Buy article online - an online subscription or single-article purchase is required to access this article.
The structure of the title compound, 2,3-dihydroxysuccinic acid monohydrate, C
4H
6O
6·H
2O, (I), was first determined (to
R = 0.22) and reported by Parry [
Acta Cryst. (1951),
4, 131-138]. We present here a redetermination with significantly improved accuracy. In the centrosymmetric crystals (space group
P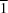
,
Z = 2), the enantiomeric molecules with both
D- and
L-configurations co-exist. An extensive intermolecular hydrogen-bond system involves hydrogen bonds formed by carboxyl and hydroxyl groups of the tartaric acid molecule, as well as the crystalline water molecule.
Supporting information
CCDC reference: 165657
Key indicators
- Single-crystal X-ray study
- T = 293 K
- Mean
![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.003 Å
(C-C) = 0.003 Å
- R factor = 0.040
- wR factor = 0.114
- Data-to-parameter ratio = 11.2
checkCIF results
No syntax errors found
ADDSYM reports no extra symmetry
Single crystals of racemic tartaric acid monohydrate (Parry, 1951) were obtained
from an aqueous solution containing L-camphoramic acid and
D,L-tartaric acid in a 1:1 molar ratio when we tried to separate the
racemic acid by L-camphoramic acid.
H atoms were located from a difference Fourier map. They were included in the
final cycles of least-squares refinement with fixed coordinates and
Uiso (0.08 Å2).
Data collection: MSC/AFC Diffractometer Control Software
(Molecular Structure Corporation, 1992a); cell refinement: MSC/AFC Diffractometer Control Software; data reduction: TEXSAN (Molecular Structure Corporation, 1992b); program(s) used to solve structure: SHELXS93 (Sheldrick, 1993); program(s) used to refine structure: SHELXL93 (Sheldrick, 1993); molecular graphics: XP (Siemens, 1994).
2,3-Dihydroxysuccinic acid monohydrate
top
Crystal data top
| C4H6O6·H2O | Z = 2 |
| Mr = 168.10 | F(000) = 176 |
| Triclinic, P1 | Dx = 1.699 Mg m−3 |
| a = 4.869 (5) Å | Mo Kα radiation, λ = 0.71069 Å |
| b = 8.052 (5) Å | Cell parameters from 16 reflections |
| c = 9.153 (5) Å | θ = 4.7–9.9° |
| α = 109.260 (5)° | µ = 0.17 mm−1 |
| β = 99.862 (5)° | T = 293 K |
| γ = 96.108 (5)° | Prism, colourless |
| V = 328.6 (4) Å3 | 0.70 × 0.60 × 0.40 mm |
Data collection top
Rigaku AFC-7S
diffractometer | 1040 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.000 |
| Graphite monochromator | θmax = 25.0°, θmin = 2.9° |
| ω–2θ scans | h = −5→5 |
Absorption correction: ψ scan
(North et al., 1968) | k = 0→9 |
| Tmin = 0.888, Tmax = 0.934 | l = −10→9 |
| 1145 measured reflections | 3 standard reflections every 150 reflections |
| 1145 independent reflections | intensity decay: 1.4% |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | H-atom parameters not refined |
| wR(F2) = 0.114 | w = 1/[σ2(Fo2) + (0.073P)2 + 0.1479P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.07 | (Δ/σ)max < 0.001 |
| 1145 reflections | Δρmax = 0.34 e Å−3 |
| 102 parameters | Δρmin = −0.16 e Å−3 |
| 0 restraints | Extinction correction: SHELXL93 (Sheldrick, 1993), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.64 (7) |
Crystal data top
| C4H6O6·H2O | γ = 96.108 (5)° |
| Mr = 168.10 | V = 328.6 (4) Å3 |
| Triclinic, P1 | Z = 2 |
| a = 4.869 (5) Å | Mo Kα radiation |
| b = 8.052 (5) Å | µ = 0.17 mm−1 |
| c = 9.153 (5) Å | T = 293 K |
| α = 109.260 (5)° | 0.70 × 0.60 × 0.40 mm |
| β = 99.862 (5)° | |
Data collection top
Rigaku AFC-7S
diffractometer | 1040 reflections with I > 2σ(I) |
Absorption correction: ψ scan
(North et al., 1968) | Rint = 0.000 |
| Tmin = 0.888, Tmax = 0.934 | 3 standard reflections every 150 reflections |
| 1145 measured reflections | intensity decay: 1.4% |
| 1145 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.040 | 0 restraints |
| wR(F2) = 0.114 | H-atom parameters not refined |
| S = 1.07 | Δρmax = 0.34 e Å−3 |
| 1145 reflections | Δρmin = −0.16 e Å−3 |
| 102 parameters | |
Special details top
Experimental. Data collection were performed with a scan width of Δω = (1.63 + 0.35 tan
θ)o and a scan rate of less than 16°min-1 in ω. |
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement on F2 for ALL reflections except for 1 with very negative
F2 or flagged by the user for potential systematic errors. Weighted
R-factors wR and all goodnesses of fit S are based on F2,
conventional R-factors R are based on F, with F
set to zero for negative F2. The observed criterion of F2 >
σ(F2) is used only for calculating_refine_ls_R_factor_gt etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R-factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.2091 (3) | 0.4357 (2) | 0.3708 (2) | 0.0406 (4) | |
| O2 | −0.0110 (3) | 0.2696 (2) | 0.4835 (2) | 0.0403 (5) | |
| O3 | 0.4040 (3) | 0.1621 (2) | 0.19547 (15) | 0.0307 (4) | |
| O4 | −0.1667 (3) | −0.0229 (2) | 0.14076 (15) | 0.0340 (4) | |
| O5 | 0.1126 (3) | −0.2886 (2) | 0.0393 (2) | 0.0430 (5) | |
| O6 | 0.3946 (3) | −0.2208 (2) | 0.2797 (2) | 0.0378 (4) | |
| OW | 0.6175 (3) | 0.5052 (2) | 0.1831 (2) | 0.0450 (5) | |
| C1 | 0.1529 (4) | 0.2952 (2) | 0.3925 (2) | 0.0268 (4) | |
| C2 | 0.2829 (3) | 0.1338 (2) | 0.3169 (2) | 0.0260 (5) | |
| C3 | 0.0594 (4) | −0.0375 (2) | 0.2524 (2) | 0.0272 (5) | |
| C4 | 0.1918 (4) | −0.1979 (2) | 0.1776 (2) | 0.0287 (5) | |
| H2 | −0.0691 | 0.3666 | 0.5268 | 0.080* | |
| H2C | 0.4305 | 0.1231 | 0.3964 | 0.080* | |
| H3 | 0.5380 | 0.1021 | 0.1814 | 0.080* | |
| H3C | −0.0136 | −0.0532 | 0.3395 | 0.080* | |
| H4 | −0.1660 | −0.0954 | 0.0391 | 0.080* | |
| H6 | 0.4477 | −0.3273 | 0.2410 | 0.080* | |
| H1W | 0.7133 | 0.4556 | 0.0990 | 0.080* | |
| H2W | 0.5156 | 0.4088 | 0.2015 | 0.080* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0561 (9) | 0.0253 (7) | 0.0478 (9) | 0.0146 (6) | 0.0253 (7) | 0.0135 (6) |
| O2 | 0.0543 (9) | 0.0266 (7) | 0.0416 (8) | 0.0103 (6) | 0.0268 (7) | 0.0051 (6) |
| O3 | 0.0352 (7) | 0.0278 (7) | 0.0337 (7) | 0.0128 (5) | 0.0151 (5) | 0.0108 (5) |
| O4 | 0.0334 (7) | 0.0343 (7) | 0.0291 (7) | 0.0128 (5) | 0.0047 (5) | 0.0034 (5) |
| O5 | 0.0548 (9) | 0.0314 (8) | 0.0342 (8) | 0.0158 (6) | 0.0069 (6) | −0.0007 (6) |
| O6 | 0.0482 (8) | 0.0288 (7) | 0.0372 (8) | 0.0177 (6) | 0.0089 (6) | 0.0099 (6) |
| OW | 0.0588 (9) | 0.0295 (8) | 0.0555 (10) | 0.0183 (6) | 0.0282 (7) | 0.0156 (7) |
| C1 | 0.0308 (9) | 0.0229 (9) | 0.0232 (8) | 0.0054 (7) | 0.0046 (7) | 0.0039 (7) |
| C2 | 0.0306 (9) | 0.0234 (9) | 0.0239 (8) | 0.0077 (7) | 0.0060 (7) | 0.0071 (7) |
| C3 | 0.0321 (9) | 0.0236 (9) | 0.0269 (9) | 0.0080 (7) | 0.0091 (7) | 0.0079 (7) |
| C4 | 0.0343 (9) | 0.0206 (8) | 0.0326 (10) | 0.0052 (7) | 0.0119 (7) | 0.0089 (7) |
Geometric parameters (Å, º) top
| O1—C1 | 1.223 (2) | O6—H6 | 0.8980 |
| O2—C1 | 1.296 (2) | OW—H1W | 0.9641 |
| O2—H2 | 0.8536 | OW—H2W | 0.9535 |
| O3—C2 | 1.413 (2) | C1—C2 | 1.516 (2) |
| O3—H3 | 0.8545 | C2—C3 | 1.536 (3) |
| O4—C3 | 1.411 (2) | C2—H2C | 0.964 |
| O4—H4 | 0.9217 | C3—C4 | 1.516 (2) |
| O5—C4 | 1.203 (2) | C3—H3C | 0.965 |
| O6—C4 | 1.311 (2) | | |
| | | |
| C1—O2—H2 | 109.1 | C1—C2—H2C | 108.6 |
| C2—O3—H3 | 109.5 | C3—C2—H2C | 108.7 |
| C3—O4—H4 | 111.4 | O4—C3—C4 | 110.43 (14) |
| C4—O6—H6 | 111.9 | O4—C3—C2 | 110.15 (14) |
| H1W—OW—H2W | 108.2 | C4—C3—C2 | 110.20 (15) |
| O1—C1—O2 | 124.9 (2) | O4—C3—H3C | 108.6 |
| O1—C1—C2 | 121.6 (2) | C4—C3—H3C | 108.8 |
| O2—C1—C2 | 113.54 (15) | C2—C3—H3C | 108.7 |
| O3—C2—C1 | 108.47 (14) | O5—C4—O6 | 126.4 (2) |
| O3—C2—C3 | 111.51 (14) | O5—C4—C3 | 121.3 (2) |
| C1—C2—C3 | 110.67 (15) | O6—C4—C3 | 112.24 (15) |
| O3—C2—H2C | 108.8 | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···O1i | 0.854 | 1.828 | 2.680 (2) | 175.0 |
| O3—H3···O4ii | 0.854 | 1.861 | 2.714 (3) | 176.4 |
| O6—H6···OWiii | 0.898 | 1.643 | 2.526 (2) | 167.0 |
| OW—H2W···O3 | 0.954 | 1.983 | 2.891 (2) | 158.5 |
| OW—H1W···O5iv | 0.964 | 1.902 | 2.824 (2) | 159.2 |
| Symmetry codes: (i) −x, −y+1, −z+1; (ii) x+1, y, z; (iii) x, y−1, z; (iv) −x+1, −y, −z. |
Experimental details
| Crystal data |
| Chemical formula | C4H6O6·H2O |
| Mr | 168.10 |
| Crystal system, space group | Triclinic, P1 |
| Temperature (K) | 293 |
| a, b, c (Å) | 4.869 (5), 8.052 (5), 9.153 (5) |
| α, β, γ (°) | 109.260 (5), 99.862 (5), 96.108 (5) |
| V (Å3) | 328.6 (4) |
| Z | 2 |
| Radiation type | Mo Kα |
| µ (mm−1) | 0.17 |
| Crystal size (mm) | 0.70 × 0.60 × 0.40 |
| |
| Data collection |
| Diffractometer | Rigaku AFC-7S
diffractometer |
| Absorption correction | ψ scan
(North et al., 1968) |
| Tmin, Tmax | 0.888, 0.934 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 1145, 1145, 1040 |
| Rint | 0.000 |
| (sin θ/λ)max (Å−1) | 0.595 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.040, 0.114, 1.07 |
| No. of reflections | 1145 |
| No. of parameters | 102 |
| H-atom treatment | H-atom parameters not refined |
| Δρmax, Δρmin (e Å−3) | 0.34, −0.16 |
Selected geometric parameters (Å, º) top| O1—C1 | 1.223 (2) | O5—C4 | 1.203 (2) |
| O2—C1 | 1.296 (2) | O6—C4 | 1.311 (2) |
| O3—C2 | 1.413 (2) | C1—C2 | 1.516 (2) |
| O4—C3 | 1.411 (2) | C2—C3 | 1.536 (3) |
| | | |
| O1—C1—O2 | 124.9 (2) | O4—C3—C4 | 110.43 (14) |
| O3—C2—C1 | 108.47 (14) | O4—C3—C2 | 110.15 (14) |
| O3—C2—C3 | 111.51 (14) | C4—C3—C2 | 110.20 (15) |
| C1—C2—C3 | 110.67 (15) | O5—C4—O6 | 126.4 (2) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···O1i | 0.854 | 1.828 | 2.680 (2) | 175.0 |
| O3—H3···O4ii | 0.854 | 1.861 | 2.714 (3) | 176.4 |
| O6—H6···OWiii | 0.898 | 1.643 | 2.526 (2) | 167.0 |
| OW—H2W···O3 | 0.954 | 1.983 | 2.891 (2) | 158.5 |
| OW—H1W···O5iv | 0.964 | 1.902 | 2.824 (2) | 159.2 |
| Symmetry codes: (i) −x, −y+1, −z+1; (ii) x+1, y, z; (iii) x, y−1, z; (iv) −x+1, −y, −z. |

 Subscribe to Acta Crystallographica Section E: Crystallographic Communications
Subscribe to Acta Crystallographica Section E: Crystallographic Communications
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.
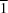 , Z = 2), the enantiomeric molecules with both D- and L-configurations co-exist. An extensive intermolecular hydrogen-bond system involves hydrogen bonds formed by carboxyl and hydroxyl groups of the tartaric acid molecule, as well as the crystalline water molecule.
, Z = 2), the enantiomeric molecules with both D- and L-configurations co-exist. An extensive intermolecular hydrogen-bond system involves hydrogen bonds formed by carboxyl and hydroxyl groups of the tartaric acid molecule, as well as the crystalline water molecule. 
 journal menu
journal menu











![[Figure 1]](http://journals.iucr.org/e/issues/2001/05/00/ya6020/ya6020fig1thm.gif)




NO COMMENT