—C18—H18 | 119.2 | C23—C28—H28 | 119.3 |
| C114—C19—C110 | 120.78 (14) | C214—C29—C210 | 120.21 (13) |
| C114—C19—N11 | 117.98 (13) | C214—C29—N21 | 118.24 (12) |
| C110—C19—N11 | 121.04 (13) | C210—C29—N21 | 121.32 (12) |
| C19—C110—C111 | 119.08 (16) | C29—C210—C211 | 119.46 (14) |
| C19—C110—H110 | 120.5 | C29—C210—H210 | 120.3 |
| C111—C110—H110 | 120.5 | C211—C210—H210 | 120.3 |
| C112—C111—C110 | 120.31 (17) | C212—C211—C210 | 120.38 (15) |
| C112—C111—H111 | 119.8 | C212—C211—H211 | 119.8 |
| C110—C111—H111 | 119.8 | C210—C211—H211 | 119.8 |
| C113—C112—C111 | 120.00 (16) | C213—C212—C211 | 119.98 (15) |
| C113—C112—H112 | 120.0 | C213—C212—H212 | 120.0 |
| C111—C112—H112 | 120.0 | C211—C212—H212 | 120.0 |
| C112—C113—C114 | 120.44 (17) | C212—C213—C214 | 120.34 (15) |
| C112—C113—H113 | 119.8 | C212—C213—H213 | 119.8 |
| C114—C113—H113 | 119.8 | C214—C213—H213 | 119.8 |
| C19—C114—C113 | 119.36 (15) | C29—C214—C213 | 119.61 (14) |
| C19—C114—H114 | 120.3 | C29—C214—H214 | 120.2 |
| C113—C114—H114 | 120.3 | C213—C214—H214 | 120.2 |
| C11—N11—C19 | 125.71 (12) | C21—N21—C29 | 126.94 (11) |
| C11—N11—H11N | 117.1 | C21—N21—H21N | 116.5 |
| C19—N11—H11N | 117.1 | C29—N21—H21N | 116.5 |
| C11—N12—N13 | 122.03 (11) | C21—N22—N23 | 123.14 (11) |
| C11—N12—H12N | 119.0 | C21—N22—H22N | 118.4 |
| N13—N12—H12N | 119.0 | N23—N22—H22N | 118.4 |
| C12—N13—N12 | 115.18 (11) | C22—N23—N22 | 114.62 (11) |
| C14—O1—H1O | 109.5 | C24—O2—H2O | 109.5 |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N11—H11N···O2 | 0.86 | 2.37 | 3.0881 (16) | 141 |
| O1—H1O···N13 | 0.82 | 2.01 | 2.7252 (16) | 145 |
| N21—H21N···O1 | 0.86 | 2.56 | 3.2806 (18) | 142 |
| O2—H2O···N23 | 0.82 | 1.98 | 2.6984 (14) | 146 |
| N12—H12N···S2i | 0.86 | 2.58 | 3.4391 (12) | 175 |
| N22—H22N···S1ii | 0.86 | 2.59 | 3.4398 (11) | 172 |
| Symmetry codes: (i) x+1, y, z; (ii) x−1, y, z. |
(II) 4-methoxysalicylaldehyde 4-phenylthiosemicarbazone
top
Crystal data top
| C15H15N3O2S | Z = 2 |
| Mr = 301.37 | F(000) = 316 |
| Triclinic, P1 | Dx = 1.353 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 4.73851 (17) Å | Cell parameters from 6731 reflections |
| b = 11.3244 (4) Å | θ = 3.7–33.8° |
| c = 14.0775 (5) Å | µ = 0.23 mm−1 |
| α = 78.390 (3)° | T = 293 K |
| β = 89.547 (3)° | Prismatic, colourless |
| γ = 89.364 (3)° | 0.40 × 0.35 × 0.30 mm |
| V = 739.90 (5) Å3 | |
Data collection top
Oxford Diffraction Xcalibur CCD
diffractometer | 2230 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.015 |
| Graphite monochromator | θmax = 25.0°, θmin = 3.8° |
| Detector resolution: 16.3426 pixels mm-1 | h = −5→5 |
| ω scans | k = −13→13 |
| 8408 measured reflections | l = −16→16 |
| 2591 independent reflections | |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.030 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.087 | H-atom parameters constrained |
| S = 1.08 | w = 1/[σ2(Fo2) + (0.0475P)2 + 0.1004P]
where P = (Fo2 + 2Fc2)/3 |
| 2591 reflections | (Δ/σ)max < 0.001 |
| 192 parameters | Δρmax = 0.16 e Å−3 |
| 0 restraints | Δρmin = −0.17 e Å−3 |
Crystal data top
| C15H15N3O2S | γ = 89.364 (3)° |
| Mr = 301.37 | V = 739.90 (5) Å3 |
| Triclinic, P1 | Z = 2 |
| a = 4.73851 (17) Å | Mo Kα radiation |
| b = 11.3244 (4) Å | µ = 0.23 mm−1 |
| c = 14.0775 (5) Å | T = 293 K |
| α = 78.390 (3)° | 0.40 × 0.35 × 0.30 mm |
| β = 89.547 (3)° | |
Data collection top
Oxford Diffraction Xcalibur CCD
diffractometer | 2230 reflections with I > 2σ(I) |
| 8408 measured reflections | Rint = 0.015 |
| 2591 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.030 | 0 restraints |
| wR(F2) = 0.087 | H-atom parameters constrained |
| S = 1.08 | Δρmax = 0.16 e Å−3 |
| 2591 reflections | Δρmin = −0.17 e Å−3 |
| 192 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.5985 (3) | 0.07055 (12) | 0.32843 (9) | 0.0415 (3) | |
| C2 | 1.0970 (3) | −0.16180 (12) | 0.39226 (10) | 0.0426 (3) | |
| H2 | 1.0946 | −0.1625 | 0.4584 | 0.051* | |
| C3 | 1.2862 (3) | −0.24382 (11) | 0.35530 (10) | 0.0391 (3) | |
| C4 | 1.3019 (3) | −0.25023 (12) | 0.25711 (10) | 0.0437 (3) | |
| C5 | 1.4833 (3) | −0.33096 (13) | 0.22599 (10) | 0.0504 (4) | |
| H5 | 1.4925 | −0.3340 | 0.1605 | 0.060* | |
| C6 | 1.6512 (3) | −0.40717 (12) | 0.29157 (11) | 0.0454 (3) | |
| C7 | 1.6416 (3) | −0.40270 (12) | 0.38908 (11) | 0.0463 (3) | |
| H7 | 1.7551 | −0.4537 | 0.4334 | 0.056* | |
| C8 | 1.4602 (3) | −0.32108 (12) | 0.41937 (10) | 0.0445 (3) | |
| H8 | 1.4541 | −0.3177 | 0.4848 | 0.053* | |
| C9 | 0.4741 (3) | 0.16797 (14) | 0.16067 (10) | 0.0533 (4) | |
| C10 | 0.4706 (4) | 0.28905 (17) | 0.15462 (13) | 0.0741 (5) | |
| H10 | 0.5715 | 0.3230 | 0.1986 | 0.089* | |
| C11 | 0.3151 (6) | 0.3616 (2) | 0.08225 (17) | 0.1031 (8) | |
| H11 | 0.3100 | 0.4445 | 0.0786 | 0.124* | |
| C12 | 0.1710 (6) | 0.3140 (3) | 0.01704 (17) | 0.1147 (11) | |
| H12 | 0.0684 | 0.3637 | −0.0315 | 0.138* | |
| C13 | 0.1767 (5) | 0.1937 (3) | 0.02275 (16) | 0.1095 (9) | |
| H13 | 0.0779 | 0.1608 | −0.0224 | 0.131* | |
| C14 | 0.3273 (5) | 0.1185 (2) | 0.09482 (13) | 0.0801 (5) | |
| H14 | 0.3291 | 0.0355 | 0.0986 | 0.096* | |
| C15 | 1.9818 (4) | −0.57121 (15) | 0.31744 (14) | 0.0673 (5) | |
| H15A | 2.1107 | −0.5307 | 0.3520 | 0.101* | |
| H15B | 2.0859 | −0.6204 | 0.2810 | 0.101* | |
| H15C | 1.8588 | −0.6211 | 0.3627 | 0.101* | |
| N1 | 0.6337 (3) | 0.08887 (13) | 0.23256 (9) | 0.0638 (4) | |
| H1N | 0.7673 | 0.0485 | 0.2118 | 0.077* | |
| N2 | 0.7574 (2) | −0.01767 (10) | 0.38096 (8) | 0.0444 (3) | |
| H2N | 0.7494 | −0.0302 | 0.4432 | 0.053* | |
| N3 | 0.9333 (2) | −0.08853 (10) | 0.33659 (8) | 0.0436 (3) | |
| O1 | 1.1397 (3) | −0.17907 (11) | 0.18946 (7) | 0.0654 (3) | |
| H1O | 1.0391 | −0.1355 | 0.2157 | 0.098* | |
| O2 | 1.8185 (2) | −0.48431 (10) | 0.25302 (8) | 0.0623 (3) | |
| S1 | 0.37321 (8) | 0.14895 (3) | 0.38499 (3) | 0.05153 (15) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1 | 0.0420 (7) | 0.0420 (7) | 0.0416 (7) | 0.0050 (6) | 0.0008 (5) | −0.0114 (6) |
| C2 | 0.0418 (7) | 0.0435 (7) | 0.0431 (7) | 0.0055 (6) | 0.0023 (6) | −0.0103 (6) |
| C3 | 0.0365 (7) | 0.0367 (7) | 0.0447 (7) | 0.0003 (5) | 0.0032 (5) | −0.0098 (5) |
| C4 | 0.0455 (7) | 0.0412 (7) | 0.0429 (7) | 0.0056 (6) | 0.0012 (6) | −0.0054 (6) |
| C5 | 0.0577 (9) | 0.0515 (8) | 0.0429 (8) | 0.0089 (7) | 0.0071 (6) | −0.0124 (6) |
| C6 | 0.0415 (7) | 0.0391 (7) | 0.0570 (8) | 0.0027 (6) | 0.0083 (6) | −0.0136 (6) |
| C7 | 0.0425 (7) | 0.0418 (7) | 0.0542 (8) | 0.0076 (6) | −0.0038 (6) | −0.0089 (6) |
| C8 | 0.0460 (7) | 0.0455 (8) | 0.0430 (7) | 0.0051 (6) | −0.0014 (6) | −0.0118 (6) |
| C9 | 0.0574 (9) | 0.0637 (10) | 0.0365 (7) | 0.0174 (7) | 0.0066 (6) | −0.0062 (6) |
| C10 | 0.0951 (13) | 0.0663 (11) | 0.0599 (10) | 0.0216 (10) | 0.0044 (9) | −0.0119 (8) |
| C11 | 0.145 (2) | 0.0838 (15) | 0.0679 (14) | 0.0565 (15) | 0.0207 (14) | 0.0094 (11) |
| C12 | 0.1098 (19) | 0.161 (3) | 0.0539 (13) | 0.0687 (19) | 0.0109 (12) | 0.0215 (15) |
| C13 | 0.0959 (17) | 0.176 (3) | 0.0514 (12) | 0.0026 (18) | −0.0183 (11) | −0.0108 (15) |
| C14 | 0.0984 (14) | 0.0874 (14) | 0.0524 (10) | −0.0047 (11) | −0.0009 (10) | −0.0086 (9) |
| C15 | 0.0602 (10) | 0.0570 (10) | 0.0842 (12) | 0.0223 (8) | 0.0091 (9) | −0.0144 (9) |
| N1 | 0.0761 (9) | 0.0729 (9) | 0.0407 (7) | 0.0394 (7) | 0.0047 (6) | −0.0097 (6) |
| N2 | 0.0461 (6) | 0.0470 (7) | 0.0403 (6) | 0.0141 (5) | 0.0034 (5) | −0.0100 (5) |
| N3 | 0.0427 (6) | 0.0422 (6) | 0.0467 (6) | 0.0085 (5) | 0.0049 (5) | −0.0117 (5) |
| O1 | 0.0807 (8) | 0.0710 (7) | 0.0427 (6) | 0.0338 (6) | −0.0051 (5) | −0.0086 (5) |
| O2 | 0.0645 (7) | 0.0565 (6) | 0.0679 (7) | 0.0223 (5) | 0.0098 (5) | −0.0188 (5) |
| S1 | 0.0608 (2) | 0.0498 (2) | 0.0441 (2) | 0.02026 (17) | 0.00687 (16) | −0.01125 (16) |
Geometric parameters (Å, º) top
| C1—N1 | 1.3331 (18) | C9—N1 | 1.4249 (19) |
| C1—N2 | 1.3441 (17) | C10—C11 | 1.385 (3) |
| C1—S1 | 1.6768 (13) | C10—H10 | 0.9300 |
| C2—N3 | 1.2797 (17) | C11—C12 | 1.348 (4) |
| C2—C3 | 1.4514 (17) | C11—H11 | 0.9300 |
| C2—H2 | 0.9300 | C12—C13 | 1.348 (4) |
| C3—C8 | 1.3916 (18) | C12—H12 | 0.9300 |
| C3—C4 | 1.4000 (19) | C13—C14 | 1.383 (3) |
| C4—O1 | 1.3551 (17) | C13—H13 | 0.9300 |
| C4—C5 | 1.3802 (19) | C14—H14 | 0.9300 |
| C5—C6 | 1.381 (2) | C15—O2 | 1.423 (2) |
| C5—H5 | 0.9300 | C15—H15A | 0.9600 |
| C6—O2 | 1.3618 (16) | C15—H15B | 0.9600 |
| C6—C7 | 1.384 (2) | C15—H15C | 0.9600 |
| C7—C8 | 1.3823 (19) | N1—H1N | 0.8600 |
| C7—H7 | 0.9300 | N2—N3 | 1.3812 (15) |
| C8—H8 | 0.9300 | N2—H2N | 0.8600 |
| C9—C10 | 1.356 (2) | O1—H1O | 0.8200 |
| C9—C14 | 1.373 (2) | | |
| | | |
| N1—C1—N2 | 115.97 (12) | C11—C10—H10 | 120.3 |
| N1—C1—S1 | 124.46 (10) | C12—C11—C10 | 121.0 (2) |
| N2—C1—S1 | 119.57 (10) | C12—C11—H11 | 119.5 |
| N3—C2—C3 | 122.02 (12) | C10—C11—H11 | 119.5 |
| N3—C2—H2 | 119.0 | C11—C12—C13 | 119.5 (2) |
| C3—C2—H2 | 119.0 | C11—C12—H12 | 120.2 |
| C8—C3—C4 | 117.58 (12) | C13—C12—H12 | 120.2 |
| C8—C3—C2 | 119.39 (12) | C12—C13—C14 | 120.9 (2) |
| C4—C3—C2 | 123.03 (12) | C12—C13—H13 | 119.5 |
| O1—C4—C5 | 117.53 (12) | C14—C13—H13 | 119.5 |
| O1—C4—C3 | 121.81 (12) | C9—C14—C13 | 119.1 (2) |
| C5—C4—C3 | 120.66 (13) | C9—C14—H14 | 120.4 |
| C4—C5—C6 | 120.32 (13) | C13—C14—H14 | 120.4 |
| C4—C5—H5 | 119.8 | O2—C15—H15A | 109.5 |
| C6—C5—H5 | 119.8 | O2—C15—H15B | 109.5 |
| O2—C6—C5 | 115.28 (13) | H15A—C15—H15B | 109.5 |
| O2—C6—C7 | 124.30 (13) | O2—C15—H15C | 109.5 |
| C5—C6—C7 | 120.42 (12) | H15A—C15—H15C | 109.5 |
| C8—C7—C6 | 118.76 (13) | H15B—C15—H15C | 109.5 |
| C8—C7—H7 | 120.6 | C1—N1—C9 | 127.14 (12) |
| C6—C7—H7 | 120.6 | C1—N1—H1N | 116.4 |
| C7—C8—C3 | 122.26 (13) | C9—N1—H1N | 116.4 |
| C7—C8—H8 | 118.9 | C1—N2—N3 | 121.11 (11) |
| C3—C8—H8 | 118.9 | C1—N2—H2N | 119.4 |
| C10—C9—C14 | 120.08 (16) | N3—N2—H2N | 119.4 |
| C10—C9—N1 | 121.90 (15) | C2—N3—N2 | 116.46 (11) |
| C14—C9—N1 | 118.00 (16) | C4—O1—H1O | 109.5 |
| C9—C10—C11 | 119.3 (2) | C6—O2—C15 | 118.18 (13) |
| C9—C10—H10 | 120.3 | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1O···N3 | 0.82 | 1.94 | 2.6642 (15) | 146 |
| C13—H13···O1i | 0.93 | 2.54 | 3.387 (2) | 151 |
| C14—H14···O1ii | 0.93 | 2.67 | 3.487 (3) | 147 |
| N2—H2N···S1iii | 0.86 | 2.58 | 3.3862 (12) | 156 |
| Symmetry codes: (i) −x+1, −y, −z; (ii) x−1, y, z; (iii) −x+1, −y, −z+1. |
Experimental details
| | (I) | (II) |
| Crystal data |
| Chemical formula | C14H13N3OS | C15H15N3O2S |
| Mr | 271.34 | 301.37 |
| Crystal system, space group | Triclinic, P1 | Triclinic, P1 |
| Temperature (K) | 293 | 293 |
| a, b, c (Å) | 10.6328 (3), 11.1114 (3), 12.8839 (10) | 4.73851 (17), 11.3244 (4), 14.0775 (5) |
| α, β, γ (°) | 75.251 (5), 65.652 (2), 81.539 (2) | 78.390 (3), 89.547 (3), 89.364 (3) |
| V (Å3) | 1339.56 (12) | 739.90 (5) |
| Z | 4 | 2 |
| Radiation type | Mo Kα | Mo Kα |
| µ (mm−1) | 0.24 | 0.23 |
| Crystal size (mm) | 0.79 × 0.59 × 0.20 | 0.40 × 0.35 × 0.30 |
| |
| Data collection |
| Diffractometer | Oxford Diffraction Xcalibur CCD
diffractometer | Oxford Diffraction Xcalibur CCD
diffractometer |
| Absorption correction | Analytical
(CrysAlis RED; Oxford Diffraction, 2003) | – |
| Tmin, Tmax | 0.856, 0.936 | – |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 14306, 4676, 4184 | 8408, 2591, 2230 |
| Rint | 0.012 | 0.015 |
| (sin θ/λ)max (Å−1) | 0.595 | 0.595 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.031, 0.079, 1.03 | 0.030, 0.087, 1.08 |
| No. of reflections | 4676 | 2591 |
| No. of parameters | 345 | 192 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.15, −0.20 | 0.16, −0.17 |
Selected bond lengths (Å) for (I) top| C11—N11 | 1.3341 (17) | C21—N21 | 1.3361 (17) |
| C11—N12 | 1.3515 (17) | C21—N22 | 1.3416 (17) |
| C11—S1 | 1.6758 (14) | C21—S2 | 1.6827 (13) |
| C12—N13 | 1.2816 (17) | C22—N23 | 1.2790 (17) |
| C12—C13 | 1.447 (2) | C22—C23 | 1.4470 (18) |
| C13—C14 | 1.400 (2) | C23—C24 | 1.4019 (18) |
| C14—O1 | 1.3563 (17) | C24—O2 | 1.3575 (16) |
| C19—N11 | 1.4300 (17) | C29—N21 | 1.4251 (17) |
| N12—N13 | 1.3770 (16) | N22—N23 | 1.3764 (15) |
Hydrogen-bond geometry (Å, º) for (I) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N11—H11N···O2 | 0.86 | 2.37 | 3.0881 (16) | 140.7 |
| O1—H1O···N13 | 0.82 | 2.01 | 2.7252 (16) | 144.9 |
| N21—H21N···O1 | 0.86 | 2.56 | 3.2806 (18) | 142.4 |
| O2—H2O···N23 | 0.82 | 1.98 | 2.6984 (14) | 145.9 |
| N12—H12N···S2i | 0.86 | 2.58 | 3.4391 (12) | 174.8 |
| N22—H22N···S1ii | 0.86 | 2.59 | 3.4398 (11) | 172.0 |
| Symmetry codes: (i) x+1, y, z; (ii) x−1, y, z. |
Selected bond lengths (Å) for (II) top| C1—N1 | 1.3331 (18) | C3—C4 | 1.4000 (19) |
| C1—N2 | 1.3441 (17) | C4—O1 | 1.3551 (17) |
| C1—S1 | 1.6768 (13) | C9—N1 | 1.4249 (19) |
| C2—N3 | 1.2797 (17) | N2—N3 | 1.3812 (15) |
| C2—C3 | 1.4514 (17) | | |
Hydrogen-bond geometry (Å, º) for (II) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1O···N3 | 0.82 | 1.94 | 2.6642 (15) | 146 |
| C13—H13···O1i | 0.93 | 2.54 | 3.387 (2) | 151 |
| C14—H14···O1ii | 0.93 | 2.67 | 3.487 (3) | 147 |
| N2—H2N···S1iii | 0.86 | 2.58 | 3.3862 (12) | 156 |
| Symmetry codes: (i) −x+1, −y, −z; (ii) x−1, y, z; (iii) −x+1, −y, −z+1. |
 S=C type. These dimers are connected through π–π stacking and T-shaped C—H
S=C type. These dimers are connected through π–π stacking and T-shaped C—H π interactions into three-dimensional networks.
π interactions into three-dimensional networks.
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2008/10/00/av3157/av3157fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2008/10/00/av3157/av3157fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2008/10/00/av3157/av3157fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/c/issues/2008/10/00/av3157/av3157fig4thm.gif)
![[Figure 5]](http://journals.iucr.org/c/issues/2008/10/00/av3157/av3157fig5thm.gif)




Thiosemicarbazones are a class of organic molecules which belong to a large family of thiourea derivatives (Casas et al., 2000). Widespread interest in the chemistry of thiosemicarbazones is associated with their broad spectrum of biological activities with potential uses in antibacterial, antiviral and antitumour treatments (Bharti et al., 2002; Smee & Sidwell, 2003; Hu et al., 2006; Oliveira et al., 2007). Many of their properties are a function of the parent aldehyde or ketone, and these properties can therefore be elegantly tuned by the appropriate choice of parent. From a structural perspective, thiosemicarbazones attract attention as interesting ligands due to their tendency to undergo tautomerism and form planar highly rigid Schiff bases capable of imposing a variety of mixed-donor coordination environments about metal cations. Thiosemicarbazones derived from salicylaldehyde can exist in several tautomeric forms but the most interesting are those denoted A and B in scheme 1. Such molecular isomerization can result in different binding modes. In form A, the molecule can act as a monoanionic ligand after losing the H atom from the hydroxyl group, or as a dianionic ligand after losing H atoms from the mercapto and hydroxyl groups of form B.
We present here the crystal structures of two closely related thiosemicarbazones derived from salicylaldehyde: salicylaldehyde 4-phenylthiosemicarbazone, (I), and 4-methoxysalicylaldehyde 4-phenylthiosemicarbazone, (II). The structure of (I) has been reported previously (Seena et al., 2008). The compound crystallized in the same space group (P1) and we refer to it as the α-polymorph (obtained from ethanol). We have now obtained a second triclinic polymorph of (I), denoted the β-polymorph (obtained from methanol). Although both polymorphs crystallize in space group No. 2, they have different unit-cell volumes at 293 K and their unit-cell parameters are different [in particular, V = 2002.1 (5) Å3 for the α-polymorph and V = 1339.6 (1) Å3 for the β-polymorph]. These two forms of (I) have different numbers of molecules in the asymmetric unit (three and two, respectively). The asymmetric unit of (I) consists of two crystallographically independent molecules, (Ia) and (Ib) (Fig. 1). The molecular structures of (I) (in both polymorphs) and (II) (Fig. 2) are very similiar, but not equal. The crystal structures are characterized by a great variety of interactions, especially of the N—H···S type, and these will be discussed later.
The atom-labelling schemes are almost the same. In the following discussion, the order of the compounds for a given value is (Ia) and (Ib) of (I), then (II). Selected bond lengths and bond angles for (I) and (II) are given in Tables 1 and 3. Similar to many uncomplexed and unprotonated thiosemicarbazones (Soriano-García et al., 1986; Dinçer et al., 2005; Seena et al., 2008), a trans configuration of atom S1 with respect to atom N3 is observed [S1—C1—N2—N3 = -178.51 (10), -178.04 (10) and 176.30 (9)°] and atom N1 is in a cis configuration with respect to atom N3. The C1—S1 bond distance of 1.6758 (14) Å [1.6827 (13) and 1.6768 (13) Å] does not differ significantly from that found in thiourea (Truter, 1967). This bond, together with the C1—N2 bond of 1.3515 (17) Å [1.3416 (17) and 1.3441 (17) Å] and the C1—N1 bond of 1.3341 (17) Å [1.3361 (17) and 1.3331 (18) Å], indicate that the molecules in the crystal structures of (I) and (II) are in the keto (thione) form. This is also in agreement with the absence of a strong band in the IR spectra centred at 2500 cm-1 [ν(S—H) stretching mode].
The molecular structures of (I) and (II) consist of three structural fragments: salicyl and phenyl parts separated by the central thiosemicarbazone part. The salicyl–thiosemicarbazone parts of molecules are almost planar. Maximum deviations from the plane in molecule (Ia) are -0.214 (1), 0.183 (1) and -0.138 (2) Å for N11, O1 and C17, respectively [-0.192 (1), 0.107 (1) and 0.097 (1) Å for N21, N22 and O2 in (Ib) and 0.285 (2), -0.140 (2), -0.092 (1) Å for N1, C15 and C1 in (II)]. The dihedral angle between the planes of the salicyl–thiosemicarbazone parts and the phenyl ring on the N1 substituents of 64.44 (4)° [and 53.07 (4)° and 52.16 (5)°] indicates non-planarity of the ligands, although the thiosemicarbazone parts themselves are planar. Such an arrangement allows delocalization of the π-electrons in the N1—C1—N2—N3—C2 moieties. The salicylaldimine parts of the molecules in the crystal structures of (I) and (II) are characterized by a six-membered pseudoaromatic rings(N3—C2—C3—C4—O1—H1O). Rings are formed by an intramolecular hydrogen bond [S(6)] which is enhanced by π-electron delocalization as can be seen easily from the bond lengths within the rings (Tables 1 and 3). Such resonance-assisted hydrogen bonds seem to be the general features of the crystal structures of thiosemicarbazones and Schiff bases derived from salicylaldehyde (Soriano-García et al., 1986; Popović et al., 2002). The C4—O1 bond lengths are comparable with those found in phenols: 1.3563 (17), 1.3575 (16) and 1.3551 (17) Å (Allen et al., 1987). In contrast, C2—N3 [1.2816 (17), 1.2790 (17), 1.2797 (17) Å] is assigned as a double bond. The endocyclic bond distances and angles found in the phenyl rings, with the exception of that at C3, do not differ from those in normal sp2 hybridized moieties. The measured collapse of the C4—C3—C8 angles together with good electron donor properties of the O atom may be a consequence of the conjugation of the phenyl ring with the thiosemicarbazone parts of the molecules.
The crystal packings are characterized by a great variety of hydrogen bonds (presented in Tables 2 and 4). A close inspection of the unit cells (Figs. 3 and 4) and two-dimensional fingerprint plots derived from Hirshfeld surfaces (McKinnon et al., 2004) of both polymorphs of (I) and (II) revealed very specific N—H···S hydrogen bonding (spikes centred around 2.6 Å). This type of structural motif [denoted R22(8); Etter et al., 1990] is very common in the crystal structures of thiosemicarbazones (Soriano-García et al., 1986; Sampath et al., 2003; Dinçer et al., 2005).
Figs. 5(a) and 5(b) unambiguously show that different intermolecular interactions are present in the polymorphs α and β, which results in different packing modes. In polymorph α, the shortest contacts corespond to the very close H···H contacts [H25···H25i and H3···H27ii are 2.25 (4) and 2.25 (6) Å; symmetry codes: (i) -x, -y, 2 - z ; (ii) 1 - x, -y, 1 - z]. In plots of (I) and (II) (Figs. 5b and 5c) `wings' are more pronounced, which resemble [indicate?] aromatic interactions [π···π stacking, C14···C214 and C114···C24 distances are 3.237 (2) and 3.267 (2) Å in (I)] and there are T-shaped C—H···π contacts between the molecules in (II) [C12···H5iii and C10···H15Biv = 2.93 and 2.86 Å; symmetry codes: (iii) 2 - x, -y, -z ; (iv) -2 + x, 1 + y, z)].