
The carboxylic acid group is an example of a functional group which possess a good hydrogen-bond donor (–OH) and acceptor (C=O). For this reason, carboxylic acids have a tendency to self-assembly by the formation of hydrogen bonds between the donor and acceptor sites. We present here the crystal structure of
N-tosyl-
L-proline (TPOH) benzene hemisolvate {systematic name: (2
S)-1-[(4-methylbenzene)sulfonyl]pyrrolidine-2-carboxylic acid benzene hemisolvate}, C
12H
15NO
4S·0.5C
6H
6, (I), in which a cyclic
R22(8) hydrogen-bonded carboxylic acid dimer with a strong O—(
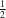
H)

(
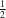
H)—O hydrogen bond is observed. The compound was characterized by single-crystal X-ray diffraction and NMR spectroscopy, and crystallizes in the space group
I2 with half a benzene molecule and one TPOH molecule in the asymmetric unit. The H atom of the carboxyl OH group is disordered over a twofold axis. An analysis of the intermolecular interactions using the noncovalent interaction (NCI) index showed that the TPOH molecules form dimers due to the strong O—(
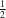
H)

(
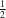
H)—O hydrogen bond, while the packing of the benzene solvent molecules is governed by weak dispersive interactions. A search of the Cambridge Structural Database revealed that the disordered dimeric motif observed in (I) was found previously only in six crystal structures.
Supporting information
CCDC reference: 1921956
Data collection: CrysAlis PRO (Rigaku OD, 2015); cell refinement: CrysAlis PRO (Rigaku OD, 2015); data reduction: CrysAlis PRO (Rigaku OD, 2015); program(s) used to solve structure: SHELXT2014 (Sheldrick, 2015a); program(s) used to refine structure: SHELXL2018 (Sheldrick, 2015b); molecular graphics: Mercury (Macrae et al., 2008); software used to prepare material for publication: SHELXL2018 (Sheldrick, 2015b).
(2
S)-1-[(4-Methylbenzene)sulfonyl]pyrrolidine-2-carboxylic acid
benzene hemisolvate
top
Crystal data top
| C12H15NO4S·0.5C6H6 | F(000) = 652 |
| Mr = 308.36 | Dx = 1.362 Mg m−3 |
| Monoclinic, I2 | Cu Kα radiation, λ = 1.54184 Å |
| a = 7.6965 (1) Å | Cell parameters from 7445 reflections |
| b = 10.9470 (2) Å | θ = 4.7–74.5° |
| c = 18.0806 (3) Å | µ = 2.05 mm−1 |
| β = 99.098 (2)° | T = 130 K |
| V = 1504.19 (4) Å3 | Block, colourless |
| Z = 4 | 0.20 × 0.10 × 0.06 mm |
Data collection top
Rigaku OD SuperNova Dual source
diffractometer with an Atlas detector | 2947 independent reflections |
| Radiation source: micro-focus sealed X-ray tube, SuperNova (Cu) X-ray Source | 2794 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.042 |
| ω scans | θmax = 72.0°, θmin = 4.7° |
Absorption correction: multi-scan
(CrysAlis PRO; Rigaku OD, 2015) | h = −9→9 |
| Tmin = 0.763, Tmax = 1.000 | k = −13→13 |
| 14284 measured reflections | l = −22→22 |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.035 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.094 | w = 1/[σ2(Fo2) + (0.0583P)2 + 0.7682P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max = 0.001 |
| 2947 reflections | Δρmax = 0.16 e Å−3 |
| 198 parameters | Δρmin = −0.25 e Å−3 |
| 3 restraints | Absolute structure: Flack x determined using 1252 quotients [(I+)-(I-)]/[(I+)+(I-)]
(Parsons et al., 2013) |
| Primary atom site location: difference Fourier map | Absolute structure parameter: −0.014 (19) |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| S1 | 0.20625 (9) | 0.54940 (8) | 0.73058 (4) | 0.02313 (18) | |
| O1 | 0.1232 (3) | 0.4554 (2) | 0.76792 (13) | 0.0297 (5) | |
| O2 | 0.1349 (3) | 0.6710 (2) | 0.72600 (14) | 0.0320 (5) | |
| O12 | 0.5122 (4) | 0.3556 (2) | 0.92834 (13) | 0.0393 (6) | |
| H12 | 0.517 (13) | 0.365 (8) | 0.9747 (12) | 0.047* | 0.5 |
| O11 | 0.4971 (3) | 0.5589 (3) | 0.92622 (12) | 0.0339 (5) | |
| H11 | 0.521 (11) | 0.566 (8) | 0.9728 (10) | 0.041* | 0.5 |
| N1 | 0.4078 (3) | 0.5602 (3) | 0.77370 (13) | 0.0246 (5) | |
| C2 | 0.2229 (4) | 0.3773 (3) | 0.62250 (18) | 0.0263 (6) | |
| H2 | 0.222288 | 0.318171 | 0.660931 | 0.032* | |
| C1 | 0.2151 (4) | 0.5004 (3) | 0.63819 (17) | 0.0236 (6) | |
| C11 | 0.5009 (4) | 0.4571 (3) | 0.89442 (17) | 0.0250 (6) | |
| C4 | 0.2359 (4) | 0.4272 (4) | 0.49284 (18) | 0.0320 (8) | |
| C4A | 0.000000 | 0.5847 (5) | 1.000000 | 0.0361 (12) | |
| H4A | 0.000002 | 0.671449 | 1.000001 | 0.043* | |
| C6 | 0.2147 (5) | 0.5878 (3) | 0.58237 (19) | 0.0314 (8) | |
| H6 | 0.207082 | 0.672224 | 0.593558 | 0.038* | |
| C10 | 0.4966 (4) | 0.4514 (3) | 0.81035 (17) | 0.0260 (6) | |
| H10 | 0.436951 | 0.375012 | 0.789299 | 0.031* | |
| C7 | 0.5324 (4) | 0.6431 (3) | 0.74423 (19) | 0.0303 (7) | |
| H7A | 0.513636 | 0.728981 | 0.758247 | 0.036* | |
| H7B | 0.520593 | 0.637140 | 0.689024 | 0.036* | |
| C3 | 0.2318 (4) | 0.3414 (3) | 0.54953 (19) | 0.0325 (8) | |
| H3 | 0.235221 | 0.256847 | 0.538021 | 0.039* | |
| C9 | 0.6836 (5) | 0.4603 (4) | 0.7903 (2) | 0.0343 (8) | |
| H9A | 0.771809 | 0.425754 | 0.830650 | 0.041* | |
| H9B | 0.691292 | 0.416629 | 0.742923 | 0.041* | |
| C3A | 0.0565 (5) | 0.5215 (4) | 0.9418 (2) | 0.0372 (9) | |
| H3A | 0.094508 | 0.564869 | 0.901746 | 0.045* | |
| C5 | 0.2256 (4) | 0.5497 (5) | 0.50985 (17) | 0.0356 (7) | |
| H5 | 0.226067 | 0.608817 | 0.471443 | 0.043* | |
| C2A | 0.0574 (5) | 0.3960 (4) | 0.9420 (2) | 0.0382 (8) | |
| H2A | 0.097383 | 0.352768 | 0.902346 | 0.046* | |
| C8 | 0.7111 (5) | 0.5968 (4) | 0.7818 (2) | 0.0356 (9) | |
| H8A | 0.802599 | 0.613004 | 0.750239 | 0.043* | |
| H8B | 0.746309 | 0.635938 | 0.831257 | 0.043* | |
| C1A | 0.000000 | 0.3321 (6) | 1.000000 | 0.0423 (12) | |
| H1A | 0.000000 | 0.245352 | 1.000000 | 0.051* | |
| C41 | 0.2484 (6) | 0.3864 (5) | 0.4139 (2) | 0.0471 (10) | |
| H41A | 0.200724 | 0.450222 | 0.378462 | 0.071* | |
| H41B | 0.180651 | 0.310980 | 0.402717 | 0.071* | |
| H41C | 0.371862 | 0.371399 | 0.409532 | 0.071* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| S1 | 0.0251 (3) | 0.0248 (4) | 0.0200 (3) | 0.0011 (3) | 0.0049 (2) | −0.0010 (3) |
| O1 | 0.0314 (12) | 0.0333 (12) | 0.0264 (11) | −0.0036 (10) | 0.0106 (9) | −0.0002 (10) |
| O2 | 0.0351 (13) | 0.0289 (13) | 0.0318 (12) | 0.0083 (10) | 0.0048 (10) | −0.0029 (10) |
| O12 | 0.0606 (17) | 0.0302 (14) | 0.0245 (12) | −0.0026 (12) | −0.0011 (11) | 0.0011 (10) |
| O11 | 0.0509 (13) | 0.0296 (13) | 0.0207 (9) | 0.0040 (12) | 0.0040 (9) | −0.0053 (11) |
| N1 | 0.0264 (11) | 0.0257 (14) | 0.0212 (11) | −0.0005 (13) | 0.0018 (9) | 0.0006 (12) |
| C2 | 0.0269 (15) | 0.0267 (17) | 0.0261 (15) | 0.0016 (13) | 0.0069 (12) | 0.0017 (13) |
| C1 | 0.0242 (15) | 0.0278 (15) | 0.0187 (15) | −0.0002 (12) | 0.0027 (11) | −0.0004 (12) |
| C11 | 0.0240 (14) | 0.0270 (15) | 0.0239 (15) | 0.0006 (12) | 0.0039 (11) | −0.0005 (13) |
| C4 | 0.0263 (16) | 0.048 (2) | 0.0222 (15) | 0.0003 (14) | 0.0043 (12) | −0.0062 (14) |
| C4A | 0.037 (3) | 0.035 (3) | 0.038 (3) | 0.000 | 0.013 (2) | 0.000 |
| C6 | 0.0371 (18) | 0.0299 (19) | 0.0263 (17) | −0.0025 (13) | 0.0021 (14) | 0.0014 (13) |
| C10 | 0.0311 (15) | 0.0254 (16) | 0.0209 (14) | 0.0022 (13) | 0.0021 (11) | −0.0024 (12) |
| C7 | 0.0298 (17) | 0.0308 (18) | 0.0308 (17) | −0.0053 (14) | 0.0062 (13) | 0.0001 (14) |
| C3 | 0.0321 (17) | 0.035 (2) | 0.0306 (18) | 0.0023 (14) | 0.0061 (13) | −0.0077 (14) |
| C9 | 0.0348 (18) | 0.042 (2) | 0.0274 (17) | 0.0086 (16) | 0.0092 (13) | −0.0029 (16) |
| C3A | 0.0336 (17) | 0.051 (3) | 0.0284 (17) | 0.0022 (15) | 0.0097 (13) | 0.0028 (14) |
| C5 | 0.0388 (16) | 0.0447 (19) | 0.0232 (14) | 0.000 (2) | 0.0045 (12) | 0.0100 (19) |
| C2A | 0.0342 (18) | 0.043 (2) | 0.038 (2) | 0.0021 (16) | 0.0072 (15) | −0.0075 (16) |
| C8 | 0.0277 (17) | 0.048 (2) | 0.0325 (18) | −0.0038 (15) | 0.0079 (14) | −0.0031 (16) |
| C1A | 0.037 (3) | 0.038 (3) | 0.051 (3) | 0.000 | 0.006 (2) | 0.000 |
| C41 | 0.043 (2) | 0.075 (3) | 0.0234 (17) | 0.007 (2) | 0.0060 (14) | −0.0087 (18) |
Geometric parameters (Å, º) top
| S1—O1 | 1.435 (3) | C6—H6 | 0.9500 |
| S1—O2 | 1.437 (3) | C10—C9 | 1.543 (5) |
| S1—N1 | 1.626 (2) | C10—H10 | 1.0000 |
| S1—C1 | 1.766 (3) | C7—C8 | 1.522 (5) |
| O12—C11 | 1.265 (4) | C7—H7A | 0.9900 |
| O12—H12 | 0.840 (14) | C7—H7B | 0.9900 |
| O11—C11 | 1.257 (4) | C3—H3 | 0.9500 |
| O11—H11 | 0.837 (14) | C9—C8 | 1.520 (6) |
| N1—C10 | 1.477 (4) | C9—H9A | 0.9900 |
| N1—C7 | 1.480 (4) | C9—H9B | 0.9900 |
| C2—C1 | 1.381 (5) | C3A—C2A | 1.373 (5) |
| C2—C3 | 1.388 (5) | C3A—H3A | 0.9500 |
| C2—H2 | 0.9500 | C5—H5 | 0.9500 |
| C1—C6 | 1.390 (5) | C2A—C1A | 1.389 (5) |
| C11—C10 | 1.516 (4) | C2A—H2A | 0.9500 |
| C4—C5 | 1.380 (6) | C8—H8A | 0.9900 |
| C4—C3 | 1.394 (5) | C8—H8B | 0.9900 |
| C4—C41 | 1.513 (5) | C1A—H1A | 0.9500 |
| C4A—C3Ai | 1.386 (4) | C41—H41A | 0.9800 |
| C4A—C3A | 1.386 (4) | C41—H41B | 0.9800 |
| C4A—H4A | 0.9500 | C41—H41C | 0.9800 |
| C6—C5 | 1.391 (5) | | |
| | | |
| O1—S1—O2 | 119.78 (15) | C8—C7—H7A | 111.1 |
| O1—S1—N1 | 106.76 (14) | N1—C7—H7B | 111.1 |
| O2—S1—N1 | 106.67 (16) | C8—C7—H7B | 111.1 |
| O1—S1—C1 | 108.31 (15) | H7A—C7—H7B | 109.1 |
| O2—S1—C1 | 107.34 (15) | C2—C3—C4 | 121.2 (3) |
| N1—S1—C1 | 107.41 (13) | C2—C3—H3 | 119.4 |
| C11—O12—H12 | 112 (6) | C4—C3—H3 | 119.4 |
| C11—O11—H11 | 122 (6) | C8—C9—C10 | 103.6 (3) |
| C10—N1—C7 | 112.1 (3) | C8—C9—H9A | 111.0 |
| C10—N1—S1 | 119.7 (2) | C10—C9—H9A | 111.0 |
| C7—N1—S1 | 119.7 (2) | C8—C9—H9B | 111.0 |
| C1—C2—C3 | 118.8 (3) | C10—C9—H9B | 111.0 |
| C1—C2—H2 | 120.6 | H9A—C9—H9B | 109.0 |
| C3—C2—H2 | 120.6 | C2A—C3A—C4A | 120.0 (4) |
| C2—C1—C6 | 121.2 (3) | C2A—C3A—H3A | 120.0 |
| C2—C1—S1 | 120.0 (2) | C4A—C3A—H3A | 120.0 |
| C6—C1—S1 | 118.8 (3) | C4—C5—C6 | 121.0 (4) |
| O11—C11—O12 | 124.2 (3) | C4—C5—H5 | 119.5 |
| O11—C11—C10 | 119.7 (3) | C6—C5—H5 | 119.5 |
| O12—C11—C10 | 116.1 (3) | C3A—C2A—C1A | 120.2 (4) |
| C5—C4—C3 | 118.8 (3) | C3A—C2A—H2A | 119.9 |
| C5—C4—C41 | 120.8 (4) | C1A—C2A—H2A | 119.9 |
| C3—C4—C41 | 120.4 (4) | C9—C8—C7 | 104.0 (3) |
| C3Ai—C4A—C3A | 120.1 (5) | C9—C8—H8A | 111.0 |
| C3Ai—C4A—H4A | 120.0 | C7—C8—H8A | 111.0 |
| C3A—C4A—H4A | 120.0 | C9—C8—H8B | 111.0 |
| C1—C6—C5 | 119.0 (4) | C7—C8—H8B | 111.0 |
| C1—C6—H6 | 120.5 | H8A—C8—H8B | 109.0 |
| C5—C6—H6 | 120.5 | C2Ai—C1A—C2A | 119.6 (6) |
| N1—C10—C11 | 110.6 (3) | C2Ai—C1A—H1A | 120.2 |
| N1—C10—C9 | 103.1 (3) | C2A—C1A—H1A | 120.2 |
| C11—C10—C9 | 111.1 (3) | C4—C41—H41A | 109.5 |
| N1—C10—H10 | 110.6 | C4—C41—H41B | 109.5 |
| C11—C10—H10 | 110.6 | H41A—C41—H41B | 109.5 |
| C9—C10—H10 | 110.6 | C4—C41—H41C | 109.5 |
| N1—C7—C8 | 103.1 (3) | H41A—C41—H41C | 109.5 |
| N1—C7—H7A | 111.1 | H41B—C41—H41C | 109.5 |
| | | |
| O1—S1—N1—C10 | 29.6 (3) | O11—C11—C10—N1 | −26.5 (4) |
| O2—S1—N1—C10 | 158.8 (2) | O12—C11—C10—N1 | 155.0 (3) |
| C1—S1—N1—C10 | −86.4 (2) | O11—C11—C10—C9 | 87.4 (4) |
| O1—S1—N1—C7 | 175.0 (2) | O12—C11—C10—C9 | −91.1 (4) |
| O2—S1—N1—C7 | −55.8 (3) | C10—N1—C7—C8 | −14.0 (3) |
| C1—S1—N1—C7 | 59.0 (3) | S1—N1—C7—C8 | −161.8 (2) |
| C3—C2—C1—C6 | 0.5 (5) | C1—C2—C3—C4 | 1.1 (5) |
| C3—C2—C1—S1 | −179.3 (2) | C5—C4—C3—C2 | −1.9 (5) |
| O1—S1—C1—C2 | −28.4 (3) | C41—C4—C3—C2 | 179.1 (3) |
| O2—S1—C1—C2 | −159.1 (3) | N1—C10—C9—C8 | 28.9 (3) |
| N1—S1—C1—C2 | 86.6 (3) | C11—C10—C9—C8 | −89.7 (3) |
| O1—S1—C1—C6 | 151.9 (3) | C3Ai—C4A—C3A—C2A | −0.4 (3) |
| O2—S1—C1—C6 | 21.2 (3) | C3—C4—C5—C6 | 1.2 (5) |
| N1—S1—C1—C6 | −93.2 (3) | C41—C4—C5—C6 | −179.8 (3) |
| C2—C1—C6—C5 | −1.2 (5) | C1—C6—C5—C4 | 0.3 (5) |
| S1—C1—C6—C5 | 178.6 (3) | C4A—C3A—C2A—C1A | 0.7 (6) |
| C7—N1—C10—C11 | 109.6 (3) | C10—C9—C8—C7 | −38.1 (3) |
| S1—N1—C10—C11 | −102.6 (3) | N1—C7—C8—C9 | 31.9 (3) |
| C7—N1—C10—C9 | −9.3 (3) | C3A—C2A—C1A—C2Ai | −0.4 (3) |
| S1—N1—C10—C9 | 138.5 (2) | | |
| Symmetry code: (i) −x, y, −z+2. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O12—H12···O12ii | 0.84 (1) | 1.81 (3) | 2.628 (5) | 166 (9) |
| O11—H11···O11ii | 0.84 (1) | 1.85 (3) | 2.661 (4) | 162 (8) |
| C10—H10···O2iii | 1.00 | 2.31 | 3.267 (4) | 161 |
| Symmetry codes: (ii) −x+1, y, −z+2; (iii) −x+1/2, y−1/2, −z+3/2. |
Number of crystal structures found in the CSD versus the symmetry of
the carboxyl cyclic dimer (compare with Fig. 6a) top| Symmetry element (with orientation) | Number of structures |
| -1 | 3066 |
| 1 | 931 |
| 2 (z) | 87 |
| 2 (y) | 6 |
| 2/m (z) | 6 |
| Sum | 4096* |
| Note: (*) the sum of the observed symmetry dependance of the carboxyl cyclic
dimer is bigger than the total number of entries retrieved from the CSD
(4089) as in several crystal structures, more than one dimer fragment was
observed. |
 (
( (
(
 journal menu
journal menu











