Buy article online - an online subscription or single-article purchase is required to access this article.

The title complexes, hexaaquacobalt(II) bis(
![[mu]](/logos/entities/mu_rmgif.gif)
-pyridine-2,6-dicarboxylato)bis[(pyridine-2,6-dicarboxylato)bismuthate(III)] dihydrate, [Co(H
2O)
6][Bi
2(C
7H
4NO
4)
4]·2H
2O, (I), and hexaaquanickel(II) bis(
![[mu]](/logos/entities/mu_rmgif.gif)
-pyridine-2,6-dicarboxylato)bis[(pyridine-2,6-dicarboxylato)bismuthate(III)] dihydrate, [Ni(H
2O)
6][Bi
2(C
7H
4NO
4)
4]·2H
2O, (II), are isomorphous and crystallize in the triclinic space group
P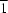
. The transition metal ions are located on the inversion centre and adopt slightly distorted
MO
6 (
M = Co or Ni) octahedral geometries. Two [Bi(pydc)
2]
- units (pydc is pyridine-2,6-dicarboxylate) are linked
via bridging carboxylate groups into centrosymmetric [Bi
2(pydc)
4]
2- dianions. The crystal packing reveals that the [
M(H
2O)
6]
2+ cations, [Bi
2(pydc)
4]
2- anions and solvent water molecules form multiple hydrogen bonds to generate a supramolecular three-dimensional network. The formation of secondary Bi

O bonds between adjacent [Bi
2(pydc)
4]
2- dimers provides an additional supramolecular synthon that directs and facilitates the crystal packing of both (I) and (II).
Supporting information
CCDC references: 819289; 819290
Pyridine-2,6-dicarboxylic acid and all other reactants were obtained
commercially. Bismuth(III) oxide (466 mg, 1.0 mmol) and
pyridine-2,6-dicarboxylic acid (668 mg, 4.0 mmol) were stirred at reflux in
water (Volume?) until the dissolution of most of the oxide. The
filtered solution was reacted with freshly prepared cobalt(II) or nickel(II)
basic carbonate, obtained upon reaction of aqueous solutions of the transition
metal nitrates with excess sodium carbonate. The solutions of the
CoII–BiIII and NiII–BiIII compounds were filtered and allowed to
stand for crystallization at ambient temperature. Light-pink crystals of (I)
and light-green crystals of (II) were obtained after 3–4 weeks.
C-bound H atoms were located in calculated positions and constrained to ride on
their parent atoms, with C—H = 0.93 Å and Uiso(H) =
1.2Ueq(C). Water H atoms were found in a difference Fourier map and
included in the refinement with the restraints O—H = 0.84 (1) Å and H···H
>= 1.33 (1) Å, and with Uiso(H) = 1.2Ueq(O).
For both compounds, data collection: XSCANS (Bruker, 1997); cell refinement: XSCANS (Bruker, 1997); data reduction: SHELXTL (Sheldrick, 2008); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008); molecular graphics: SHELXTL (Sheldrick, 2008); software used to prepare material for publication: SHELXTL (Sheldrick, 2008).
(I) hexaaquacobalt(II) bis(µ-pyridine-2,6-dicarboxylato)bis[(pyridine-2,6-
dicarboxylato)bismuthate(III)] dihydrate
top
Crystal data top
| [Co(H2O)6][Bi2(C7H4NO4)4]·2H2O | Z = 1 |
| Mr = 1281.43 | F(000) = 609 |
| Triclinic, P1 | Dx = 2.385 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.2966 (15) Å | Cell parameters from 9400 reflections |
| b = 11.184 (2) Å | θ = 2.8–28.7° |
| c = 12.004 (2) Å | µ = 10.40 mm−1 |
| α = 112.99 (3)° | T = 293 K |
| β = 90.67 (3)° | Plate, light-pink |
| γ = 97.44 (3)° | 0.21 × 0.16 × 0.11 mm |
| V = 892.1 (3) Å3 | |
Data collection top
Bruker CCD 1000 area-detector
diffractometer | 4176 independent reflections |
| Radiation source: fine-focus sealed tube | 3914 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.035 |
| ω scans | θmax = 28.5°, θmin = 2.0° |
Absorption correction: multi-scan
(SADABS; Sheldrick, 1996) | h = −9→9 |
| Tmin = 0.219, Tmax = 0.394 | k = −14→14 |
| 10685 measured reflections | l = −15→15 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.024 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.063 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0304P)2 + 1.1642P]
where P = (Fo2 + 2Fc2)/3 |
| 4176 reflections | (Δ/σ)max = 0.001 |
| 292 parameters | Δρmax = 1.10 e Å−3 |
| 12 restraints | Δρmin = −1.73 e Å−3 |
Crystal data top
| [Co(H2O)6][Bi2(C7H4NO4)4]·2H2O | γ = 97.44 (3)° |
| Mr = 1281.43 | V = 892.1 (3) Å3 |
| Triclinic, P1 | Z = 1 |
| a = 7.2966 (15) Å | Mo Kα radiation |
| b = 11.184 (2) Å | µ = 10.40 mm−1 |
| c = 12.004 (2) Å | T = 293 K |
| α = 112.99 (3)° | 0.21 × 0.16 × 0.11 mm |
| β = 90.67 (3)° | |
Data collection top
Bruker CCD 1000 area-detector
diffractometer | 4176 independent reflections |
Absorption correction: multi-scan
(SADABS; Sheldrick, 1996) | 3914 reflections with I > 2σ(I) |
| Tmin = 0.219, Tmax = 0.394 | Rint = 0.035 |
| 10685 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.024 | 12 restraints |
| wR(F2) = 0.063 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.07 | Δρmax = 1.10 e Å−3 |
| 4176 reflections | Δρmin = −1.73 e Å−3 |
| 292 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Bi1 | 0.389333 (19) | 0.311936 (13) | 0.371001 (12) | 0.02241 (6) | |
| N1 | 0.3445 (5) | 0.2102 (3) | 0.5221 (3) | 0.0242 (7) | |
| N2 | 0.5785 (5) | 0.2340 (3) | 0.2057 (3) | 0.0234 (7) | |
| O11 | 0.4431 (5) | 0.4703 (3) | 0.6070 (3) | 0.0318 (7) | |
| O12 | 0.4744 (5) | 0.4851 (3) | 0.7978 (3) | 0.0353 (7) | |
| O13 | 0.3068 (5) | 0.0871 (3) | 0.2822 (3) | 0.0331 (7) | |
| O14 | 0.2018 (6) | −0.1019 (3) | 0.2943 (3) | 0.0454 (9) | |
| O21 | 0.2421 (4) | 0.2833 (3) | 0.1716 (3) | 0.0288 (6) | |
| O22 | 0.2624 (5) | 0.2020 (4) | −0.0287 (3) | 0.0467 (9) | |
| O23 | 0.6765 (4) | 0.2977 (3) | 0.4349 (3) | 0.0325 (7) | |
| O24 | 0.9639 (5) | 0.2571 (4) | 0.3941 (3) | 0.0455 (9) | |
| C11 | 0.4371 (6) | 0.4224 (4) | 0.6881 (4) | 0.0269 (8) | |
| C12 | 0.3773 (6) | 0.2763 (4) | 0.6420 (4) | 0.0253 (8) | |
| C13 | 0.3537 (7) | 0.2131 (4) | 0.7204 (4) | 0.0319 (9) | |
| H13A | 0.3811 | 0.2596 | 0.8033 | 0.038* | |
| C14 | 0.2895 (8) | 0.0816 (5) | 0.6751 (4) | 0.0428 (12) | |
| H14A | 0.2685 | 0.0384 | 0.7271 | 0.051* | |
| C15 | 0.2561 (8) | 0.0134 (5) | 0.5511 (4) | 0.0387 (11) | |
| H15A | 0.2141 | −0.0763 | 0.5183 | 0.046* | |
| C16 | 0.2866 (6) | 0.0815 (4) | 0.4770 (4) | 0.0272 (8) | |
| C17 | 0.2610 (6) | 0.0155 (4) | 0.3405 (4) | 0.0280 (8) | |
| C21 | 0.3229 (6) | 0.2311 (4) | 0.0764 (4) | 0.0288 (9) | |
| C22 | 0.5142 (6) | 0.2002 (4) | 0.0925 (4) | 0.0259 (8) | |
| C23 | 0.6174 (6) | 0.1385 (4) | −0.0042 (4) | 0.0305 (9) | |
| H23A | 0.5724 | 0.1174 | −0.0834 | 0.037* | |
| C24 | 0.7881 (6) | 0.1089 (4) | 0.0187 (4) | 0.0331 (9) | |
| H24A | 0.8581 | 0.0655 | −0.0451 | 0.040* | |
| C25 | 0.8540 (6) | 0.1445 (4) | 0.1380 (4) | 0.0304 (9) | |
| H25A | 0.9681 | 0.1250 | 0.1556 | 0.036* | |
| C26 | 0.7457 (6) | 0.2100 (4) | 0.2303 (4) | 0.0263 (8) | |
| C27 | 0.8038 (6) | 0.2583 (4) | 0.3617 (4) | 0.0304 (9) | |
| Co1 | 0.0000 | 0.5000 | 0.0000 | 0.02622 (16) | |
| O1 | −0.2898 (5) | 0.4621 (4) | −0.0162 (3) | 0.0439 (8) | |
| H1A | −0.359 (6) | 0.463 (6) | 0.039 (3) | 0.053* | |
| H1B | −0.356 (6) | 0.471 (6) | −0.069 (4) | 0.053* | |
| O2 | −0.0030 (5) | 0.5428 (3) | −0.1541 (3) | 0.0376 (7) | |
| H2A | −0.022 (7) | 0.478 (3) | −0.221 (3) | 0.045* | |
| H2B | −0.071 (6) | 0.596 (4) | −0.159 (4) | 0.045* | |
| O3 | −0.0090 (5) | 0.6937 (3) | 0.1035 (3) | 0.0387 (8) | |
| H3A | −0.077 (6) | 0.730 (4) | 0.074 (5) | 0.046* | |
| H3B | 0.066 (6) | 0.756 (3) | 0.153 (4) | 0.046* | |
| O4 | −0.0665 (17) | 0.3260 (6) | 0.6517 (5) | 0.146 (4) | |
| H4A | −0.077 (19) | 0.250 (5) | 0.652 (10) | 0.176* | |
| H4B | −0.058 (19) | 0.315 (11) | 0.579 (3) | 0.176* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Bi1 | 0.03055 (9) | 0.02192 (8) | 0.01466 (8) | 0.00454 (5) | 0.00155 (5) | 0.00691 (6) |
| N1 | 0.0319 (17) | 0.0214 (16) | 0.0178 (16) | 0.0038 (13) | 0.0022 (13) | 0.0061 (13) |
| N2 | 0.0285 (16) | 0.0241 (16) | 0.0196 (16) | 0.0038 (12) | 0.0016 (13) | 0.0106 (13) |
| O11 | 0.0517 (19) | 0.0236 (14) | 0.0202 (14) | 0.0036 (13) | 0.0044 (13) | 0.0091 (12) |
| O12 | 0.0520 (19) | 0.0312 (16) | 0.0185 (15) | 0.0001 (14) | 0.0000 (13) | 0.0071 (13) |
| O13 | 0.0528 (19) | 0.0251 (15) | 0.0174 (14) | 0.0006 (13) | 0.0037 (13) | 0.0057 (12) |
| O14 | 0.073 (3) | 0.0279 (17) | 0.0243 (17) | −0.0103 (16) | −0.0036 (16) | 0.0042 (14) |
| O21 | 0.0298 (14) | 0.0358 (16) | 0.0219 (14) | 0.0076 (12) | 0.0035 (11) | 0.0117 (12) |
| O22 | 0.056 (2) | 0.066 (2) | 0.0207 (16) | 0.0290 (19) | −0.0018 (15) | 0.0140 (17) |
| O23 | 0.0341 (16) | 0.0446 (18) | 0.0195 (14) | 0.0073 (13) | −0.0027 (12) | 0.0129 (13) |
| O24 | 0.0348 (17) | 0.069 (3) | 0.0341 (19) | 0.0090 (16) | −0.0046 (14) | 0.0222 (18) |
| C11 | 0.031 (2) | 0.027 (2) | 0.022 (2) | 0.0053 (15) | 0.0054 (16) | 0.0086 (16) |
| C12 | 0.0298 (19) | 0.029 (2) | 0.0170 (18) | 0.0028 (15) | 0.0035 (15) | 0.0097 (16) |
| C13 | 0.049 (3) | 0.030 (2) | 0.0178 (19) | 0.0012 (18) | 0.0002 (17) | 0.0115 (17) |
| C14 | 0.074 (4) | 0.034 (2) | 0.023 (2) | −0.002 (2) | 0.003 (2) | 0.017 (2) |
| C15 | 0.066 (3) | 0.023 (2) | 0.024 (2) | −0.005 (2) | −0.002 (2) | 0.0093 (18) |
| C16 | 0.035 (2) | 0.0236 (19) | 0.0190 (19) | 0.0002 (15) | −0.0008 (16) | 0.0056 (16) |
| C17 | 0.037 (2) | 0.025 (2) | 0.0194 (19) | 0.0007 (16) | −0.0006 (16) | 0.0082 (16) |
| C21 | 0.037 (2) | 0.029 (2) | 0.024 (2) | 0.0082 (17) | −0.0002 (16) | 0.0131 (17) |
| C22 | 0.033 (2) | 0.0246 (19) | 0.023 (2) | 0.0047 (15) | 0.0003 (16) | 0.0119 (16) |
| C23 | 0.044 (2) | 0.029 (2) | 0.0180 (19) | 0.0067 (17) | 0.0054 (17) | 0.0081 (16) |
| C24 | 0.036 (2) | 0.029 (2) | 0.030 (2) | 0.0059 (17) | 0.0120 (18) | 0.0062 (18) |
| C25 | 0.026 (2) | 0.032 (2) | 0.032 (2) | 0.0052 (16) | 0.0058 (17) | 0.0110 (18) |
| C26 | 0.030 (2) | 0.0255 (19) | 0.025 (2) | 0.0051 (15) | 0.0033 (16) | 0.0121 (17) |
| C27 | 0.034 (2) | 0.032 (2) | 0.029 (2) | 0.0017 (17) | −0.0024 (17) | 0.0165 (18) |
| Co1 | 0.0294 (4) | 0.0281 (4) | 0.0203 (4) | 0.0072 (3) | −0.0014 (3) | 0.0079 (3) |
| O1 | 0.0298 (16) | 0.069 (3) | 0.0351 (19) | 0.0098 (16) | 0.0007 (14) | 0.0227 (19) |
| O2 | 0.055 (2) | 0.0348 (18) | 0.0242 (16) | 0.0111 (15) | −0.0012 (14) | 0.0119 (14) |
| O3 | 0.049 (2) | 0.0292 (16) | 0.0310 (18) | 0.0143 (14) | −0.0128 (14) | 0.0023 (14) |
| O4 | 0.343 (12) | 0.050 (3) | 0.031 (3) | −0.008 (5) | −0.044 (5) | 0.013 (2) |
Geometric parameters (Å, º) top
| Bi1—O23 | 2.269 (3) | C15—C16 | 1.382 (6) |
| Bi1—O13 | 2.307 (3) | C15—H15A | 0.9300 |
| Bi1—N2 | 2.372 (3) | C16—C17 | 1.508 (6) |
| Bi1—O11i | 2.493 (3) | C21—C22 | 1.508 (6) |
| Bi1—N1 | 2.494 (3) | C22—C23 | 1.386 (6) |
| Bi1—O21 | 2.498 (3) | C23—C24 | 1.380 (7) |
| Bi1—O11 | 2.682 (3) | C23—H23A | 0.9300 |
| N1—C16 | 1.331 (5) | C24—C25 | 1.389 (7) |
| N1—C12 | 1.338 (5) | C24—H24A | 0.9300 |
| N2—C22 | 1.323 (5) | C25—C26 | 1.387 (6) |
| N2—C26 | 1.337 (5) | C25—H25A | 0.9300 |
| O11—C11 | 1.281 (5) | C26—C27 | 1.490 (6) |
| O11—Bi1i | 2.493 (3) | Co1—O3ii | 2.044 (3) |
| O12—C11 | 1.233 (5) | Co1—O3 | 2.044 (3) |
| O13—C17 | 1.272 (5) | Co1—O2ii | 2.084 (3) |
| O14—C17 | 1.223 (5) | Co1—O2 | 2.084 (3) |
| O21—C21 | 1.256 (5) | Co1—O1ii | 2.094 (3) |
| O22—C21 | 1.234 (5) | Co1—O1 | 2.094 (3) |
| O23—C27 | 1.285 (5) | O1—H1A | 0.836 (10) |
| O24—C27 | 1.230 (5) | O1—H1B | 0.835 (10) |
| C11—C12 | 1.507 (6) | O2—H2A | 0.836 (10) |
| C12—C13 | 1.381 (6) | O2—H2B | 0.837 (10) |
| C13—C14 | 1.367 (7) | O3—H3A | 0.839 (10) |
| C13—H13A | 0.9300 | O3—H3B | 0.841 (10) |
| C14—C15 | 1.383 (7) | O4—H4A | 0.840 (10) |
| C14—H14A | 0.9300 | O4—H4B | 0.839 (10) |
| | | |
| O23—Bi1—O13 | 93.19 (12) | C15—C16—C17 | 122.7 (4) |
| O23—Bi1—N2 | 68.79 (11) | O14—C17—O13 | 125.0 (4) |
| O13—Bi1—N2 | 73.57 (12) | O14—C17—C16 | 118.3 (4) |
| O23—Bi1—O11i | 79.71 (12) | O13—C17—C16 | 116.7 (4) |
| O13—Bi1—O11i | 156.24 (11) | O22—C21—O21 | 127.0 (4) |
| N2—Bi1—O11i | 82.74 (11) | O22—C21—C22 | 116.5 (4) |
| O23—Bi1—N1 | 73.56 (11) | O21—C21—C22 | 116.5 (4) |
| O13—Bi1—N1 | 67.02 (11) | N2—C22—C23 | 121.1 (4) |
| N2—Bi1—N1 | 122.67 (11) | N2—C22—C21 | 115.9 (4) |
| O11i—Bi1—N1 | 130.55 (11) | C23—C22—C21 | 123.0 (4) |
| O23—Bi1—O21 | 134.27 (10) | C24—C23—C22 | 119.1 (4) |
| O13—Bi1—O21 | 78.84 (11) | C24—C23—H23A | 120.5 |
| N2—Bi1—O21 | 65.75 (11) | C22—C23—H23A | 120.5 |
| O11i—Bi1—O21 | 89.88 (11) | C23—C24—C25 | 119.3 (4) |
| N1—Bi1—O21 | 137.88 (11) | C23—C24—H24A | 120.4 |
| O23—Bi1—O11 | 72.66 (11) | C25—C24—H24A | 120.4 |
| O13—Bi1—O11 | 129.08 (10) | C26—C25—C24 | 118.5 (4) |
| N2—Bi1—O11 | 136.25 (11) | C26—C25—H25A | 120.8 |
| O11i—Bi1—O11 | 70.63 (11) | C24—C25—H25A | 120.8 |
| N1—Bi1—O11 | 62.07 (10) | N2—C26—C25 | 121.1 (4) |
| O21—Bi1—O11 | 144.46 (10) | N2—C26—C27 | 115.1 (4) |
| C16—N1—C12 | 119.8 (4) | C25—C26—C27 | 123.7 (4) |
| C16—N1—Bi1 | 116.0 (3) | O24—C27—O23 | 124.1 (4) |
| C12—N1—Bi1 | 124.2 (3) | O24—C27—C26 | 120.3 (4) |
| C22—N2—C26 | 120.9 (4) | O23—C27—C26 | 115.6 (4) |
| C22—N2—Bi1 | 121.7 (3) | O3ii—Co1—O3 | 180.0 (3) |
| C26—N2—Bi1 | 116.9 (3) | O3ii—Co1—O2ii | 88.90 (14) |
| C11—O11—Bi1i | 126.3 (3) | O3—Co1—O2ii | 91.10 (14) |
| C11—O11—Bi1 | 120.5 (3) | O3ii—Co1—O2 | 91.10 (14) |
| Bi1i—O11—Bi1 | 109.37 (11) | O3—Co1—O2 | 88.90 (14) |
| C17—O13—Bi1 | 124.3 (3) | O2ii—Co1—O2 | 180.00 (17) |
| C21—O21—Bi1 | 119.5 (3) | O3ii—Co1—O1ii | 90.42 (16) |
| C27—O23—Bi1 | 122.6 (3) | O3—Co1—O1ii | 89.58 (16) |
| O12—C11—O11 | 125.7 (4) | O2ii—Co1—O1ii | 89.79 (15) |
| O12—C11—C12 | 118.9 (4) | O2—Co1—O1ii | 90.21 (15) |
| O11—C11—C12 | 115.4 (4) | O3ii—Co1—O1 | 89.58 (16) |
| N1—C12—C13 | 121.1 (4) | O3—Co1—O1 | 90.42 (16) |
| N1—C12—C11 | 117.4 (4) | O2ii—Co1—O1 | 90.21 (15) |
| C13—C12—C11 | 121.4 (4) | O2—Co1—O1 | 89.79 (15) |
| C14—C13—C12 | 119.4 (4) | O1ii—Co1—O1 | 180.0 |
| C14—C13—H13A | 120.3 | Co1—O1—H1A | 126 (4) |
| C12—C13—H13A | 120.3 | Co1—O1—H1B | 124 (3) |
| C13—C14—C15 | 119.3 (4) | H1A—O1—H1B | 105.9 (17) |
| C13—C14—H14A | 120.3 | Co1—O2—H2A | 116 (4) |
| C15—C14—H14A | 120.3 | Co1—O2—H2B | 119 (4) |
| C16—C15—C14 | 118.6 (4) | H2A—O2—H2B | 105.6 (17) |
| C16—C15—H15A | 120.7 | Co1—O3—H3A | 115 (3) |
| C14—C15—H15A | 120.7 | Co1—O3—H3B | 135 (3) |
| N1—C16—C15 | 121.7 (4) | H3A—O3—H3B | 104.3 (16) |
| N1—C16—C17 | 115.6 (4) | H4A—O4—H4B | 105.0 (17) |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x, −y+1, −z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···O12iii | 0.84 (1) | 2.04 (2) | 2.848 (5) | 163 (6) |
| O1—H1B···O12iv | 0.84 (1) | 2.08 (1) | 2.913 (5) | 177 (6) |
| O2—H2A···O4v | 0.84 (1) | 1.78 (1) | 2.607 (6) | 171 (6) |
| O2—H2B···O21ii | 0.84 (1) | 2.00 (1) | 2.839 (5) | 178 (5) |
| O3—H3A···O22ii | 0.84 (1) | 1.81 (2) | 2.631 (5) | 167 (6) |
| O3—H3B···O14vi | 0.84 (1) | 1.96 (2) | 2.785 (5) | 167 (6) |
| O4—H4A···O14vii | 0.84 (1) | 2.11 (6) | 2.885 (7) | 153 (12) |
| O4—H4B···O24viii | 0.84 (1) | 2.07 (2) | 2.898 (7) | 171 (11) |
| Symmetry codes: (ii) −x, −y+1, −z; (iii) −x, −y+1, −z+1; (iv) x−1, y, z−1; (v) x, y, z−1; (vi) x, y+1, z; (vii) −x, −y, −z+1; (viii) x−1, y, z. |
(II) hexaaquacobalt(II) bis(µ-pyridine-2,6-dicarboxylato)bis[(pyridine-2,6-
dicarboxylato)nickelate(III)] dihydrate
top
Crystal data top
| [Ni(H2O)6][Bi2(C7H4NO4)4]·2H2O | Z = 1 |
| Mr = 1281.21 | F(000) = 610 |
| Triclinic, P1 | Dx = 2.388 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.2615 (15) Å | Cell parameters from 8595 reflections |
| b = 11.210 (2) Å | θ = 2.8–28.3° |
| c = 12.014 (2) Å | µ = 10.48 mm−1 |
| α = 112.96 (3)° | T = 293 K |
| β = 90.79 (3)° | Plate, light-green |
| γ = 97.33 (3)° | 0.17 × 0.14 × 0.12 mm |
| V = 891.0 (3) Å3 | |
Data collection top
Bruker CCD 1000 area-detector
diffractometer | 4214 independent reflections |
| Radiation source: fine-focus sealed tube | 3837 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.030 |
| ω scans | θmax = 28.3°, θmin = 1.9° |
Absorption correction: multi-scan
(SADABS; Sheldrick, 1996) | h = −9→9 |
| Tmin = 0.269, Tmax = 0.366 | k = −14→14 |
| 10920 measured reflections | l = −15→15 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.021 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.050 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0218P)2 + 0.3727P]
where P = (Fo2 + 2Fc2)/3 |
| 4214 reflections | (Δ/σ)max = 0.001 |
| 292 parameters | Δρmax = 1.06 e Å−3 |
| 12 restraints | Δρmin = −0.85 e Å−3 |
Crystal data top
| [Ni(H2O)6][Bi2(C7H4NO4)4]·2H2O | γ = 97.33 (3)° |
| Mr = 1281.21 | V = 891.0 (3) Å3 |
| Triclinic, P1 | Z = 1 |
| a = 7.2615 (15) Å | Mo Kα radiation |
| b = 11.210 (2) Å | µ = 10.48 mm−1 |
| c = 12.014 (2) Å | T = 293 K |
| α = 112.96 (3)° | 0.17 × 0.14 × 0.12 mm |
| β = 90.79 (3)° | |
Data collection top
Bruker CCD 1000 area-detector
diffractometer | 4214 independent reflections |
Absorption correction: multi-scan
(SADABS; Sheldrick, 1996) | 3837 reflections with I > 2σ(I) |
| Tmin = 0.269, Tmax = 0.366 | Rint = 0.030 |
| 10920 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.021 | 12 restraints |
| wR(F2) = 0.050 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | Δρmax = 1.06 e Å−3 |
| 4214 reflections | Δρmin = −0.85 e Å−3 |
| 292 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Bi1 | 0.388113 (17) | 0.312794 (11) | 0.371459 (11) | 0.02065 (5) | |
| N1 | 0.3443 (4) | 0.2100 (3) | 0.5218 (3) | 0.0220 (6) | |
| N2 | 0.5790 (4) | 0.2347 (3) | 0.2063 (3) | 0.0203 (6) | |
| O11 | 0.4418 (4) | 0.4700 (2) | 0.6081 (2) | 0.0299 (6) | |
| O12 | 0.4719 (4) | 0.4843 (3) | 0.7984 (2) | 0.0341 (6) | |
| O13 | 0.3067 (4) | 0.0883 (2) | 0.2805 (2) | 0.0316 (6) | |
| O14 | 0.2033 (5) | −0.1015 (3) | 0.2917 (3) | 0.0442 (8) | |
| O21 | 0.2411 (3) | 0.2857 (2) | 0.1712 (2) | 0.0269 (5) | |
| O22 | 0.2625 (4) | 0.2032 (3) | −0.0288 (3) | 0.0465 (8) | |
| O23 | 0.6780 (4) | 0.2988 (3) | 0.4358 (2) | 0.0311 (6) | |
| O24 | 0.9667 (4) | 0.2579 (3) | 0.3959 (3) | 0.0431 (7) | |
| C11 | 0.4356 (5) | 0.4221 (3) | 0.6886 (3) | 0.0232 (7) | |
| C12 | 0.3764 (5) | 0.2756 (3) | 0.6416 (3) | 0.0231 (7) | |
| C13 | 0.3529 (6) | 0.2119 (4) | 0.7198 (3) | 0.0319 (8) | |
| H13A | 0.3797 | 0.2577 | 0.8028 | 0.038* | |
| C14 | 0.2898 (7) | 0.0810 (4) | 0.6733 (4) | 0.0428 (11) | |
| H14A | 0.2699 | 0.0373 | 0.7248 | 0.051* | |
| C15 | 0.2553 (6) | 0.0131 (4) | 0.5498 (4) | 0.0380 (10) | |
| H15A | 0.2125 | −0.0763 | 0.5171 | 0.046* | |
| C16 | 0.2860 (5) | 0.0812 (3) | 0.4756 (3) | 0.0248 (7) | |
| C17 | 0.2612 (5) | 0.0158 (4) | 0.3397 (3) | 0.0278 (8) | |
| C21 | 0.3233 (5) | 0.2324 (4) | 0.0757 (3) | 0.0257 (7) | |
| C22 | 0.5159 (5) | 0.2006 (3) | 0.0925 (3) | 0.0221 (7) | |
| C23 | 0.6191 (6) | 0.1392 (4) | −0.0034 (3) | 0.0293 (8) | |
| H23A | 0.5747 | 0.1186 | −0.0827 | 0.035* | |
| C24 | 0.7902 (5) | 0.1090 (4) | 0.0206 (3) | 0.0312 (8) | |
| H24A | 0.8606 | 0.0651 | −0.0427 | 0.037* | |
| C25 | 0.8557 (5) | 0.1447 (4) | 0.1398 (4) | 0.0285 (8) | |
| H25A | 0.9699 | 0.1247 | 0.1577 | 0.034* | |
| C26 | 0.7483 (5) | 0.2104 (3) | 0.2310 (3) | 0.0220 (7) | |
| C27 | 0.8054 (5) | 0.2587 (4) | 0.3627 (3) | 0.0267 (8) | |
| Ni1 | 0.0000 | 0.5000 | 0.0000 | 0.02368 (13) | |
| O1 | −0.2866 (4) | 0.4586 (3) | −0.0175 (3) | 0.0400 (7) | |
| H1A | −0.345 (5) | 0.467 (5) | 0.043 (2) | 0.048* | |
| H1B | −0.358 (5) | 0.470 (5) | −0.066 (3) | 0.048* | |
| O2 | 0.0047 (4) | 0.5436 (3) | −0.1505 (2) | 0.0341 (6) | |
| H2A | −0.030 (5) | 0.476 (2) | −0.213 (3) | 0.041* | |
| H2B | −0.072 (5) | 0.594 (3) | −0.149 (4) | 0.041* | |
| O3 | −0.0126 (4) | 0.6901 (3) | 0.1053 (3) | 0.0363 (7) | |
| H3A | −0.083 (4) | 0.731 (3) | 0.082 (4) | 0.044* | |
| H3B | 0.079 (4) | 0.745 (3) | 0.142 (4) | 0.044* | |
| O4 | −0.0812 (13) | 0.3289 (4) | 0.6558 (4) | 0.144 (3) | |
| H4A | −0.080 (15) | 0.251 (3) | 0.646 (8) | 0.173* | |
| H4B | −0.067 (14) | 0.332 (8) | 0.588 (4) | 0.173* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Bi1 | 0.02727 (7) | 0.02004 (7) | 0.01416 (7) | 0.00388 (5) | 0.00127 (5) | 0.00609 (5) |
| N1 | 0.0271 (15) | 0.0231 (14) | 0.0161 (14) | 0.0029 (12) | 0.0014 (11) | 0.0084 (12) |
| N2 | 0.0248 (14) | 0.0184 (13) | 0.0178 (14) | 0.0024 (11) | 0.0016 (11) | 0.0074 (11) |
| O11 | 0.0475 (16) | 0.0214 (12) | 0.0191 (13) | 0.0008 (11) | 0.0042 (11) | 0.0075 (10) |
| O12 | 0.0497 (17) | 0.0287 (14) | 0.0189 (13) | 0.0003 (12) | 0.0004 (12) | 0.0057 (11) |
| O13 | 0.0497 (17) | 0.0237 (13) | 0.0187 (13) | 0.0008 (12) | 0.0047 (11) | 0.0067 (11) |
| O14 | 0.072 (2) | 0.0229 (14) | 0.0285 (15) | −0.0125 (14) | −0.0055 (14) | 0.0062 (12) |
| O21 | 0.0273 (13) | 0.0330 (14) | 0.0226 (13) | 0.0071 (10) | 0.0022 (10) | 0.0123 (11) |
| O22 | 0.0536 (18) | 0.066 (2) | 0.0228 (15) | 0.0286 (16) | −0.0023 (13) | 0.0152 (15) |
| O23 | 0.0325 (14) | 0.0416 (15) | 0.0189 (13) | 0.0071 (12) | 0.0004 (11) | 0.0112 (12) |
| O24 | 0.0299 (15) | 0.064 (2) | 0.0345 (17) | 0.0088 (14) | −0.0063 (12) | 0.0184 (15) |
| C11 | 0.0268 (17) | 0.0269 (18) | 0.0171 (17) | 0.0048 (14) | 0.0037 (13) | 0.0097 (14) |
| C12 | 0.0286 (17) | 0.0256 (17) | 0.0157 (16) | 0.0020 (14) | 0.0024 (13) | 0.0092 (14) |
| C13 | 0.048 (2) | 0.030 (2) | 0.0167 (17) | 0.0006 (17) | 0.0026 (16) | 0.0098 (15) |
| C14 | 0.071 (3) | 0.034 (2) | 0.028 (2) | −0.003 (2) | 0.002 (2) | 0.0196 (19) |
| C15 | 0.060 (3) | 0.0222 (19) | 0.030 (2) | −0.0047 (18) | 0.0011 (19) | 0.0115 (17) |
| C16 | 0.0313 (18) | 0.0220 (17) | 0.0200 (17) | 0.0005 (14) | 0.0006 (14) | 0.0081 (14) |
| C17 | 0.0336 (19) | 0.0252 (18) | 0.0220 (18) | 0.0013 (15) | 0.0029 (15) | 0.0074 (15) |
| C21 | 0.0321 (18) | 0.0259 (18) | 0.0221 (18) | 0.0061 (14) | −0.0014 (15) | 0.0123 (15) |
| C22 | 0.0305 (18) | 0.0191 (16) | 0.0163 (16) | 0.0023 (13) | 0.0026 (13) | 0.0069 (13) |
| C23 | 0.042 (2) | 0.0289 (19) | 0.0149 (17) | 0.0047 (16) | 0.0045 (15) | 0.0068 (15) |
| C24 | 0.036 (2) | 0.0277 (19) | 0.0255 (19) | 0.0064 (16) | 0.0119 (16) | 0.0049 (16) |
| C25 | 0.0277 (18) | 0.0259 (18) | 0.032 (2) | 0.0050 (15) | 0.0035 (15) | 0.0111 (16) |
| C26 | 0.0235 (16) | 0.0193 (16) | 0.0231 (18) | 0.0006 (13) | 0.0018 (13) | 0.0088 (14) |
| C27 | 0.0316 (19) | 0.0274 (18) | 0.0228 (18) | 0.0035 (15) | −0.0005 (15) | 0.0120 (15) |
| Ni1 | 0.0257 (3) | 0.0256 (3) | 0.0189 (3) | 0.0061 (2) | −0.0012 (2) | 0.0073 (3) |
| O1 | 0.0290 (14) | 0.064 (2) | 0.0296 (16) | 0.0088 (14) | 0.0014 (12) | 0.0212 (15) |
| O2 | 0.0460 (17) | 0.0341 (15) | 0.0225 (14) | 0.0121 (13) | −0.0003 (12) | 0.0098 (12) |
| O3 | 0.0440 (16) | 0.0268 (14) | 0.0314 (16) | 0.0103 (12) | −0.0123 (13) | 0.0032 (12) |
| O4 | 0.333 (9) | 0.051 (3) | 0.034 (2) | −0.008 (4) | −0.058 (4) | 0.013 (2) |
Geometric parameters (Å, º) top
| Bi1—O23 | 2.277 (3) | C15—C16 | 1.385 (5) |
| Bi1—O13 | 2.310 (3) | C15—H15A | 0.9300 |
| Bi1—N2 | 2.378 (3) | C16—C17 | 1.503 (5) |
| Bi1—N1 | 2.497 (3) | C21—C22 | 1.517 (5) |
| Bi1—O11i | 2.504 (3) | C22—C23 | 1.379 (5) |
| Bi1—O21 | 2.510 (3) | C23—C24 | 1.384 (6) |
| Bi1—O11 | 2.688 (3) | C23—H23A | 0.9300 |
| N1—C16 | 1.337 (4) | C24—C25 | 1.386 (5) |
| N1—C12 | 1.337 (4) | C24—H24A | 0.9300 |
| N2—C22 | 1.325 (4) | C25—C26 | 1.375 (5) |
| N2—C26 | 1.346 (4) | C25—H25A | 0.9300 |
| O11—C11 | 1.275 (4) | C26—C27 | 1.492 (5) |
| O11—Bi1i | 2.504 (3) | Ni1—O3ii | 2.024 (3) |
| O12—C11 | 1.234 (4) | Ni1—O3 | 2.024 (3) |
| O13—C17 | 1.289 (4) | Ni1—O2ii | 2.049 (3) |
| O14—C17 | 1.225 (4) | Ni1—O2 | 2.049 (3) |
| O21—C21 | 1.266 (4) | Ni1—O1 | 2.063 (3) |
| O22—C21 | 1.227 (4) | Ni1—O1ii | 2.063 (3) |
| O23—C27 | 1.285 (4) | O1—H1A | 0.832 (10) |
| O24—C27 | 1.235 (4) | O1—H1B | 0.835 (10) |
| C11—C12 | 1.514 (5) | O2—H2A | 0.839 (10) |
| C12—C13 | 1.386 (5) | O2—H2B | 0.838 (10) |
| C13—C14 | 1.363 (6) | O3—H3A | 0.839 (10) |
| C13—H13A | 0.9300 | O3—H3B | 0.835 (10) |
| C14—C15 | 1.380 (6) | O4—H4A | 0.835 (10) |
| C14—H14A | 0.9300 | O4—H4B | 0.837 (10) |
| | | |
| O23—Bi1—O13 | 93.28 (10) | C15—C16—C17 | 122.8 (3) |
| O23—Bi1—N2 | 68.68 (9) | O14—C17—O13 | 123.9 (4) |
| O13—Bi1—N2 | 72.91 (10) | O14—C17—C16 | 119.2 (3) |
| O23—Bi1—N1 | 73.46 (10) | O13—C17—C16 | 116.9 (3) |
| O13—Bi1—N1 | 67.39 (9) | O22—C21—O21 | 126.7 (3) |
| N2—Bi1—N1 | 122.22 (9) | O22—C21—C22 | 116.8 (3) |
| O23—Bi1—O11i | 79.49 (10) | O21—C21—C22 | 116.5 (3) |
| O13—Bi1—O11i | 155.11 (9) | N2—C22—C23 | 121.7 (3) |
| N2—Bi1—O11i | 82.30 (9) | N2—C22—C21 | 115.5 (3) |
| N1—Bi1—O11i | 131.06 (9) | C23—C22—C21 | 122.8 (3) |
| O23—Bi1—O21 | 134.06 (9) | C22—C23—C24 | 118.6 (3) |
| O13—Bi1—O21 | 78.75 (9) | C22—C23—H23A | 120.7 |
| N2—Bi1—O21 | 65.72 (9) | C24—C23—H23A | 120.7 |
| N1—Bi1—O21 | 138.26 (9) | C23—C24—C25 | 119.5 (3) |
| O11i—Bi1—O21 | 89.20 (9) | C23—C24—H24A | 120.3 |
| O23—Bi1—O11 | 72.58 (9) | C25—C24—H24A | 120.3 |
| O13—Bi1—O11 | 129.42 (9) | C26—C25—C24 | 118.6 (3) |
| N2—Bi1—O11 | 136.28 (9) | C26—C25—H25A | 120.7 |
| N1—Bi1—O11 | 62.04 (8) | C24—C25—H25A | 120.7 |
| O11i—Bi1—O11 | 71.33 (9) | N2—C26—C25 | 121.3 (3) |
| O21—Bi1—O11 | 144.48 (8) | N2—C26—C27 | 114.6 (3) |
| C16—N1—C12 | 120.1 (3) | C25—C26—C27 | 124.1 (3) |
| C16—N1—Bi1 | 115.8 (2) | O24—C27—O23 | 123.8 (3) |
| C12—N1—Bi1 | 124.1 (2) | O24—C27—C26 | 120.1 (3) |
| C22—N2—C26 | 120.2 (3) | O23—C27—C26 | 116.1 (3) |
| C22—N2—Bi1 | 122.2 (2) | O3ii—Ni1—O3 | 180.0 (2) |
| C26—N2—Bi1 | 117.1 (2) | O3ii—Ni1—O2ii | 89.91 (12) |
| C11—O11—Bi1i | 126.7 (2) | O3—Ni1—O2ii | 90.09 (12) |
| C11—O11—Bi1 | 120.7 (2) | O3ii—Ni1—O2 | 90.09 (12) |
| Bi1i—O11—Bi1 | 108.67 (9) | O3—Ni1—O2 | 89.91 (12) |
| C17—O13—Bi1 | 123.6 (2) | O2ii—Ni1—O2 | 180.00 (14) |
| C21—O21—Bi1 | 119.2 (2) | O3ii—Ni1—O1 | 89.10 (13) |
| C27—O23—Bi1 | 122.6 (2) | O3—Ni1—O1 | 90.90 (13) |
| O12—C11—O11 | 125.9 (3) | O2ii—Ni1—O1 | 88.32 (12) |
| O12—C11—C12 | 118.8 (3) | O2—Ni1—O1 | 91.68 (12) |
| O11—C11—C12 | 115.3 (3) | O3ii—Ni1—O1ii | 90.90 (13) |
| N1—C12—C13 | 121.1 (3) | O3—Ni1—O1ii | 89.10 (13) |
| N1—C12—C11 | 117.6 (3) | O2ii—Ni1—O1ii | 91.68 (12) |
| C13—C12—C11 | 121.3 (3) | O2—Ni1—O1ii | 88.32 (12) |
| C14—C13—C12 | 118.9 (4) | O1—Ni1—O1ii | 180.0 |
| C14—C13—H13A | 120.5 | Ni1—O1—H1A | 120 (3) |
| C12—C13—H13A | 120.5 | Ni1—O1—H1B | 127 (3) |
| C13—C14—C15 | 120.1 (4) | H1A—O1—H1B | 106.1 (16) |
| C13—C14—H14A | 119.9 | Ni1—O2—H2A | 110 (3) |
| C15—C14—H14A | 119.9 | Ni1—O2—H2B | 112 (3) |
| C14—C15—C16 | 118.4 (4) | H2A—O2—H2B | 104.6 (16) |
| C14—C15—H15A | 120.8 | Ni1—O3—H3A | 119 (3) |
| C16—C15—H15A | 120.8 | Ni1—O3—H3B | 125 (3) |
| N1—C16—C15 | 121.2 (3) | H3A—O3—H3B | 105.1 (16) |
| N1—C16—C17 | 115.9 (3) | H4A—O4—H4B | 105.8 (17) |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x, −y+1, −z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···O12iii | 0.83 (1) | 2.03 (1) | 2.856 (4) | 172 (5) |
| O1—H1B···O12iv | 0.84 (1) | 2.10 (1) | 2.927 (4) | 174 (4) |
| O2—H2A···O4v | 0.84 (1) | 1.77 (1) | 2.608 (5) | 175 (4) |
| O2—H2B···O21ii | 0.84 (1) | 2.02 (1) | 2.852 (4) | 170 (4) |
| O3—H3A···O22ii | 0.84 (1) | 1.80 (1) | 2.634 (4) | 170 (4) |
| O3—H3B···O14vi | 0.84 (1) | 2.04 (3) | 2.796 (4) | 151 (4) |
| O4—H4A···O14vii | 0.84 (1) | 2.19 (6) | 2.895 (6) | 142 (8) |
| O4—H4B···O24viii | 0.84 (1) | 2.15 (4) | 2.944 (6) | 157 (8) |
| Symmetry codes: (ii) −x, −y+1, −z; (iii) −x, −y+1, −z+1; (iv) x−1, y, z−1; (v) x, y, z−1; (vi) x, y+1, z; (vii) −x, −y, −z+1; (viii) x−1, y, z. |
Experimental details
| | (I) | (II) |
| Crystal data |
| Chemical formula | [Co(H2O)6][Bi2(C7H4NO4)4]·2H2O | [Ni(H2O)6][Bi2(C7H4NO4)4]·2H2O |
| Mr | 1281.43 | 1281.21 |
| Crystal system, space group | Triclinic, P1 | Triclinic, P1 |
| Temperature (K) | 293 | 293 |
| a, b, c (Å) | 7.2966 (15), 11.184 (2), 12.004 (2) | 7.2615 (15), 11.210 (2), 12.014 (2) |
| α, β, γ (°) | 112.99 (3), 90.67 (3), 97.44 (3) | 112.96 (3), 90.79 (3), 97.33 (3) |
| V (Å3) | 892.1 (3) | 891.0 (3) |
| Z | 1 | 1 |
| Radiation type | Mo Kα | Mo Kα |
| µ (mm−1) | 10.40 | 10.48 |
| Crystal size (mm) | 0.21 × 0.16 × 0.11 | 0.17 × 0.14 × 0.12 |
| |
| Data collection |
| Diffractometer | Bruker CCD 1000 area-detector
diffractometer | Bruker CCD 1000 area-detector
diffractometer |
| Absorption correction | Multi-scan
(SADABS; Sheldrick, 1996) | Multi-scan
(SADABS; Sheldrick, 1996) |
| Tmin, Tmax | 0.219, 0.394 | 0.269, 0.366 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 10685, 4176, 3914 | 10920, 4214, 3837 |
| Rint | 0.035 | 0.030 |
| (sin θ/λ)max (Å−1) | 0.672 | 0.667 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.024, 0.063, 1.07 | 0.021, 0.050, 1.06 |
| No. of reflections | 4176 | 4214 |
| No. of parameters | 292 | 292 |
| No. of restraints | 12 | 12 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 1.10, −1.73 | 1.06, −0.85 |
Hydrogen-bond geometry (Å, º) for (I) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···O12i | 0.836 (10) | 2.04 (2) | 2.848 (5) | 163 (6) |
| O1—H1B···O12ii | 0.835 (10) | 2.079 (12) | 2.913 (5) | 177 (6) |
| O2—H2A···O4iii | 0.836 (10) | 1.778 (14) | 2.607 (6) | 171 (6) |
| O2—H2B···O21iv | 0.837 (10) | 2.002 (11) | 2.839 (5) | 178 (5) |
| O3—H3A···O22iv | 0.839 (10) | 1.807 (17) | 2.631 (5) | 167 (6) |
| O3—H3B···O14v | 0.841 (10) | 1.958 (17) | 2.785 (5) | 167 (6) |
| O4—H4A···O14vi | 0.840 (10) | 2.11 (6) | 2.885 (7) | 153 (12) |
| O4—H4B···O24vii | 0.839 (10) | 2.07 (2) | 2.898 (7) | 171 (11) |
| Symmetry codes: (i) −x, −y+1, −z+1; (ii) x−1, y, z−1; (iii) x, y, z−1; (iv) −x, −y+1, −z; (v) x, y+1, z; (vi) −x, −y, −z+1; (vii) x−1, y, z. |
Hydrogen-bond geometry (Å, º) for (II) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···O12i | 0.832 (10) | 2.030 (13) | 2.856 (4) | 172 (5) |
| O1—H1B···O12ii | 0.835 (10) | 2.095 (11) | 2.927 (4) | 174 (4) |
| O2—H2A···O4iii | 0.839 (10) | 1.771 (11) | 2.608 (5) | 175 (4) |
| O2—H2B···O21iv | 0.838 (10) | 2.023 (13) | 2.852 (4) | 170 (4) |
| O3—H3A···O22iv | 0.839 (10) | 1.803 (11) | 2.634 (4) | 170 (4) |
| O3—H3B···O14v | 0.835 (10) | 2.04 (3) | 2.796 (4) | 151 (4) |
| O4—H4A···O14vi | 0.835 (10) | 2.19 (6) | 2.895 (6) | 142 (8) |
| O4—H4B···O24vii | 0.837 (10) | 2.15 (4) | 2.944 (6) | 157 (8) |
| Symmetry codes: (i) −x, −y+1, −z+1; (ii) x−1, y, z−1; (iii) x, y, z−1; (iv) −x, −y+1, −z; (v) x, y+1, z; (vi) −x, −y, −z+1; (vii) x−1, y, z. |
Selected geometric parameters for (I) and (II) (Å, °) top| Bond/angle | (I) | (II) |
| M1—O1 | 2.093 (3) | 2.063 (3) |
| M1—O2 | 2.083 (3) | 2.047 (3) |
| M1—O3 | 2.045 (3) | 2.023 (3) |
| Bi1—N1 | 2.495 (3) | 2.497 (3) |
| Bi1—N2 | 2.373 (3) | 2.376 (3) |
| Bi1—O11 | 2.682 (3) | 2.687 (3) |
| Bi1—O13 | 2.306 (3) | 2.308 (2) |
| Bi1—O21 | 2.497 (3) | 2.510 (2) |
| Bi1—O23 | 2.269 (3) | 2.278 (2) |
| Bi1—O11i | 2.492 (3) | 2.505 (2) |
| Bi—O24ii | 3.121 (4) | 3.084 (3) |
| | |
| N1–Bi1–O11 | 62.1 (1) | 62.03 (9) |
| N1–Bi1–O13 | 67.0 (1) | 67.37 (9) |
| N1–Bi1–O23 | 73.6 (1) | 73.46 (9) |
| N2–Bi1–O21 | 65.7 (1) | 65.71 (8) |
| N2–Bi1–O23 | 68.8 (1) | 68.70 (9) |
| O11–Bi1–O23 | 72.7 (1) | 79.50 (9) |
| O13–Bi1–O21 | 78.9 (1) | 78.78 (9) |
| O13–Bi1–O23 | 93.2 (1) | 93.2 (1) |
| Note: M = Co or Ni.
Symmetry codes: (i) -x + 1, -y + 1, -z + 1; (ii) -x, -y - 1, -z. |
Hydrogen-bond geometry (Å, °) for (I) and (II) top| D—H···A | D—H | | H···A | | D···A | | D—H···A | |
| (I) | (II) | (I) | (II) | (I) | (II) | (I) | (II) |
| O1—H1A···O12i | 0.84 (7) | 0.84 (4) | 2.03 (3) | 2.02 (2) | 163 (8) | 177 (7) | 2.845 (7) | 2.850 (6) |
| O1—H1B···O12ii | 0.84 (6) | 0.84 (5) | 2.08 (3) | 2.10 (2) | 169 (9) | 174 (8) | 2.911 (6) | 2.936 (5) |
| O2—H2A···O4 | 0.84 (5) | 0.84 (4) | 1.80 (2) | 1.79 (2) | 165 (7) | 167 (7) | 2.615 (9) | 2.616 (7) |
| O2—H2B···O21iii | 0.84 (7) | 0.83 (5) | 2.01 (2) | 2.03 (2) | 170 (6) | 167 (5) | 2.840 (6) | 2.851 (5) |
| O3—H3A···O22iii | 0.84 (6) | 0.84 (5) | 1.82 (3) | 1.79 (2) | 164 (7) | 174 (6) | 2.638 (6) | 2.627 (5) |
| O3—H3B···O14iv | 0.84 (6) | 0.84 (5) | 1.98 (3) | 2.05 (1) | 161 (8) | 158 (7) | 2.788 (8) | 2.800 (6) |
| O4—H4A···O14 | 0.84 (12) | 0.84 (8) | 2.03 (1) | 2.04 (1) | 176 (9) | 160 (8) | 2.878 (9) | 2.892 (7) |
| O4—H4B···O24ii | 0.84 (3) | 0.84 (8) | 2.07 (3) | 2.14 (2) | 171 (9) | 159 (8) | 2.905 (9) | 2.947 (7) |
| Symmetry codes: (i) x, y - 1, z - 1; (ii) -x + 1, -y, -z + 1;
(iii) x, y - 1, z; (iv) -x, -y - 1, -z.
Table added in Chester from old Fig. 3. Data not checked. In bond 8, H3B
changed to H4B. Please check carefully. |

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.
![[mu]](/logos/entities/mu_rmgif.gif) -pyridine-2,6-dicarboxylato)bis[(pyridine-2,6-dicarboxylato)bismuthate(III)] dihydrate, [Co(H2O)6][Bi2(C7H4NO4)4]·2H2O, (I), and hexaaquanickel(II) bis(
-pyridine-2,6-dicarboxylato)bis[(pyridine-2,6-dicarboxylato)bismuthate(III)] dihydrate, [Co(H2O)6][Bi2(C7H4NO4)4]·2H2O, (I), and hexaaquanickel(II) bis(![[mu]](/logos/entities/mu_rmgif.gif) -pyridine-2,6-dicarboxylato)bis[(pyridine-2,6-dicarboxylato)bismuthate(III)] dihydrate, [Ni(H2O)6][Bi2(C7H4NO4)4]·2H2O, (II), are isomorphous and crystallize in the triclinic space group P
-pyridine-2,6-dicarboxylato)bis[(pyridine-2,6-dicarboxylato)bismuthate(III)] dihydrate, [Ni(H2O)6][Bi2(C7H4NO4)4]·2H2O, (II), are isomorphous and crystallize in the triclinic space group P O bonds between adjacent [Bi2(pydc)4]2- dimers provides an additional supramolecular synthon that directs and facilitates the crystal packing of both (I) and (II).
O bonds between adjacent [Bi2(pydc)4]2- dimers provides an additional supramolecular synthon that directs and facilitates the crystal packing of both (I) and (II).
 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2011/03/00/sq3274/sq3274fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2011/03/00/sq3274/sq3274fig2thm.gif)

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry



Compounds of bismuth(III) with aminopolycarboxylate ligands have been of interest over the past few decades, mainly due to their high stability in aqueous solutions and their rich structural chemistry (Stavila et al., 2006). The bismuth(III) centre is highly acidic and can achieve coordination numbers as high as 10 (Briand & Burford, 2000; Stavila et al., 2006). Pyridine-2,6-dicarboxylic acid (H2pydc) is a versatile chelating ligand and its metal complexes have been intensively explored in the context of their biological activity (Hwang et al., 2003) and rich coordination chemistry (Serezhkin et al., 2009). Bismuth(III) is known to form stable chelate complexes with H2pydc (or its anions), and several crystal structures have been reported to date (Agbabozorg et al., 2008; Ranjbar et al., 2003, 2001; Sheshmani et al., 2005). In these structures, H2pydc acts as an O,N,O-donor ligand to the metal ion to form five-membered metallocycles. This high coordination capacity, coupled with the ability to link to metal centres in a variety of bridging modes, makes aminopolycarboxylate ligands attractive for the assembly of mixed-metal complexes. Previously, we have described several transition metal–bismuth complexes based on ethylenediaminetetraacetate and nitrilotriacetate ligands (Bachman et al., 2003; Stavila et al., 2000, 2002, 2003; Stavila, Gulea, Shova et al. 2004; Stavila, Gulea, Popa et al., 2004). Here, we report the crystal structures of the first transition metal–bismuth pyridine-2,6-dicarboxylates, namely hexaaquacobalt(II) bis(µ-pyridine-2,6-dicarboxylato)bis[(pyridine-2,6-dicarboxylato)bismuthate(III)] dihydrate, (I), and hexaaquanickel(II) bis(µ-pyridine-2,6-dicarboxylato)bis[(pyridine-2,6-dicarboxylato)bismuthate(III)] dihydrate, (II).
Compounds (I) and (II) were obtained under slightly acidic reaction conditions by dissolution of freshly prepared cobalt(II) or nickel(II) hydroxycarbonates in a saturated solution of Bi(Hpydc)(pydc). Both compounds crystallize in the triclinic system (space group P1) and they are essentially isomorphous. The transition metal rests on the inversion centre and is coordinated to six water molecules in a slightly distorted octahedral coordination geometry with typical Co—O and Ni—O distances (Fig. 1, Table 1) (Guo et al., 2008; Morzyk-Ociepa, 2007). The primary coordination environment of BiIII includes seven donor atoms, six from two tridentate pydc2- ligands and one bridging O atom from an adjacent symmetry-related complex. The pydc2- ligand is coordinated to the BiIII centre in a conventional O,N,O-tridentate fashion via the N atom and two O atoms, one from each of the two carboxylate groups. Atom Bi1 and its symmetry counterpart constitute the anionic part of the molecule, forming a [Bi2(pydc)4]2- dimer (Fig. 1).
The bridging carboxylate group C11—O11—O12 in (I) and (II) displays a monoatomic bidentate η0:η2:µ2-type coordination, which represents the most common bridging motif in structurally characterized BiIII complexes with dipicolinate ligands (Stavila et al., 2006). According to the nomenclature proposed by Serezhkin et al. (2009), the coordination type of the bridging aminocarboxylate ligand in (I) and (II) can be represented as T101, indicating that one pydc2- ion acts as a tridentate ligand towards one BiIII atom and as a monodentate ligand towards the other. This type of coordination of the pydc2- ligand is also found in complexes of TlI, PbII and some lanthanides (Serezhkin et al., 2009). The non-bridging pydc2- group has a coordination of type T001, which represents the most frequent coordination mode in dipicolinate complexes and is typically adopted by the bis(pydc2-) complexes of the first- and second-row transition metals. The crystal structures of other BiIII complexes with H2pydc acid exhibit similar coordination features. The dimeric structure of [Bi(Hpydc)(pydc)(dmso)]2 (Zevaco et al., 1992) (dmso is dimethylsulfoxide) displays octa-coordinated BiIII atoms surrounded by six donor atoms from the Hnpydc(2-n)- ions, a bridging O atom from an adjacent complex and one O atom from a dmso molecule. [{BiCl(H2O)(pydc)}2]n (Ranjbar et al., 2001) contains only one pydc2- group per BiIII atom, with an additional Cl- ligand to complete a pentagonal–bipyramidal environment. In [pydaH]2[Bi2(pydc)4(H2O)2]·4H2O (Ranjbar et al., 2003) (pyda is pyridine-2,6-diamine) and (phenH)2[Bi2(pydc)4(H2O)2]·5H2O (Sheshmani et al., 2005) (phen is 1,10-phenanthroline), the BiIII centres are coordinated by two pydc2- ions, a bridging carboxylate O atom and a water molecule. Interestingly enough, no BiIII–H2O bonds were found in [creatH]2[Bi2(pydc)4]·4H2O (Agbabozorg et al., 2008) (creatH = creatinine, 2-amino-1-methyl-5H-imidazol-4-one). We have recently synthesized and determined the crystal structures of two bismuth(III) dipicolinate complexes with thiourea and thiosemicarbazide (Stavila et al., 2009), [Bi6(pydc)8(Hpydc)2(tu)8] and [Bi2(pydc)3(tsc)(H2O)2]·H2O (tu is thiourea and tsc is thiosemicarbazide), which contain Bi—S bonds. In [Bi6(pydc)8(Hpydc)2(tu)8], there are three independent BiIII atoms connected by means of bridging carboxylate groups into hexanuclear Bi6 units, while [Bi2(pydc)3(tsc)(H2O)2]·H2O is a coordination polymer generated by bridging carboxylate groups.
The Bi···Bi distances in the [Bi(pydc)2]22- dimers here are 4.225 (4) and 4.220 (4) Å for (I) and (II), respectively, which is somewhat shorter than the sum of the van der Waals radii (4.8 Å; Standard reference?). The transition metal and BiIII atoms in (I) and (II) are separated by 6.394 (5) and 6.381 (4) Å, respectively. The interatomic distances and bond angles in (I) and (II) are statistically about the same (Table 1). The Bi—N distances found in the [Bi2(pydc)4]2- dimer correlate well with those in other structurally characterized BiIII complexes with pydc2- ligands (Stavila et al., 2006). The Bi—O distances are asymmetric, with the longest distance being to the bridging atom O11 [2.682 (3) and 2.687 (3) for (I) and (II), respectively], which is situated closer to the adjacent BiIII atom of the dimer at 2.492 (3) and 2.505 (2) Å, respectively.
The extended three-dimensional structure of (I) and (II) shows layers of anions alternating with layers of cations along the b axis. Each anion layer contains dimeric [Bi2(pydc)4]2- complexes joined into chains by secondary Bi···O24ii bonds [3.121 (4) and 3.084 (3) Å for (I) and (II), respectively; symmetry code: (ii) -x, -y - 1, -z]. The layers are held together by a network of nearly linear O—H···O hydrogen bonds with medium to long O···O separations (Fig. 2, Table 2). The H atoms of all water molecules in the structure are involved in hydrogen bonds with the O atoms of other water molecules or carboxylate O atoms. The hydrogen bonds, secondary Bi—O bonds and electrostatic interactions between the cations and anions are the major packing forces that stabilize the crystal structures of compounds (I) and (II).