The title compound,
meso-5,7,7,12,14,14-hexamethyl-4,11-diaza-1,8-diazoniacyclotetradecane bis(3-carboxy-5-nitrobenzoate), C
16H
38N
42+·2C
8H
4NO
6-, is a salt in which the cation is present as two configurational isomers, disordered across a common centre of inversion in
P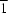
, with occupancies of 0.847 (3) and 0.153 (3). The anions are linked into chains by a single O-H

O hydrogen bond [H

O 1.71 Å, O

O 2.5063 (15) Å and O-H

O 156°] and the cations link these anion chains into sheets by means of a range of N-H

O hydrogen bonds [H

O 1.81-2.53 Å, N

O 2.718 (5)-3.3554 (19) Å and N-H

O 146-171°].
Supporting information
CCDC reference: 193423
Stoichiometric quantities of the two components were individually dissolved in
methanol. The solutions were mixed and the mixture was set aside to
crystallize, yielding microcrystalline (I). Analysis, found: C 54.7, H 6.5, N
11.8%; C32H46N6O12 requires: C 54.4, H 6.6, N 11.9%. Despite many
preparations, the crystallites proved to be too small for single-crystal X-ray
diffractometry using conventional laboratory sources, but an excellent data
set was obtained using synchrotron radiation, collected at the Daresbury
synchrotron radiation source, Station 9.8 (Cernik et al., 1997; Clegg
et al., 1998).
Compound (I) is triclinic, and space group P1 was assumed and confirmed
by the subsequent analysis. It was apparent from an early stage that the
cation was disordered over two sets of sites; with the minor occupancy atoms
refined isotropically, the site-occupancy factors refined to 0.847 (3) and
0.153 (3) for the major and minor sites, respectively. All the H atoms of the
anion and the major isomer of the cation were located from difference maps;
the locations of the 0.153 (3) occupancy H atoms bonded to N in the minor
isomer of the cation were inferred from the non-bonded contact distances
involving N21 and N24. All H atoms were treated as riding, with distances
O—H 0.84 Å, N—H 0.92 Å and C—H 0.95 (aromatic), 0.98 (CH3), 0.99
(CH2) or 1.00 Å (aliphatic CH). In the final difference map, bonding
electron density was clearly apparent in all of the C—C, C—N, C—O and
N—O bonds of the anion; the largest densities were in bonds involving C11.
Data collection: SMART (Bruker, 1998); cell refinement: SAINT (Bruker, 2000); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 1998); program(s) used to refine structure: SHELXTL and SHELXL97 (Sheldrick, 1997); molecular graphics: PLATON (Spek, 2002); software used to prepare material for publication: SHELXL97 and PRPKAPPA (Ferguson, 1999).
meso-5,7,7,12,14,14-hexamethyl-4,11-diaza-1,8-diazoniacyclotetradecane
bis(3-carboxy-5-nitrobenzoate)
top
Crystal data top
| C16H38N42+·2C8H4NO6− | Z = 1 |
| Mr = 706.46 | F(000) = 376 |
| Triclinic, P1 | Dx = 1.367 Mg m−3 |
| Hall symbol: -P 1 | Synchrotron radiation, λ = 0.6867 Å |
| a = 8.5183 (6) Å | Cell parameters from 5312 reflections |
| b = 10.2278 (8) Å | θ = 2.4–29.3° |
| c = 11.3272 (9) Å | µ = 0.11 mm−1 |
| α = 66.635 (1)° | T = 150 K |
| β = 71.387 (1)° | Block, colourless |
| γ = 82.976 (1)° | 0.09 × 0.06 × 0.03 mm |
| V = 858.54 (11) Å3 | |
Data collection top
Bruker SMART 1K CCD area-detector
diffractometer | 4702 independent reflections |
| Radiation source: Daresbury SRS station 9.8 | 4097 reflections with I > 2σ(I) |
| Silicon 111 monochromator | Rint = 0.027 |
| ω rotation with narrow frames scans | θmax = 29.3°, θmin = 2.0° |
Absorption correction: multi-scan
(SADABS; Bruker, 2000) | h = −11→11 |
| Tmin = 0.991, Tmax = 0.997 | k = −14→13 |
| 8877 measured reflections | l = −16→15 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.062 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.165 | H-atom parameters constrained |
| S = 1.13 | w = 1/[σ2(Fo2) + (0.0776P)2 + 0.2773P]
where P = (Fo2 + 2Fc2)/3 |
| 4702 reflections | (Δ/σ)max < 0.001 |
| 262 parameters | Δρmax = 0.64 e Å−3 |
| 9 restraints | Δρmin = −0.27 e Å−3 |
Crystal data top
| C16H38N42+·2C8H4NO6− | γ = 82.976 (1)° |
| Mr = 706.46 | V = 858.54 (11) Å3 |
| Triclinic, P1 | Z = 1 |
| a = 8.5183 (6) Å | Synchrotron radiation, λ = 0.6867 Å |
| b = 10.2278 (8) Å | µ = 0.11 mm−1 |
| c = 11.3272 (9) Å | T = 150 K |
| α = 66.635 (1)° | 0.09 × 0.06 × 0.03 mm |
| β = 71.387 (1)° | |
Data collection top
Bruker SMART 1K CCD area-detector
diffractometer | 4702 independent reflections |
Absorption correction: multi-scan
(SADABS; Bruker, 2000) | 4097 reflections with I > 2σ(I) |
| Tmin = 0.991, Tmax = 0.997 | Rint = 0.027 |
| 8877 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.062 | 9 restraints |
| wR(F2) = 0.165 | H-atom parameters constrained |
| S = 1.13 | Δρmax = 0.64 e Å−3 |
| 4702 reflections | Δρmin = −0.27 e Å−3 |
| 262 parameters | |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| O1 | 1.49566 (14) | 1.15193 (14) | 0.06528 (15) | 0.0536 (4) | |
| O2 | 1.43084 (14) | 0.92631 (13) | 0.19819 (14) | 0.0480 (3) | |
| H2 | 1.5255 | 0.9253 | 0.2066 | 0.072* | |
| O3 | 0.88430 (14) | 0.74429 (12) | 0.36103 (14) | 0.0471 (3) | |
| O4 | 0.68298 (13) | 0.86088 (15) | 0.27784 (13) | 0.0498 (4) | |
| O5 | 0.82203 (17) | 1.34823 (17) | −0.07586 (17) | 0.0701 (5) | |
| O6 | 1.06128 (18) | 1.44045 (16) | −0.13114 (18) | 0.0726 (5) | |
| N11 | 0.96118 (17) | 1.34182 (16) | −0.06684 (15) | 0.0457 (4) | |
| C11 | 1.22518 (16) | 1.06822 (15) | 0.11204 (14) | 0.0285 (3) | |
| C12 | 1.11236 (16) | 0.95671 (15) | 0.19031 (14) | 0.0280 (3) | |
| H12 | 1.1474 | 0.8693 | 0.2470 | 0.034* | |
| C13 | 0.94868 (15) | 0.97090 (15) | 0.18704 (13) | 0.0270 (3) | |
| C14 | 0.89797 (16) | 1.09789 (16) | 0.10156 (14) | 0.0295 (3) | |
| H14 | 0.7877 | 1.1092 | 0.0964 | 0.035* | |
| C15 | 1.01319 (17) | 1.20707 (16) | 0.02434 (14) | 0.0324 (3) | |
| C16 | 1.17551 (18) | 1.19641 (17) | 0.02789 (15) | 0.0339 (3) | |
| H16 | 1.2505 | 1.2742 | −0.0255 | 0.041* | |
| C17 | 1.39859 (16) | 1.05513 (16) | 0.12086 (14) | 0.0310 (3) | |
| C18 | 0.82911 (17) | 0.84889 (16) | 0.28138 (15) | 0.0317 (3) | |
| N1 | 0.63983 (16) | 0.33334 (14) | 0.49439 (14) | 0.0272 (3) | 0.847 (3) |
| H1 | 0.5780 | 0.2735 | 0.5778 | 0.033* | 0.847 (3) |
| C2 | 0.7882 (2) | 0.3796 (2) | 0.5081 (2) | 0.0323 (4) | 0.847 (3) |
| H2A | 0.8469 | 0.4548 | 0.4221 | 0.039* | 0.847 (3) |
| H2B | 0.8644 | 0.2981 | 0.5285 | 0.039* | 0.847 (3) |
| C3 | 0.7408 (3) | 0.4364 (3) | 0.6193 (3) | 0.0343 (5) | 0.847 (3) |
| H3A | 0.6701 | 0.3662 | 0.7032 | 0.041* | 0.847 (3) |
| H3B | 0.8415 | 0.4532 | 0.6371 | 0.041* | 0.847 (3) |
| N4 | 0.6482 (6) | 0.5736 (3) | 0.5761 (4) | 0.0274 (5) | 0.847 (3) |
| H4A | 0.5603 | 0.5574 | 0.5530 | 0.033* | 0.847 (3) |
| H4B | 0.7173 | 0.6386 | 0.5003 | 0.033* | 0.847 (3) |
| C5 | 0.5825 (7) | 0.6389 (6) | 0.6830 (6) | 0.0351 (5) | 0.847 (3) |
| C51 | 0.7283 (6) | 0.6911 (5) | 0.7042 (6) | 0.0502 (11) | 0.847 (3) |
| H51A | 0.7967 | 0.6097 | 0.7400 | 0.075* | 0.847 (3) |
| H51B | 0.6864 | 0.7385 | 0.7683 | 0.075* | 0.847 (3) |
| H51C | 0.7953 | 0.7584 | 0.6180 | 0.075* | 0.847 (3) |
| C52 | 0.4772 (10) | 0.5286 (8) | 0.8141 (7) | 0.0572 (10) | 0.847 (3) |
| H52A | 0.3984 | 0.4842 | 0.7941 | 0.086* | 0.847 (3) |
| H52B | 0.4164 | 0.5757 | 0.8756 | 0.086* | 0.847 (3) |
| H52C | 0.5494 | 0.4553 | 0.8564 | 0.086* | 0.847 (3) |
| C6 | 0.4813 (7) | 0.7698 (6) | 0.6221 (7) | 0.0363 (7) | 0.847 (3) |
| H6A | 0.4514 | 0.8239 | 0.6817 | 0.044* | 0.847 (3) |
| H6B | 0.5539 | 0.8319 | 0.5342 | 0.044* | 0.847 (3) |
| C7 | 0.3217 (2) | 0.7412 (2) | 0.6003 (2) | 0.0319 (4) | 0.847 (3) |
| H7 | 0.2483 | 0.6778 | 0.6888 | 0.038* | 0.847 (3) |
| C71 | 0.2295 (7) | 0.8821 (5) | 0.5497 (7) | 0.0509 (7) | 0.847 (3) |
| H71A | 0.2235 | 0.9365 | 0.6054 | 0.076* | 0.847 (3) |
| H71B | 0.1172 | 0.8614 | 0.5558 | 0.076* | 0.847 (3) |
| H71C | 0.2892 | 0.9379 | 0.4558 | 0.076* | 0.847 (3) |
| N21 | 0.7072 (9) | 0.3868 (8) | 0.4004 (8) | 0.0267 (12)* | 0.153 (3) |
| H21 | 0.7604 | 0.4127 | 0.3098 | 0.032* | 0.153 (3) |
| C22 | 0.8227 (13) | 0.4009 (14) | 0.4656 (10) | 0.0267 (12)* | 0.153 (3) |
| H22A | 0.8990 | 0.4814 | 0.4020 | 0.032* | 0.153 (3) |
| H22B | 0.8900 | 0.3132 | 0.4858 | 0.032* | 0.153 (3) |
| C23 | 0.7419 (19) | 0.4253 (16) | 0.5923 (13) | 0.0267 (12)* | 0.153 (3) |
| H23A | 0.6626 | 0.3460 | 0.6523 | 0.032* | 0.153 (3) |
| H23B | 0.8287 | 0.4162 | 0.6359 | 0.032* | 0.153 (3) |
| N24 | 0.653 (4) | 0.558 (2) | 0.590 (3) | 0.0267 (12)* | 0.153 (3) |
| H24A | 0.5611 | 0.5495 | 0.5673 | 0.032* | 0.153 (3) |
| H24B | 0.7186 | 0.6252 | 0.5137 | 0.032* | 0.153 (3) |
| C25 | 0.588 (3) | 0.637 (3) | 0.680 (3) | 0.0267 (12)* | 0.153 (3) |
| C251 | 0.743 (3) | 0.676 (3) | 0.698 (2) | 0.0267 (12)* | 0.153 (3) |
| H25A | 0.8128 | 0.7391 | 0.6103 | 0.040* | 0.153 (3) |
| H25B | 0.8046 | 0.5892 | 0.7329 | 0.040* | 0.153 (3) |
| H25C | 0.7117 | 0.7245 | 0.7608 | 0.040* | 0.153 (3) |
| C252 | 0.488 (4) | 0.531 (3) | 0.816 (3) | 0.0267 (12)* | 0.153 (3) |
| H25D | 0.3916 | 0.4980 | 0.8053 | 0.040* | 0.153 (3) |
| H25E | 0.4497 | 0.5783 | 0.8803 | 0.040* | 0.153 (3) |
| H25F | 0.5573 | 0.4498 | 0.8489 | 0.040* | 0.153 (3) |
| C26 | 0.474 (3) | 0.757 (4) | 0.626 (3) | 0.0267 (12)* | 0.153 (3) |
| H26A | 0.5462 | 0.8422 | 0.5691 | 0.032* | 0.153 (3) |
| H26B | 0.4018 | 0.7769 | 0.7051 | 0.032* | 0.153 (3) |
| C27 | 0.3601 (13) | 0.7565 (11) | 0.5463 (12) | 0.0267 (12)* | 0.153 (3) |
| H27 | 0.4277 | 0.7803 | 0.4502 | 0.032* | 0.153 (3) |
| C271 | 0.237 (3) | 0.8766 (19) | 0.554 (2) | 0.0267 (12)* | 0.153 (3) |
| H27A | 0.1599 | 0.8815 | 0.5040 | 0.040* | 0.153 (3) |
| H27B | 0.2976 | 0.9672 | 0.5136 | 0.040* | 0.153 (3) |
| H27C | 0.1759 | 0.8585 | 0.6480 | 0.040* | 0.153 (3) |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0252 (5) | 0.0473 (7) | 0.0685 (9) | −0.0083 (5) | −0.0163 (5) | 0.0023 (6) |
| O2 | 0.0282 (5) | 0.0441 (6) | 0.0650 (8) | −0.0030 (5) | −0.0277 (5) | −0.0023 (6) |
| O3 | 0.0287 (5) | 0.0345 (6) | 0.0630 (8) | −0.0038 (4) | −0.0223 (5) | 0.0047 (5) |
| O4 | 0.0235 (5) | 0.0593 (8) | 0.0489 (7) | −0.0071 (5) | −0.0185 (5) | 0.0055 (6) |
| O5 | 0.0368 (7) | 0.0593 (9) | 0.0804 (11) | −0.0013 (6) | −0.0315 (7) | 0.0193 (8) |
| O6 | 0.0457 (8) | 0.0516 (8) | 0.0812 (11) | −0.0108 (6) | −0.0244 (7) | 0.0222 (8) |
| N11 | 0.0311 (6) | 0.0428 (8) | 0.0434 (8) | 0.0015 (6) | −0.0137 (6) | 0.0051 (6) |
| C11 | 0.0203 (6) | 0.0361 (7) | 0.0282 (6) | 0.0010 (5) | −0.0089 (5) | −0.0104 (5) |
| C12 | 0.0214 (6) | 0.0314 (6) | 0.0306 (6) | 0.0026 (5) | −0.0111 (5) | −0.0093 (5) |
| C13 | 0.0205 (6) | 0.0314 (6) | 0.0281 (6) | 0.0007 (5) | −0.0090 (5) | −0.0089 (5) |
| C14 | 0.0211 (6) | 0.0367 (7) | 0.0276 (6) | 0.0033 (5) | −0.0091 (5) | −0.0086 (5) |
| C15 | 0.0261 (6) | 0.0350 (7) | 0.0286 (6) | 0.0032 (5) | −0.0098 (5) | −0.0040 (5) |
| C16 | 0.0252 (6) | 0.0375 (7) | 0.0318 (7) | −0.0023 (5) | −0.0084 (5) | −0.0052 (6) |
| C17 | 0.0209 (6) | 0.0400 (7) | 0.0308 (6) | 0.0005 (5) | −0.0087 (5) | −0.0115 (6) |
| C18 | 0.0224 (6) | 0.0341 (7) | 0.0356 (7) | −0.0009 (5) | −0.0118 (5) | −0.0074 (6) |
| N1 | 0.0255 (6) | 0.0219 (6) | 0.0349 (7) | −0.0007 (5) | −0.0110 (5) | −0.0096 (5) |
| C2 | 0.0241 (9) | 0.0317 (10) | 0.0456 (11) | 0.0062 (7) | −0.0181 (8) | −0.0149 (9) |
| C3 | 0.0389 (9) | 0.0276 (8) | 0.0455 (13) | 0.0089 (7) | −0.0257 (10) | −0.0153 (8) |
| N4 | 0.0271 (7) | 0.0177 (9) | 0.0388 (14) | −0.0008 (7) | −0.0183 (8) | −0.0053 (9) |
| C5 | 0.0366 (10) | 0.0338 (9) | 0.0406 (10) | 0.0009 (7) | −0.0190 (8) | −0.0143 (7) |
| C51 | 0.0526 (19) | 0.0480 (19) | 0.0763 (19) | 0.0109 (13) | −0.0411 (16) | −0.0362 (13) |
| C52 | 0.046 (2) | 0.0639 (16) | 0.0441 (12) | 0.0048 (13) | −0.0167 (11) | −0.0016 (11) |
| C6 | 0.0405 (12) | 0.0286 (17) | 0.0493 (12) | 0.0029 (11) | −0.0210 (10) | −0.0191 (13) |
| C7 | 0.0319 (9) | 0.0299 (8) | 0.0369 (9) | 0.0049 (7) | −0.0131 (7) | −0.0148 (8) |
| C71 | 0.0546 (14) | 0.0400 (12) | 0.0732 (16) | 0.0215 (9) | −0.0341 (13) | −0.0312 (11) |
Geometric parameters (Å, º) top
| O1—C17 | 1.1974 (18) | C52—H52C | 0.9800 |
| O2—H2 | 0.8400 | C6—H6A | 0.9900 |
| O2—C17 | 1.3147 (18) | C6—H6B | 0.9900 |
| O3—C18 | 1.2497 (18) | C6—C7 | 1.538 (4) |
| O4—C18 | 1.2477 (17) | C7—H7 | 1.0000 |
| O5—N11 | 1.2136 (19) | C7—N1i | 1.484 (2) |
| O6—N11 | 1.227 (2) | C7—C71 | 1.542 (4) |
| N11—C15 | 1.4752 (19) | C71—H71A | 0.9800 |
| C11—C12 | 1.3902 (19) | C71—H71B | 0.9800 |
| C11—C16 | 1.392 (2) | C71—H71C | 0.9800 |
| C11—C17 | 1.4984 (18) | N21—H21 | 0.9200 |
| C12—H12 | 0.9500 | N21—C22 | 1.455 (9) |
| C12—C13 | 1.3948 (17) | C22—H22A | 0.9900 |
| C13—C14 | 1.3940 (18) | C22—H22B | 0.9900 |
| C13—C18 | 1.5149 (19) | C22—C23 | 1.485 (9) |
| C14—H14 | 0.9500 | C23—H23A | 0.9900 |
| C14—C15 | 1.386 (2) | C23—H23B | 0.9900 |
| C15—C16 | 1.3858 (19) | C23—N24 | 1.467 (10) |
| C16—H16 | 0.9500 | N24—H24A | 0.9200 |
| N1—H1 | 0.9200 | N24—H24B | 0.9200 |
| N1—C2 | 1.475 (2) | N24—C25 | 1.473 (10) |
| C2—H2A | 0.9900 | C25—C251 | 1.521 (10) |
| C2—H2B | 0.9900 | C25—C252 | 1.526 (10) |
| C2—C3 | 1.511 (3) | C25—C26 | 1.524 (10) |
| C3—H3A | 0.9900 | C251—H25A | 0.9800 |
| C3—H3B | 0.9900 | C251—H25B | 0.9800 |
| C3—N4 | 1.505 (2) | C251—H25C | 0.9800 |
| N4—H4A | 0.9200 | C252—H25D | 0.9800 |
| N4—H4B | 0.9200 | C252—H25E | 0.9800 |
| N4—C5 | 1.530 (3) | C252—H25F | 0.9800 |
| C5—C51 | 1.530 (3) | C26—H26A | 0.9900 |
| C5—C52 | 1.533 (4) | C26—H26B | 0.9900 |
| C5—C6 | 1.542 (3) | C26—C27 | 1.524 (10) |
| C51—H51A | 0.9800 | C27—H27 | 1.0000 |
| C51—H51B | 0.9800 | C27—C271 | 1.523 (10) |
| C51—H51C | 0.9800 | C271—H27A | 0.9800 |
| C52—H52A | 0.9800 | C271—H27B | 0.9800 |
| C52—H52B | 0.9800 | C271—H27C | 0.9800 |
| | | |
| H2—O2—C17 | 109.5 | H6A—C6—C7 | 108.0 |
| O5—N11—O6 | 123.48 (14) | H6B—C6—C7 | 108.0 |
| O5—N11—C15 | 117.99 (14) | C6—C7—H7 | 108.2 |
| O6—N11—C15 | 118.52 (13) | C6—C7—C71 | 110.0 (4) |
| C12—C11—C16 | 119.89 (12) | H7—C7—C71 | 108.2 |
| C12—C11—C17 | 121.02 (12) | H21—N21—C22 | 108.8 |
| C16—C11—C17 | 119.04 (13) | N21—C22—H22A | 108.8 |
| C11—C12—H12 | 119.4 | N21—C22—H22B | 108.8 |
| C11—C12—C13 | 121.17 (12) | N21—C22—C23 | 114.0 (10) |
| H12—C12—C13 | 119.4 | H22A—C22—H22B | 107.6 |
| C12—C13—C14 | 119.54 (12) | H22A—C22—C23 | 108.8 |
| C12—C13—C18 | 118.57 (12) | H22B—C22—C23 | 108.8 |
| C14—C13—C18 | 121.85 (11) | C22—C23—H23A | 107.1 |
| C13—C14—H14 | 121.0 | C22—C23—H23B | 107.1 |
| C13—C14—C15 | 118.04 (12) | C22—C23—N24 | 120.7 (15) |
| H14—C14—C15 | 121.0 | H23A—C23—H23B | 106.8 |
| N11—C15—C14 | 118.55 (12) | H23A—C23—N24 | 107.1 |
| N11—C15—C16 | 118.01 (13) | H23B—C23—N24 | 107.1 |
| C14—C15—C16 | 123.43 (13) | C23—N24—H24A | 102.5 |
| C11—C16—C15 | 117.91 (13) | C23—N24—H24B | 102.5 |
| C11—C16—H16 | 121.0 | C23—N24—C25 | 138 (2) |
| C15—C16—H16 | 121.0 | H24A—N24—H24B | 104.9 |
| O1—C17—O2 | 123.99 (13) | H24A—N24—C25 | 102.5 |
| O1—C17—C11 | 123.62 (14) | H24B—N24—C25 | 102.5 |
| O2—C17—C11 | 112.38 (12) | N24—C25—C251 | 103 (2) |
| O3—C18—O4 | 124.64 (14) | N24—C25—C252 | 107 (2) |
| O3—C18—C13 | 116.65 (12) | N24—C25—C26 | 112 (2) |
| O4—C18—C13 | 118.67 (13) | C251—C25—C252 | 108 (2) |
| H1—N1—C2 | 108.8 | C251—C25—C26 | 118 (2) |
| N1—C2—H2A | 109.5 | C252—C25—C26 | 109 (2) |
| N1—C2—H2B | 109.5 | C25—C251—H25A | 109.5 |
| N1—C2—C3 | 110.67 (17) | C25—C251—H25B | 109.5 |
| H2A—C2—H2B | 108.1 | C25—C251—H25C | 109.5 |
| H2A—C2—C3 | 109.5 | H25A—C251—H25B | 109.5 |
| H2B—C2—C3 | 109.5 | H25A—C251—H25C | 109.5 |
| C2—C3—H3A | 110.0 | H25B—C251—H25C | 109.5 |
| C2—C3—H3B | 110.0 | C25—C252—H25D | 109.5 |
| C2—C3—N4 | 108.6 (2) | C25—C252—H25E | 109.5 |
| H3A—C3—H3B | 108.3 | C25—C252—H25F | 109.5 |
| H3A—C3—N4 | 110.0 | H25D—C252—H25E | 109.5 |
| H3B—C3—N4 | 110.0 | H25D—C252—H25F | 109.5 |
| C3—N4—H4A | 108.7 | H25E—C252—H25F | 109.5 |
| C3—N4—H4B | 108.7 | C25—C26—H26A | 105.9 |
| C3—N4—C5 | 114.4 (3) | C25—C26—H26B | 105.9 |
| H4A—N4—H4B | 107.6 | C25—C26—C27 | 126 (2) |
| H4A—N4—C5 | 108.7 | H26A—C26—H26B | 106.2 |
| H4B—N4—C5 | 108.7 | H26A—C26—C27 | 105.9 |
| N4—C5—C51 | 109.3 (4) | H26B—C26—C27 | 105.9 |
| N4—C5—C52 | 109.7 (5) | C26—C27—H27 | 108.4 |
| N4—C5—C6 | 105.5 (4) | C26—C27—C271 | 105.4 (17) |
| C51—C5—C52 | 111.6 (5) | H27—C27—C271 | 108.4 |
| C51—C5—C6 | 108.1 (4) | C27—C271—H27A | 109.5 |
| C52—C5—C6 | 112.4 (5) | C27—C271—H27B | 109.5 |
| C5—C6—H6A | 108.0 | C27—C271—H27C | 109.5 |
| C5—C6—H6B | 108.0 | H27A—C271—H27B | 109.5 |
| C5—C6—C7 | 117.0 (4) | H27A—C271—H27C | 109.5 |
| H6A—C6—H6B | 107.3 | H27B—C271—H27C | 109.5 |
| | | |
| C7i—N1—C2—C3 | −172.3 (2) | C27i—N21—C22—C23 | −84 (2) |
| N1—C2—C3—N4 | −68.2 (3) | N21—C22—C23—N24 | −67 (2) |
| C2—C3—N4—C5 | 176.3 (4) | C22—C23—N24—C25 | −164 (3) |
| C3—N4—C5—C6 | −175.5 (4) | C23—N24—C25—C26 | −168 (4) |
| N4—C5—C6—C7 | 66.8 (6) | N24—C25—C26—C27 | 33 (4) |
| C5—C6—C7—N1i | −60.9 (6) | C25—C26—C27—N21i | 37 (3) |
| C6—C7—N1i—C2i | 173.2 (3) | C26—C27—N21i—C22i | 178 (2) |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···O4ii | 0.84 | 1.71 | 2.5063 (15) | 156 |
| N4—H4A···N1i | 0.92 | 2.02 | 2.764 (4) | 137 |
| N1—H1···O4i | 0.92 | 2.48 | 3.3025 (18) | 149 |
| N1—H1···O2iii | 0.92 | 2.53 | 3.3554 (19) | 149 |
| N4—H4B···O3 | 0.92 | 1.81 | 2.718 (5) | 169 |
| N24—H24A···N21i | 0.92 | 2.25 | 3.03 (3) | 141 |
| N21—H21···O6iv | 0.92 | 2.20 | 3.009 (8) | 146 |
| N24—H24B···O3 | 0.92 | 1.91 | 2.82 (3) | 171 |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x+1, y, z; (iii) −x+2, −y+1, −z+1; (iv) −x+2, −y+2, −z. |
Experimental details
| Crystal data |
| Chemical formula | C16H38N42+·2C8H4NO6− |
| Mr | 706.46 |
| Crystal system, space group | Triclinic, P1 |
| Temperature (K) | 150 |
| a, b, c (Å) | 8.5183 (6), 10.2278 (8), 11.3272 (9) |
| α, β, γ (°) | 66.635 (1), 71.387 (1), 82.976 (1) |
| V (Å3) | 858.54 (11) |
| Z | 1 |
| Radiation type | Synchrotron, λ = 0.6867 Å |
| µ (mm−1) | 0.11 |
| Crystal size (mm) | 0.09 × 0.06 × 0.03 |
| |
| Data collection |
| Diffractometer | Bruker SMART 1K CCD area-detector
diffractometer |
| Absorption correction | Multi-scan
(SADABS; Bruker, 2000) |
| Tmin, Tmax | 0.991, 0.997 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 8877, 4702, 4097 |
| Rint | 0.027 |
| (sin θ/λ)max (Å−1) | 0.713 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.062, 0.165, 1.13 |
| No. of reflections | 4702 |
| No. of parameters | 262 |
| No. of restraints | 9 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.64, −0.27 |
Selected geometric parameters (Å, º) top| O1—C17 | 1.1974 (18) | N1—C2 | 1.475 (2) |
| O2—C17 | 1.3147 (18) | C3—N4 | 1.505 (2) |
| O3—C18 | 1.2497 (18) | N4—C5 | 1.530 (3) |
| O4—C18 | 1.2477 (17) | C7—N1i | 1.484 (2) |
| | | |
| C7i—N1—C2—C3 | −172.3 (2) | C27i—N21—C22—C23 | −84 (2) |
| N1—C2—C3—N4 | −68.2 (3) | N21—C22—C23—N24 | −67 (2) |
| C2—C3—N4—C5 | 176.3 (4) | C22—C23—N24—C25 | −164 (3) |
| C3—N4—C5—C6 | −175.5 (4) | C23—N24—C25—C26 | −168 (4) |
| N4—C5—C6—C7 | 66.8 (6) | N24—C25—C26—C27 | 33 (4) |
| C5—C6—C7—N1i | −60.9 (6) | C25—C26—C27—N21i | 37 (3) |
| C6—C7—N1i—C2i | 173.2 (3) | C26—C27—N21i—C22i | 178 (2) |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···O4ii | 0.84 | 1.71 | 2.5063 (15) | 156 |
| N4—H4A···N1i | 0.92 | 2.02 | 2.764 (4) | 137 |
| N1—H1···O4i | 0.92 | 2.48 | 3.3025 (18) | 149 |
| N1—H1···O2iii | 0.92 | 2.53 | 3.3554 (19) | 149 |
| N4—H4B···O3 | 0.92 | 1.81 | 2.718 (5) | 169 |
| N24—H24A···N21i | 0.92 | 2.25 | 3.03 (3) | 141 |
| N21—H21···O6iv | 0.92 | 2.20 | 3.009 (8) | 146 |
| N24—H24B···O3 | 0.92 | 1.91 | 2.82 (3) | 171 |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x+1, y, z; (iii) −x+2, −y+1, −z+1; (iv) −x+2, −y+2, −z. |
 O hydrogen bond [H
O hydrogen bond [H O 1.71 Å, O
O 1.71 Å, O O 2.5063 (15) Å and O-H
O 2.5063 (15) Å and O-H O 156°] and the cations link these anion chains into sheets by means of a range of N-H
O 156°] and the cations link these anion chains into sheets by means of a range of N-H O hydrogen bonds [H
O hydrogen bonds [H O 1.81-2.53 Å, N
O 1.81-2.53 Å, N O 2.718 (5)-3.3554 (19) Å and N-H
O 2.718 (5)-3.3554 (19) Å and N-H O 146-171°].
O 146-171°].
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2002/08/00/tr1034/tr1034fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2002/08/00/tr1034/tr1034fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2002/08/00/tr1034/tr1034fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/c/issues/2002/08/00/tr1034/tr1034fig4thm.gif)




We report here the structure of the 1:2 salt, (I), formed between the tetraaza macrocycle meso-5,5,7,12,12,14-hexa-C-methyl-1,4,8,11-tetraazacyclotetradecane (tet-a) and 5-nitroisophthalic acid. We have recently analysed the supramolecular structures of the salt-type adducts formed between tet-a and both monocarboxylic (Gregson et al., 2000) and dicarboxylic acids (Lough et al., 2000; Burchell et al., 2000). With the trigonally trisubstituted benzenecarboxylic acid 3,5-dinitrobenzoic acid, (II), tet-a forms a one-dimensional supramolecular structure, while with 5-hydroxyisophthalic acid, (III), the rather simple hydrogen-bonded array is two-dimensional (Burchell et al., 2000). With the analogous 5-nitroisophthalic acid, whose monoanion, (IV), is isoelectronic with (II), a more complex two-dimensional supramolecular structure is formed in (I). \sch
The constitution of (I) is that of a salt, [(tet-a)H2]2+·2[O2NC6H3(COOH)COO]- (Fig. 1). The cation lies across a centre of inversion, chosen for the sake of convenience as that at (1/2,1/2,1/2), while the monoanion lies in a general position. While, in general (Gregson et al., 2000; Lough et al., 2000: Burchell et al., 2000), [(tet-a)H2]2+ cations adopt the trans-III configuration (Barefield et al., 1986), in compound (I), the cation is disordered over two sets of sites, with occupancies 0.847 (3) and 0.153 (3), such that the major form (Fig. 1a) is the usual trans-III isomer, while the minor form (Fig. 1 b) is the rather uncommon trans-IV isomer. The major differences between these configurational isomers are found in the skeletal torsion angles (Table 1), and in the location of the axial N—H bonds (Figs. 1a and 1 b). In each isomer, there are two axial N—H bonds on each face of the macrocycle, but the two bonds on a common face are separated by a C3 spacer unit in the trans-III isomer and by a C2 spacer in the trans-IV isomer. Both isomers of the cation contain the usual pair of intramolecular N—H···N hydrogen bonds (Table 2) and in the major isomer, the C—N distances clearly reflect the site of protonation at N4. Similarly, in the almost planar anion, the C—O distances clearly reflect the presence of an H atom at O2 only.
The cations and anions in (I) are linked, by a number of hydrogen bonds of the O—H···O and N—H···O types (Table 2), into sheets whose formation is readily analysed by means of the substructure approach (Gregson et al., 2000). The two isomers of the cation display rather different hydrogen-bonding behaviour, despite sharing a common site (Fig. 2), consequent upon their different arrangements of the axial N—H bonds. The anions alone form a simple C(8) chain generated by translation along the [100] direction. Carboxyl atom O2 in the anion at (x,y,z) acts as a donor to the carboxylate atom O4 in the anion at (1 + x, y, z), in a hydrogen bond whose O···O distance is very short, but where the H atom is unambiguously located adjacent to one O atom, rather than being centred between the two O sites. Two antiparallel chains, related to one another by centres of inversion, run through each unit cell, and it is the pairwise linking of such chains by the cations which generates the sheet structure.
In the major isomer of the cation centred at (1/2,1/2,1/2), atoms N4 at (x,y,z) and N1 at (1 - x, 1 - y, 1 - z) act as hydrogen-bond donors to, respectively, O3 in the anion at (x,y,z) and O2 in the anion at (x - 1, y, z), both of which are components of the anion chain lying approximately along the line (x, 1.0, 0.1). The symmetry-related N4 atom at (1 - x, 1 - y, 1 - z) and N1 at (x,y,z) in the same cation similarly act as donors to O3 and O2 in the anions at (1 - x, 1 - y, 1 - z) and (2 - x, 1 - y, 1 - z), respectively, which are components of the chain lying approximately along the line (-x, 0, 0.9). In this way, a chain of edge-fused rings, or a molecular ladder, is formed along the line (x,1/2,1/2) (Fig. 3).
Adjacent ladders are linked into an (011) sheet by the minor isomer of the cation. In the minor isomer centred at (1/2, 1/2, 1/2), atoms N24 and N21 at (x,y,z) act as hydrogen-bond donors to, respectively, carboxylate atom O3 in the anion at (x,y,z), part of the (x,1/2,1/2) ladder, and nitro atom O6 in the anion at (2 - x, 2 - y, -z), which is a component of the ladder along (x,3/2,-1/2) (Fig. 4). The symmetry-related atoms N24 and N21 in the same cation are at (1 - x, 1 - y, 1 - z), and these act as hydrogen-bond donors to, respectively, O3 in the anion at (1 - x, 1 - y, 1 - z), which is another component of the (x, 1/2, 1/2) ladder, and atom O6 in the anion at (x - 1, y - 1, 1 + z), which lies in the ladder along (x,-1/2,3/2). Hence, the [100] ladders contain the major isomer of the cation and are linked into an (011) sheet by the minor isomer.