Two polymorphs of biphenyl-4,4'-diaminium bis(3-carboxy-4-hydroxybenzenesulfonate) dihydrate, C
12H
14N
22+·2C
7H
5O
6S
-·2H
2O, have been obtained and crystallographically characterized. Polymorph (I) crystallizes in the space group
P2
1/
c with
Z' = 2 and polymorph (II) in the space group
P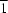
with
Z' = 0.5. The benzidinium cation in (II) is located on a crystallographic inversion centre. In both (I) and (II), the sulfonic acid H atoms are transferred to the benzidine N atoms, forming dihydrated 1:2 molecular adducts (base-acid). In the crystal packings of (I) and (II), the component ions are linked into three-dimensional networks by combinations of
X-H

O (
X = O, N and C) hydrogen bonds. In addition,
![[pi]](/logos/entities/pi_rmgif.gif)
-
![[pi]](/logos/entities/pi_rmgif.gif)
interactions are observed in (I) between inversion-related benzene rings [centroid-centroid distances = 3.632 (2) and 3.627 (2) Å]. In order to simplify the complex three-dimensional networks in (I) and (II), we also give their rationalized topological analyses.
Supporting information
CCDC references: 798600; 798601
All reagents and solvents were used as obtained without further purification.
Benzidine (0.1 mmol, 18.4 mg) and 5-sulfosalicylic acid dihydrate (0.2 mmol,
50.8 g), in a 1:2 molar ratio, were dissolved in 95% methanol (10 ml). The
mixture was stirred for half an hour at ambient temperature and then filtered.
The resulting pale-yellow solution was kept in air for two weeks. Colourless
plate and block crystals of (I) and (II), respectively, suitable for
single-crystal X-ray diffraction analysis, were grown by slow evaporation of
the solution at the bottom of the vessel. The crystals were separated
manually.
H atoms bonded to C atoms were positioned geometrically, with C—H = 0.93 Å
(aromatic), and refined in a riding mode with Uiso(H) =
1.2Ueq(aromatic C). H atoms bonded to N and O atoms were found in
difference Fourier maps and their N—H and O—H distances were refined with
restraints of N—H = 0.86 (2) and O—H = 0.82 (2) Å, and with
Uiso(H) = 1.2Ueq(N) or 1.5Ueq(O).
For both compounds, data collection: SMART (Bruker, 2001); cell refinement: SMART (Bruker, 2001); data reduction: SAINT-Plus (Bruker, 2001); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008); molecular graphics: PLATON (Spek, 2009); software used to prepare material for publication: PLATON (Spek, 2009).
(I) biphenyl-4,4'-diaminium bis(3-carboxy-4-hydroxybenzenesulfonate) dihydrate
top
Crystal data top
| C12H14N22+·2C7H5O6S−·2H2O | F(000) = 2736 |
| Mr = 656.62 | Dx = 1.547 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 2357 reflections |
| a = 15.1851 (3) Å | θ = 2.2–19.8° |
| b = 18.6584 (6) Å | µ = 0.27 mm−1 |
| c = 19.9114 (6) Å | T = 298 K |
| β = 91.300 (2)° | Plate, colourless |
| V = 5640.0 (3) Å3 | 0.40 × 0.10 × 0.04 mm |
| Z = 8 | |
Data collection top
Bruker SMART APEX CCD area-detector
diffractometer | 12875 independent reflections |
| Radiation source: fine focus sealed Siemens Mo tube | 6411 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.100 |
| 0.3° wide ω exposures scans | θmax = 27.5°, θmin = 1.3° |
Absorption correction: multi-scan
(SADABS; Sheldrick, 2003) | h = −19→19 |
| Tmin = 0.901, Tmax = 0.989 | k = −24→24 |
| 48494 measured reflections | l = −24→25 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.063 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.165 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.95 | w = 1/[σ2(Fo2) + (0.0699P)2]
where P = (Fo2 + 2Fc2)/3 |
| 12875 reflections | (Δ/σ)max < 0.001 |
| 877 parameters | Δρmax = 0.49 e Å−3 |
| 28 restraints | Δρmin = −0.27 e Å−3 |
Crystal data top
| C12H14N22+·2C7H5O6S−·2H2O | V = 5640.0 (3) Å3 |
| Mr = 656.62 | Z = 8 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 15.1851 (3) Å | µ = 0.27 mm−1 |
| b = 18.6584 (6) Å | T = 298 K |
| c = 19.9114 (6) Å | 0.40 × 0.10 × 0.04 mm |
| β = 91.300 (2)° | |
Data collection top
Bruker SMART APEX CCD area-detector
diffractometer | 12875 independent reflections |
Absorption correction: multi-scan
(SADABS; Sheldrick, 2003) | 6411 reflections with I > 2σ(I) |
| Tmin = 0.901, Tmax = 0.989 | Rint = 0.100 |
| 48494 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.063 | 28 restraints |
| wR(F2) = 0.165 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.95 | Δρmax = 0.49 e Å−3 |
| 12875 reflections | Δρmin = −0.27 e Å−3 |
| 877 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.3042 (2) | 0.26183 (18) | 0.02604 (18) | 0.0334 (9) | |
| C2 | 0.3633 (2) | 0.2065 (2) | 0.04253 (19) | 0.0382 (9) | |
| C3 | 0.3735 (2) | 0.18561 (19) | 0.10881 (19) | 0.0409 (10) | |
| H3 | 0.4121 | 0.1486 | 0.1199 | 0.049* | |
| C4 | 0.3275 (2) | 0.21865 (18) | 0.15845 (18) | 0.0345 (9) | |
| H4 | 0.3345 | 0.2036 | 0.2028 | 0.041* | |
| C5 | 0.2705 (2) | 0.27468 (17) | 0.14268 (16) | 0.0269 (8) | |
| C6 | 0.2592 (2) | 0.29555 (18) | 0.07675 (17) | 0.0312 (8) | |
| H6 | 0.2208 | 0.3329 | 0.0661 | 0.037* | |
| C7 | 0.2898 (3) | 0.2824 (2) | −0.04513 (19) | 0.0405 (10) | |
| C8 | 0.0552 (2) | 0.58948 (18) | −0.01644 (17) | 0.0300 (8) | |
| C9 | 0.1139 (2) | 0.53248 (19) | −0.02519 (17) | 0.0316 (9) | |
| C10 | 0.1466 (2) | 0.49502 (19) | 0.02988 (17) | 0.0334 (9) | |
| H10 | 0.1856 | 0.4572 | 0.0237 | 0.040* | |
| C11 | 0.1221 (2) | 0.51325 (19) | 0.09351 (17) | 0.0338 (9) | |
| H11 | 0.1440 | 0.4875 | 0.1302 | 0.041* | |
| C12 | 0.0641 (2) | 0.57036 (19) | 0.10351 (16) | 0.0307 (8) | |
| C13 | 0.0310 (2) | 0.60791 (19) | 0.04938 (17) | 0.0325 (9) | |
| H13 | −0.0076 | 0.6458 | 0.0561 | 0.039* | |
| C14 | 0.0182 (2) | 0.62554 (19) | −0.07637 (18) | 0.0324 (9) | |
| C15 | 0.4440 (2) | 0.41183 (18) | 1.01696 (17) | 0.0308 (8) | |
| C16 | 0.3858 (2) | 0.46991 (18) | 1.02549 (17) | 0.0313 (8) | |
| C17 | 0.3537 (2) | 0.50700 (19) | 0.96944 (17) | 0.0333 (9) | |
| H17 | 0.3160 | 0.5457 | 0.9751 | 0.040* | |
| C18 | 0.3769 (2) | 0.48719 (18) | 0.90612 (17) | 0.0321 (9) | |
| H18 | 0.3545 | 0.5122 | 0.8691 | 0.039* | |
| C19 | 0.4340 (2) | 0.42978 (18) | 0.89669 (17) | 0.0295 (8) | |
| C20 | 0.4675 (2) | 0.39231 (18) | 0.95174 (17) | 0.0307 (8) | |
| H20 | 0.5056 | 0.3541 | 0.9454 | 0.037* | |
| C21 | 0.4780 (2) | 0.37329 (19) | 1.07651 (19) | 0.0353 (9) | |
| C22 | 0.1946 (2) | 0.73813 (19) | 0.95454 (18) | 0.0357 (9) | |
| C23 | 0.1404 (3) | 0.7967 (2) | 0.9405 (2) | 0.0457 (10) | |
| C24 | 0.1170 (3) | 0.8123 (2) | 0.8741 (2) | 0.0527 (12) | |
| H24 | 0.0801 | 0.8509 | 0.8647 | 0.063* | |
| C25 | 0.1476 (3) | 0.7717 (2) | 0.8226 (2) | 0.0443 (10) | |
| H25 | 0.1317 | 0.7827 | 0.7785 | 0.053* | |
| C26 | 0.2025 (2) | 0.71373 (19) | 0.83652 (17) | 0.0323 (8) | |
| C27 | 0.2260 (2) | 0.69699 (18) | 0.90158 (18) | 0.0335 (9) | |
| H27 | 0.2628 | 0.6582 | 0.9104 | 0.040* | |
| C28 | 0.2176 (3) | 0.7209 (2) | 1.0252 (2) | 0.0426 (10) | |
| C29 | 0.0403 (2) | 0.4033 (2) | 0.33730 (17) | 0.0335 (9) | |
| C30 | 0.0616 (3) | 0.3437 (2) | 0.37481 (18) | 0.0472 (11) | |
| H30 | 0.0555 | 0.2981 | 0.3564 | 0.057* | |
| C31 | 0.0923 (3) | 0.3523 (2) | 0.44014 (19) | 0.0472 (11) | |
| H31 | 0.1082 | 0.3121 | 0.4651 | 0.057* | |
| C32 | 0.1000 (2) | 0.41959 (19) | 0.46946 (17) | 0.0310 (8) | |
| C33 | 0.0770 (2) | 0.4789 (2) | 0.42990 (17) | 0.0389 (9) | |
| H33 | 0.0813 | 0.5247 | 0.4481 | 0.047* | |
| C34 | 0.0479 (2) | 0.4703 (2) | 0.36413 (18) | 0.0411 (10) | |
| H34 | 0.0334 | 0.5102 | 0.3382 | 0.049* | |
| C35 | 0.1301 (2) | 0.43006 (19) | 0.54032 (17) | 0.0310 (8) | |
| C36 | 0.1143 (2) | 0.37854 (19) | 0.58952 (17) | 0.0362 (9) | |
| H36 | 0.0860 | 0.3361 | 0.5773 | 0.043* | |
| C37 | 0.1394 (2) | 0.38905 (19) | 0.65577 (18) | 0.0359 (9) | |
| H37 | 0.1276 | 0.3544 | 0.6879 | 0.043* | |
| C38 | 0.1826 (2) | 0.45188 (19) | 0.67379 (17) | 0.0304 (8) | |
| C39 | 0.1991 (2) | 0.5037 (2) | 0.62680 (17) | 0.0371 (9) | |
| H39 | 0.2274 | 0.5460 | 0.6394 | 0.045* | |
| C40 | 0.1735 (2) | 0.4925 (2) | 0.56087 (17) | 0.0395 (10) | |
| H40 | 0.1854 | 0.5276 | 0.5291 | 0.047* | |
| C41 | 0.3075 (2) | 0.53242 (19) | 0.31286 (16) | 0.0306 (8) | |
| C42 | 0.3002 (2) | 0.4759 (2) | 0.35557 (18) | 0.0390 (9) | |
| H42 | 0.2792 | 0.4320 | 0.3398 | 0.047* | |
| C43 | 0.3240 (2) | 0.4842 (2) | 0.42268 (18) | 0.0407 (10) | |
| H43 | 0.3178 | 0.4456 | 0.4518 | 0.049* | |
| C44 | 0.3569 (2) | 0.54830 (19) | 0.44746 (17) | 0.0315 (8) | |
| C45 | 0.3642 (3) | 0.6049 (2) | 0.40229 (18) | 0.0417 (10) | |
| H45 | 0.3856 | 0.6489 | 0.4175 | 0.050* | |
| C46 | 0.3403 (3) | 0.5968 (2) | 0.33545 (18) | 0.0432 (10) | |
| H46 | 0.3464 | 0.6349 | 0.3058 | 0.052* | |
| C47 | 0.3834 (2) | 0.55790 (19) | 0.51960 (17) | 0.0320 (9) | |
| C48 | 0.4174 (2) | 0.50169 (19) | 0.55751 (17) | 0.0362 (9) | |
| H48 | 0.4240 | 0.4570 | 0.5375 | 0.043* | |
| C49 | 0.4415 (2) | 0.51032 (19) | 0.62408 (18) | 0.0372 (9) | |
| H49 | 0.4636 | 0.4717 | 0.6488 | 0.045* | |
| C50 | 0.4327 (2) | 0.57585 (18) | 0.65358 (16) | 0.0293 (8) | |
| C51 | 0.4007 (2) | 0.6332 (2) | 0.61722 (18) | 0.0389 (10) | |
| H51 | 0.3958 | 0.6779 | 0.6373 | 0.047* | |
| C52 | 0.3760 (2) | 0.62386 (19) | 0.55074 (18) | 0.0392 (9) | |
| H52 | 0.3539 | 0.6627 | 0.5264 | 0.047* | |
| N1 | 0.0069 (2) | 0.3950 (2) | 0.26813 (16) | 0.0387 (8) | |
| H1A | 0.042 (2) | 0.3678 (17) | 0.2454 (16) | 0.046* | |
| H1B | −0.0448 (15) | 0.3774 (19) | 0.2661 (18) | 0.046* | |
| H1C | 0.004 (2) | 0.4373 (12) | 0.2495 (17) | 0.046* | |
| N2 | 0.2081 (2) | 0.46402 (18) | 0.74452 (17) | 0.0399 (8) | |
| H2A | 0.2485 (19) | 0.4349 (17) | 0.7596 (18) | 0.048* | |
| H2B | 0.224 (2) | 0.5083 (11) | 0.7513 (18) | 0.048* | |
| H2C | 0.1670 (19) | 0.450 (2) | 0.7708 (16) | 0.048* | |
| N3 | 0.2826 (2) | 0.52599 (18) | 0.24137 (17) | 0.0388 (8) | |
| H3A | 0.270 (2) | 0.4824 (11) | 0.2319 (18) | 0.047* | |
| H3B | 0.2331 (16) | 0.5490 (18) | 0.2383 (19) | 0.047* | |
| H3C | 0.3239 (19) | 0.5442 (19) | 0.2169 (16) | 0.047* | |
| N4 | 0.4590 (2) | 0.58621 (17) | 0.72452 (16) | 0.0338 (7) | |
| H4A | 0.486 (2) | 0.5495 (14) | 0.7440 (16) | 0.041* | |
| H4B | 0.4977 (19) | 0.6210 (15) | 0.7265 (18) | 0.041* | |
| H4C | 0.4113 (16) | 0.5994 (18) | 0.7451 (16) | 0.041* | |
| O1 | 0.2315 (2) | 0.33351 (17) | −0.05437 (13) | 0.0555 (8) | |
| H1D | 0.221 (3) | 0.341 (2) | −0.0949 (11) | 0.083* | |
| O2 | 0.32958 (19) | 0.25392 (16) | −0.09125 (13) | 0.0599 (8) | |
| O3 | 0.4114 (2) | 0.17256 (16) | −0.00403 (14) | 0.0580 (8) | |
| H3D | 0.401 (3) | 0.191 (2) | −0.0412 (14) | 0.087* | |
| O4 | 0.22168 (19) | 0.39435 (13) | 0.19512 (13) | 0.0503 (7) | |
| O5 | 0.11825 (15) | 0.29722 (14) | 0.19346 (12) | 0.0429 (7) | |
| O6 | 0.24469 (16) | 0.29409 (14) | 0.26946 (11) | 0.0417 (7) | |
| O7 | −0.04024 (18) | 0.67707 (15) | −0.06438 (12) | 0.0448 (7) | |
| H7A | −0.060 (3) | 0.690 (2) | −0.1009 (13) | 0.067* | |
| O8 | 0.03831 (16) | 0.60981 (14) | −0.13307 (12) | 0.0418 (7) | |
| O9 | 0.14137 (17) | 0.51098 (15) | −0.08618 (13) | 0.0460 (7) | |
| H9A | 0.116 (3) | 0.536 (2) | −0.1148 (17) | 0.069* | |
| O10 | −0.00481 (17) | 0.66103 (13) | 0.18600 (12) | 0.0441 (7) | |
| O11 | 0.11477 (16) | 0.58648 (14) | 0.22717 (12) | 0.0481 (7) | |
| O12 | −0.02814 (16) | 0.53518 (13) | 0.20715 (11) | 0.0410 (7) | |
| O13 | 0.53098 (19) | 0.31975 (15) | 1.06360 (13) | 0.0488 (7) | |
| H13A | 0.546 (3) | 0.298 (2) | 1.0984 (14) | 0.073* | |
| O14 | 0.45832 (18) | 0.39000 (14) | 1.13386 (13) | 0.0491 (7) | |
| O15 | 0.35960 (18) | 0.49163 (15) | 1.08649 (13) | 0.0468 (7) | |
| H15A | 0.388 (3) | 0.466 (2) | 1.1122 (18) | 0.070* | |
| O16 | 0.52587 (16) | 0.46151 (12) | 0.79163 (11) | 0.0373 (6) | |
| O17 | 0.38336 (16) | 0.40828 (15) | 0.77319 (12) | 0.0486 (7) | |
| O18 | 0.50476 (16) | 0.33708 (13) | 0.81822 (12) | 0.0419 (7) | |
| O19 | 0.2684 (2) | 0.66446 (16) | 1.03382 (13) | 0.0534 (8) | |
| H19A | 0.288 (3) | 0.660 (2) | 1.0734 (12) | 0.080* | |
| O20 | 0.1908 (2) | 0.75578 (17) | 1.07229 (14) | 0.0662 (9) | |
| O21 | 0.1082 (2) | 0.83868 (17) | 0.98914 (15) | 0.0695 (10) | |
| H21A | 0.129 (3) | 0.825 (3) | 1.0248 (16) | 0.104* | |
| O22 | 0.2886 (2) | 0.60281 (14) | 0.79251 (13) | 0.0603 (8) | |
| O23 | 0.30101 (17) | 0.71324 (14) | 0.73397 (12) | 0.0496 (7) | |
| O24 | 0.16660 (18) | 0.64683 (18) | 0.72440 (14) | 0.0712 (10) | |
| O25 | 0.18352 (19) | 0.16493 (15) | 0.32023 (14) | 0.0472 (7) | |
| H25A | 0.203 (3) | 0.2021 (15) | 0.305 (2) | 0.071* | |
| H25B | 0.1305 (13) | 0.162 (2) | 0.313 (2) | 0.071* | |
| O26 | 0.31347 (19) | 0.65699 (16) | 0.16198 (15) | 0.0487 (7) | |
| H26A | 0.291 (3) | 0.6941 (16) | 0.176 (2) | 0.073* | |
| H26B | 0.3662 (13) | 0.661 (3) | 0.169 (2) | 0.073* | |
| O27 | 0.44273 (19) | 0.75037 (17) | 0.82263 (15) | 0.0507 (8) | |
| H27A | 0.400 (2) | 0.735 (2) | 0.7993 (19) | 0.076* | |
| H27B | 0.486 (2) | 0.747 (3) | 0.799 (2) | 0.076* | |
| O28 | 0.43106 (19) | 0.20264 (16) | 0.78836 (17) | 0.0561 (8) | |
| H28A | 0.3763 (14) | 0.197 (3) | 0.792 (2) | 0.084* | |
| H28B | 0.444 (3) | 0.2429 (15) | 0.805 (2) | 0.084* | |
| S1 | 0.21036 (6) | 0.31845 (5) | 0.20480 (4) | 0.0305 (2) | |
| S2 | 0.03450 (6) | 0.59040 (5) | 0.18703 (4) | 0.0323 (2) | |
| S3 | 0.46353 (6) | 0.40726 (5) | 0.81379 (4) | 0.0313 (2) | |
| S4 | 0.24176 (6) | 0.66487 (5) | 0.76758 (5) | 0.0347 (2) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1 | 0.037 (2) | 0.031 (2) | 0.032 (2) | −0.0046 (17) | 0.0000 (17) | −0.0026 (17) |
| C2 | 0.041 (2) | 0.035 (2) | 0.039 (2) | −0.0005 (18) | 0.0074 (19) | −0.0109 (19) |
| C3 | 0.045 (2) | 0.034 (2) | 0.044 (3) | 0.0107 (18) | 0.000 (2) | −0.0022 (19) |
| C4 | 0.040 (2) | 0.032 (2) | 0.031 (2) | 0.0011 (17) | −0.0058 (18) | 0.0005 (17) |
| C5 | 0.0259 (19) | 0.0300 (19) | 0.025 (2) | −0.0025 (15) | 0.0003 (15) | −0.0003 (15) |
| C6 | 0.035 (2) | 0.030 (2) | 0.029 (2) | 0.0014 (16) | −0.0015 (17) | 0.0023 (16) |
| C7 | 0.046 (3) | 0.041 (2) | 0.035 (2) | −0.008 (2) | 0.005 (2) | −0.0011 (19) |
| C8 | 0.030 (2) | 0.031 (2) | 0.028 (2) | −0.0056 (16) | −0.0069 (16) | 0.0008 (16) |
| C9 | 0.032 (2) | 0.041 (2) | 0.022 (2) | −0.0057 (17) | 0.0005 (16) | −0.0006 (17) |
| C10 | 0.030 (2) | 0.036 (2) | 0.034 (2) | 0.0004 (16) | 0.0005 (17) | 0.0011 (17) |
| C11 | 0.033 (2) | 0.039 (2) | 0.030 (2) | 0.0017 (17) | −0.0075 (17) | 0.0047 (17) |
| C12 | 0.032 (2) | 0.038 (2) | 0.022 (2) | −0.0057 (17) | −0.0056 (16) | 0.0039 (16) |
| C13 | 0.031 (2) | 0.036 (2) | 0.030 (2) | −0.0014 (16) | −0.0017 (17) | −0.0017 (17) |
| C14 | 0.030 (2) | 0.038 (2) | 0.029 (2) | −0.0033 (17) | −0.0070 (17) | 0.0000 (18) |
| C15 | 0.032 (2) | 0.029 (2) | 0.031 (2) | −0.0050 (16) | −0.0034 (17) | 0.0013 (16) |
| C16 | 0.029 (2) | 0.035 (2) | 0.030 (2) | −0.0039 (17) | 0.0020 (17) | −0.0020 (17) |
| C17 | 0.033 (2) | 0.034 (2) | 0.034 (2) | 0.0053 (16) | −0.0014 (17) | 0.0002 (17) |
| C18 | 0.033 (2) | 0.034 (2) | 0.029 (2) | −0.0008 (17) | −0.0046 (17) | 0.0044 (17) |
| C19 | 0.0272 (19) | 0.035 (2) | 0.026 (2) | −0.0047 (16) | 0.0009 (16) | 0.0020 (16) |
| C20 | 0.030 (2) | 0.028 (2) | 0.034 (2) | 0.0024 (16) | −0.0033 (17) | −0.0003 (16) |
| C21 | 0.039 (2) | 0.032 (2) | 0.034 (2) | −0.0066 (18) | −0.0042 (19) | 0.0020 (18) |
| C22 | 0.040 (2) | 0.030 (2) | 0.037 (2) | −0.0018 (17) | −0.0015 (18) | −0.0047 (17) |
| C23 | 0.051 (3) | 0.041 (2) | 0.046 (3) | 0.002 (2) | 0.013 (2) | −0.006 (2) |
| C24 | 0.066 (3) | 0.043 (3) | 0.050 (3) | 0.024 (2) | 0.004 (2) | 0.006 (2) |
| C25 | 0.052 (3) | 0.044 (2) | 0.037 (2) | 0.007 (2) | 0.004 (2) | 0.010 (2) |
| C26 | 0.034 (2) | 0.035 (2) | 0.028 (2) | −0.0034 (17) | 0.0069 (17) | 0.0031 (17) |
| C27 | 0.037 (2) | 0.026 (2) | 0.038 (2) | 0.0000 (16) | 0.0022 (18) | 0.0014 (17) |
| C28 | 0.048 (3) | 0.040 (2) | 0.041 (3) | −0.006 (2) | 0.005 (2) | −0.004 (2) |
| C29 | 0.030 (2) | 0.045 (2) | 0.026 (2) | 0.0012 (17) | −0.0003 (16) | −0.0033 (18) |
| C30 | 0.074 (3) | 0.037 (2) | 0.031 (2) | 0.009 (2) | −0.007 (2) | −0.0099 (19) |
| C31 | 0.071 (3) | 0.039 (2) | 0.031 (2) | 0.015 (2) | −0.011 (2) | 0.0027 (18) |
| C32 | 0.034 (2) | 0.037 (2) | 0.0214 (19) | 0.0000 (17) | 0.0013 (16) | 0.0004 (16) |
| C33 | 0.051 (3) | 0.035 (2) | 0.031 (2) | −0.0008 (18) | −0.0034 (19) | −0.0017 (17) |
| C34 | 0.050 (3) | 0.042 (2) | 0.032 (2) | 0.0025 (19) | −0.0024 (19) | 0.0060 (18) |
| C35 | 0.031 (2) | 0.037 (2) | 0.025 (2) | 0.0042 (17) | 0.0021 (16) | 0.0022 (17) |
| C36 | 0.046 (2) | 0.033 (2) | 0.030 (2) | −0.0001 (18) | −0.0071 (18) | −0.0043 (17) |
| C37 | 0.046 (2) | 0.034 (2) | 0.028 (2) | 0.0010 (18) | −0.0030 (18) | 0.0063 (17) |
| C38 | 0.031 (2) | 0.039 (2) | 0.021 (2) | 0.0032 (17) | −0.0037 (16) | −0.0011 (16) |
| C39 | 0.044 (2) | 0.038 (2) | 0.029 (2) | −0.0113 (18) | −0.0011 (18) | −0.0020 (17) |
| C40 | 0.049 (2) | 0.044 (2) | 0.026 (2) | −0.0083 (19) | −0.0016 (18) | 0.0057 (18) |
| C41 | 0.034 (2) | 0.038 (2) | 0.0192 (19) | 0.0019 (17) | −0.0030 (16) | 0.0002 (16) |
| C42 | 0.046 (2) | 0.034 (2) | 0.037 (2) | −0.0075 (18) | −0.0046 (19) | −0.0056 (18) |
| C43 | 0.054 (3) | 0.034 (2) | 0.034 (2) | −0.0029 (19) | −0.001 (2) | 0.0064 (18) |
| C44 | 0.032 (2) | 0.036 (2) | 0.027 (2) | 0.0019 (17) | 0.0018 (16) | 0.0001 (17) |
| C45 | 0.059 (3) | 0.033 (2) | 0.033 (2) | −0.0027 (19) | −0.005 (2) | −0.0001 (18) |
| C46 | 0.065 (3) | 0.035 (2) | 0.030 (2) | −0.007 (2) | −0.004 (2) | 0.0062 (18) |
| C47 | 0.034 (2) | 0.036 (2) | 0.025 (2) | 0.0021 (17) | 0.0013 (16) | 0.0033 (17) |
| C48 | 0.045 (2) | 0.030 (2) | 0.034 (2) | 0.0020 (18) | −0.0008 (18) | −0.0042 (17) |
| C49 | 0.048 (2) | 0.032 (2) | 0.032 (2) | 0.0025 (18) | −0.0047 (18) | 0.0049 (17) |
| C50 | 0.032 (2) | 0.035 (2) | 0.0214 (19) | −0.0024 (16) | 0.0007 (16) | −0.0017 (16) |
| C51 | 0.053 (3) | 0.035 (2) | 0.029 (2) | 0.0076 (19) | 0.0000 (19) | −0.0078 (17) |
| C52 | 0.050 (3) | 0.035 (2) | 0.032 (2) | 0.0106 (18) | −0.0029 (19) | 0.0049 (18) |
| N1 | 0.035 (2) | 0.057 (2) | 0.0238 (19) | 0.0043 (18) | −0.0039 (16) | −0.0033 (16) |
| N2 | 0.051 (2) | 0.041 (2) | 0.0271 (19) | −0.0021 (18) | −0.0076 (16) | 0.0032 (16) |
| N3 | 0.045 (2) | 0.039 (2) | 0.032 (2) | −0.0048 (17) | −0.0049 (17) | 0.0007 (16) |
| N4 | 0.035 (2) | 0.039 (2) | 0.0276 (18) | 0.0005 (15) | 0.0001 (15) | −0.0015 (15) |
| O1 | 0.073 (2) | 0.067 (2) | 0.0265 (16) | 0.0154 (17) | −0.0016 (16) | 0.0070 (15) |
| O2 | 0.074 (2) | 0.074 (2) | 0.0326 (16) | 0.0043 (17) | 0.0140 (16) | −0.0096 (15) |
| O3 | 0.075 (2) | 0.0506 (19) | 0.0492 (19) | 0.0172 (16) | 0.0158 (18) | −0.0115 (15) |
| O4 | 0.078 (2) | 0.0309 (15) | 0.0419 (17) | 0.0029 (14) | 0.0050 (15) | −0.0012 (12) |
| O5 | 0.0337 (15) | 0.0606 (18) | 0.0343 (15) | 0.0067 (13) | −0.0031 (12) | −0.0031 (13) |
| O6 | 0.0444 (16) | 0.0575 (17) | 0.0228 (14) | 0.0071 (13) | −0.0043 (12) | 0.0067 (12) |
| O7 | 0.0510 (18) | 0.0522 (18) | 0.0311 (16) | 0.0129 (14) | −0.0033 (14) | 0.0045 (14) |
| O8 | 0.0445 (16) | 0.0551 (17) | 0.0257 (15) | 0.0027 (13) | −0.0010 (13) | −0.0003 (13) |
| O9 | 0.0442 (17) | 0.065 (2) | 0.0292 (16) | 0.0137 (14) | 0.0019 (13) | −0.0019 (14) |
| O10 | 0.0545 (17) | 0.0401 (16) | 0.0380 (16) | 0.0097 (13) | 0.0027 (13) | −0.0049 (12) |
| O11 | 0.0447 (17) | 0.0675 (19) | 0.0315 (15) | 0.0074 (14) | −0.0131 (13) | −0.0127 (14) |
| O12 | 0.0482 (16) | 0.0461 (16) | 0.0289 (15) | −0.0036 (13) | 0.0062 (12) | 0.0024 (12) |
| O13 | 0.0596 (19) | 0.0472 (18) | 0.0393 (17) | 0.0174 (15) | −0.0047 (15) | 0.0101 (14) |
| O14 | 0.0631 (19) | 0.0567 (18) | 0.0274 (16) | 0.0083 (15) | 0.0008 (14) | 0.0018 (13) |
| O15 | 0.0488 (18) | 0.062 (2) | 0.0294 (16) | 0.0165 (14) | 0.0029 (13) | −0.0055 (14) |
| O16 | 0.0480 (16) | 0.0367 (15) | 0.0274 (14) | −0.0019 (12) | 0.0052 (12) | 0.0021 (11) |
| O17 | 0.0412 (16) | 0.070 (2) | 0.0339 (16) | 0.0056 (14) | −0.0094 (13) | −0.0089 (14) |
| O18 | 0.0455 (16) | 0.0344 (15) | 0.0459 (16) | 0.0050 (12) | 0.0017 (13) | −0.0056 (12) |
| O19 | 0.066 (2) | 0.064 (2) | 0.0296 (16) | 0.0135 (16) | −0.0078 (15) | 0.0005 (15) |
| O20 | 0.092 (2) | 0.068 (2) | 0.0387 (18) | 0.0130 (18) | 0.0056 (17) | −0.0151 (16) |
| O21 | 0.098 (3) | 0.056 (2) | 0.055 (2) | 0.0347 (18) | 0.016 (2) | −0.0115 (17) |
| O22 | 0.092 (2) | 0.0467 (18) | 0.0427 (17) | 0.0267 (16) | 0.0143 (16) | 0.0061 (14) |
| O23 | 0.0559 (18) | 0.0527 (17) | 0.0410 (17) | −0.0088 (14) | 0.0176 (14) | 0.0041 (14) |
| O24 | 0.0402 (18) | 0.116 (3) | 0.057 (2) | −0.0176 (17) | 0.0040 (15) | −0.0384 (19) |
| O25 | 0.0568 (19) | 0.0483 (18) | 0.0363 (17) | −0.0099 (16) | −0.0046 (16) | −0.0008 (14) |
| O26 | 0.0544 (19) | 0.0502 (18) | 0.0412 (17) | 0.0076 (16) | −0.0039 (16) | −0.0020 (14) |
| O27 | 0.048 (2) | 0.0560 (19) | 0.048 (2) | 0.0007 (16) | −0.0025 (14) | 0.0080 (15) |
| O28 | 0.0440 (17) | 0.0517 (19) | 0.072 (2) | 0.0109 (16) | −0.0088 (17) | −0.0197 (16) |
| S1 | 0.0352 (5) | 0.0336 (5) | 0.0226 (5) | 0.0034 (4) | −0.0021 (4) | 0.0015 (4) |
| S2 | 0.0368 (5) | 0.0382 (5) | 0.0216 (5) | 0.0034 (4) | −0.0023 (4) | −0.0017 (4) |
| S3 | 0.0346 (5) | 0.0339 (5) | 0.0254 (5) | 0.0013 (4) | −0.0031 (4) | −0.0025 (4) |
| S4 | 0.0354 (5) | 0.0381 (5) | 0.0307 (5) | −0.0020 (4) | 0.0043 (4) | −0.0012 (4) |
Geometric parameters (Å, º) top
| C1—C6 | 1.383 (5) | C36—H36 | 0.9300 |
| C1—C2 | 1.401 (5) | C37—C38 | 1.386 (5) |
| C1—C7 | 1.479 (5) | C37—H37 | 0.9300 |
| C2—O3 | 1.351 (4) | C38—C39 | 1.373 (5) |
| C2—C3 | 1.382 (5) | C38—N2 | 1.470 (5) |
| C3—C4 | 1.370 (5) | C39—C40 | 1.377 (5) |
| C3—H3 | 0.9300 | C39—H39 | 0.9300 |
| C4—C5 | 1.389 (4) | C40—H40 | 0.9300 |
| C4—H4 | 0.9300 | C41—C42 | 1.361 (5) |
| C5—C6 | 1.376 (4) | C41—C46 | 1.373 (5) |
| C5—S1 | 1.755 (3) | C41—N3 | 1.469 (5) |
| C6—H6 | 0.9300 | C42—C43 | 1.385 (5) |
| C7—O2 | 1.231 (4) | C42—H42 | 0.9300 |
| C7—O1 | 1.313 (5) | C43—C44 | 1.384 (5) |
| C8—C9 | 1.402 (5) | C43—H43 | 0.9300 |
| C8—C13 | 1.411 (4) | C44—C45 | 1.394 (5) |
| C8—C14 | 1.470 (5) | C44—C47 | 1.494 (5) |
| C9—O9 | 1.354 (4) | C45—C46 | 1.380 (5) |
| C9—C10 | 1.383 (5) | C45—H45 | 0.9300 |
| C10—C11 | 1.372 (4) | C46—H46 | 0.9300 |
| C10—H10 | 0.9300 | C47—C52 | 1.384 (5) |
| C11—C12 | 1.399 (5) | C47—C48 | 1.385 (5) |
| C11—H11 | 0.9300 | C48—C49 | 1.376 (5) |
| C12—C13 | 1.372 (4) | C48—H48 | 0.9300 |
| C12—S2 | 1.772 (3) | C49—C50 | 1.364 (5) |
| C13—H13 | 0.9300 | C49—H49 | 0.9300 |
| C14—O8 | 1.212 (4) | C50—C51 | 1.374 (5) |
| C14—O7 | 1.334 (4) | C50—N4 | 1.472 (4) |
| C15—C20 | 1.402 (4) | C51—C52 | 1.379 (5) |
| C15—C16 | 1.411 (5) | C51—H51 | 0.9300 |
| C15—C21 | 1.470 (5) | C52—H52 | 0.9300 |
| C16—O15 | 1.349 (4) | N1—H1A | 0.872 (18) |
| C16—C17 | 1.392 (5) | N1—H1B | 0.851 (18) |
| C17—C18 | 1.368 (4) | N1—H1C | 0.874 (18) |
| C17—H17 | 0.9300 | N2—H2A | 0.867 (18) |
| C18—C19 | 1.393 (5) | N2—H2B | 0.872 (18) |
| C18—H18 | 0.9300 | N2—H2C | 0.863 (18) |
| C19—C20 | 1.387 (4) | N3—H3A | 0.854 (18) |
| C19—S3 | 1.771 (3) | N3—H3B | 0.867 (18) |
| C20—H20 | 0.9300 | N3—H3C | 0.872 (18) |
| C21—O14 | 1.227 (4) | N4—H4A | 0.883 (18) |
| C21—O13 | 1.312 (4) | N4—H4B | 0.877 (18) |
| C22—C23 | 1.392 (5) | N4—H4C | 0.876 (18) |
| C22—C27 | 1.397 (5) | O1—H1D | 0.831 (19) |
| C22—C28 | 1.478 (5) | O3—H3D | 0.830 (19) |
| C23—O21 | 1.345 (5) | O4—S1 | 1.440 (3) |
| C23—C24 | 1.393 (5) | O5—S1 | 1.466 (3) |
| C24—C25 | 1.364 (5) | O6—S1 | 1.451 (2) |
| C24—H24 | 0.9300 | O7—H7A | 0.814 (18) |
| C25—C26 | 1.389 (5) | O9—H9A | 0.83 (4) |
| C25—H25 | 0.9300 | O10—S2 | 1.447 (2) |
| C26—C27 | 1.372 (5) | O11—S2 | 1.444 (2) |
| C26—S4 | 1.763 (4) | O12—S2 | 1.464 (2) |
| C27—H27 | 0.9300 | O13—H13A | 0.826 (19) |
| C28—O20 | 1.219 (4) | O15—H15A | 0.82 (4) |
| C28—O19 | 1.313 (5) | O16—S3 | 1.461 (2) |
| C29—C34 | 1.363 (5) | O17—S3 | 1.446 (2) |
| C29—C30 | 1.374 (5) | O18—S3 | 1.453 (2) |
| C29—N1 | 1.465 (5) | O19—H19A | 0.840 (19) |
| C30—C31 | 1.381 (5) | O21—H21A | 0.81 (4) |
| C30—H30 | 0.9300 | O22—S4 | 1.441 (3) |
| C31—C32 | 1.388 (5) | O23—S4 | 1.449 (3) |
| C31—H31 | 0.9300 | O24—S4 | 1.453 (3) |
| C32—C33 | 1.397 (5) | O25—H25A | 0.815 (19) |
| C32—C35 | 1.486 (5) | O25—H25B | 0.816 (19) |
| C33—C34 | 1.382 (5) | O26—H26A | 0.820 (19) |
| C33—H33 | 0.9300 | O26—H26B | 0.813 (19) |
| C34—H34 | 0.9300 | O27—H27A | 0.836 (19) |
| C35—C40 | 1.395 (5) | O27—H27B | 0.817 (19) |
| C35—C36 | 1.397 (5) | O28—H28A | 0.842 (19) |
| C36—C37 | 1.379 (5) | O28—H28B | 0.839 (19) |
| | | |
| C6—C1—C2 | 119.2 (3) | C36—C37—C38 | 119.1 (3) |
| C6—C1—C7 | 121.1 (3) | C36—C37—H37 | 120.4 |
| C2—C1—C7 | 119.7 (3) | C38—C37—H37 | 120.4 |
| O3—C2—C3 | 118.2 (3) | C39—C38—C37 | 120.8 (3) |
| O3—C2—C1 | 122.5 (4) | C39—C38—N2 | 119.7 (3) |
| C3—C2—C1 | 119.3 (3) | C37—C38—N2 | 119.4 (3) |
| C4—C3—C2 | 120.9 (4) | C38—C39—C40 | 119.4 (3) |
| C4—C3—H3 | 119.5 | C38—C39—H39 | 120.3 |
| C2—C3—H3 | 119.5 | C40—C39—H39 | 120.3 |
| C3—C4—C5 | 120.1 (3) | C39—C40—C35 | 121.9 (3) |
| C3—C4—H4 | 120.0 | C39—C40—H40 | 119.1 |
| C5—C4—H4 | 120.0 | C35—C40—H40 | 119.1 |
| C6—C5—C4 | 119.5 (3) | C42—C41—C46 | 120.5 (3) |
| C6—C5—S1 | 119.0 (3) | C42—C41—N3 | 121.3 (3) |
| C4—C5—S1 | 121.5 (3) | C46—C41—N3 | 118.2 (3) |
| C5—C6—C1 | 121.0 (3) | C41—C42—C43 | 119.6 (3) |
| C5—C6—H6 | 119.5 | C41—C42—H42 | 120.2 |
| C1—C6—H6 | 119.5 | C43—C42—H42 | 120.2 |
| O2—C7—O1 | 123.4 (4) | C44—C43—C42 | 121.6 (3) |
| O2—C7—C1 | 122.6 (4) | C44—C43—H43 | 119.2 |
| O1—C7—C1 | 114.0 (3) | C42—C43—H43 | 119.2 |
| C9—C8—C13 | 118.7 (3) | C43—C44—C45 | 117.3 (3) |
| C9—C8—C14 | 118.6 (3) | C43—C44—C47 | 122.3 (3) |
| C13—C8—C14 | 122.7 (3) | C45—C44—C47 | 120.4 (3) |
| O9—C9—C10 | 116.7 (3) | C46—C45—C44 | 121.1 (3) |
| O9—C9—C8 | 123.1 (3) | C46—C45—H45 | 119.5 |
| C10—C9—C8 | 120.2 (3) | C44—C45—H45 | 119.5 |
| C11—C10—C9 | 120.4 (3) | C41—C46—C45 | 119.8 (3) |
| C11—C10—H10 | 119.8 | C41—C46—H46 | 120.1 |
| C9—C10—H10 | 119.8 | C45—C46—H46 | 120.1 |
| C10—C11—C12 | 120.4 (3) | C52—C47—C48 | 117.5 (3) |
| C10—C11—H11 | 119.8 | C52—C47—C44 | 121.0 (3) |
| C12—C11—H11 | 119.8 | C48—C47—C44 | 121.5 (3) |
| C13—C12—C11 | 119.9 (3) | C49—C48—C47 | 121.6 (3) |
| C13—C12—S2 | 122.2 (3) | C49—C48—H48 | 119.2 |
| C11—C12—S2 | 117.9 (3) | C47—C48—H48 | 119.2 |
| C12—C13—C8 | 120.5 (3) | C50—C49—C48 | 119.5 (3) |
| C12—C13—H13 | 119.8 | C50—C49—H49 | 120.2 |
| C8—C13—H13 | 119.8 | C48—C49—H49 | 120.2 |
| O8—C14—O7 | 121.6 (3) | C49—C50—C51 | 120.5 (3) |
| O8—C14—C8 | 123.1 (3) | C49—C50—N4 | 120.3 (3) |
| O7—C14—C8 | 115.3 (3) | C51—C50—N4 | 119.2 (3) |
| C20—C15—C16 | 118.9 (3) | C50—C51—C52 | 119.6 (3) |
| C20—C15—C21 | 121.9 (3) | C50—C51—H51 | 120.2 |
| C16—C15—C21 | 119.2 (3) | C52—C51—H51 | 120.2 |
| O15—C16—C17 | 117.9 (3) | C51—C52—C47 | 121.2 (3) |
| O15—C16—C15 | 122.5 (3) | C51—C52—H52 | 119.4 |
| C17—C16—C15 | 119.6 (3) | C47—C52—H52 | 119.4 |
| C18—C17—C16 | 120.8 (3) | C29—N1—H1A | 110 (2) |
| C18—C17—H17 | 119.6 | C29—N1—H1B | 113 (3) |
| C16—C17—H17 | 119.6 | H1A—N1—H1B | 109 (4) |
| C17—C18—C19 | 120.4 (3) | C29—N1—H1C | 109 (3) |
| C17—C18—H18 | 119.8 | H1A—N1—H1C | 110 (4) |
| C19—C18—H18 | 119.8 | H1B—N1—H1C | 107 (4) |
| C20—C19—C18 | 119.9 (3) | C38—N2—H2A | 114 (3) |
| C20—C19—S3 | 121.4 (3) | C38—N2—H2B | 111 (2) |
| C18—C19—S3 | 118.7 (3) | H2A—N2—H2B | 110 (4) |
| C19—C20—C15 | 120.3 (3) | C38—N2—H2C | 111 (3) |
| C19—C20—H20 | 119.8 | H2A—N2—H2C | 97 (3) |
| C15—C20—H20 | 119.8 | H2B—N2—H2C | 113 (4) |
| O14—C21—O13 | 122.6 (3) | C41—N3—H3A | 110 (3) |
| O14—C21—C15 | 122.5 (3) | C41—N3—H3B | 103 (3) |
| O13—C21—C15 | 114.8 (3) | H3A—N3—H3B | 106 (4) |
| C23—C22—C27 | 119.4 (3) | C41—N3—H3C | 110 (2) |
| C23—C22—C28 | 119.2 (3) | H3A—N3—H3C | 114 (4) |
| C27—C22—C28 | 121.4 (3) | H3B—N3—H3C | 114 (4) |
| O21—C23—C22 | 122.4 (4) | C50—N4—H4A | 116 (2) |
| O21—C23—C24 | 118.2 (4) | C50—N4—H4B | 108 (2) |
| C22—C23—C24 | 119.4 (4) | H4A—N4—H4B | 104 (3) |
| C25—C24—C23 | 120.9 (4) | C50—N4—H4C | 106 (2) |
| C25—C24—H24 | 119.5 | H4A—N4—H4C | 113 (3) |
| C23—C24—H24 | 119.5 | H4B—N4—H4C | 109 (3) |
| C24—C25—C26 | 119.7 (4) | C7—O1—H1D | 112 (3) |
| C24—C25—H25 | 120.1 | C2—O3—H3D | 109 (3) |
| C26—C25—H25 | 120.1 | C14—O7—H7A | 106 (3) |
| C27—C26—C25 | 120.5 (3) | C9—O9—H9A | 108 (3) |
| C27—C26—S4 | 122.2 (3) | C21—O13—H13A | 111 (3) |
| C25—C26—S4 | 117.4 (3) | C16—O15—H15A | 103 (3) |
| C26—C27—C22 | 120.1 (3) | C28—O19—H19A | 113 (3) |
| C26—C27—H27 | 119.9 | C23—O21—H21A | 108 (4) |
| C22—C27—H27 | 119.9 | H25A—O25—H25B | 111 (5) |
| O20—C28—O19 | 122.2 (4) | H26A—O26—H26B | 105 (5) |
| O20—C28—C22 | 122.7 (4) | H27A—O27—H27B | 106 (5) |
| O19—C28—C22 | 115.1 (3) | H28A—O28—H28B | 107 (4) |
| C34—C29—C30 | 120.8 (3) | O4—S1—O6 | 112.66 (15) |
| C34—C29—N1 | 119.3 (3) | O4—S1—O5 | 111.18 (16) |
| C30—C29—N1 | 119.9 (3) | O6—S1—O5 | 111.98 (15) |
| C29—C30—C31 | 119.2 (4) | O4—S1—C5 | 107.32 (16) |
| C29—C30—H30 | 120.4 | O6—S1—C5 | 107.31 (15) |
| C31—C30—H30 | 120.4 | O5—S1—C5 | 105.97 (15) |
| C30—C31—C32 | 121.7 (3) | O11—S2—O10 | 113.39 (16) |
| C30—C31—H31 | 119.2 | O11—S2—O12 | 110.99 (16) |
| C32—C31—H31 | 119.2 | O10—S2—O12 | 111.99 (15) |
| C31—C32—C33 | 117.4 (3) | O11—S2—C12 | 106.24 (16) |
| C31—C32—C35 | 122.6 (3) | O10—S2—C12 | 107.00 (16) |
| C33—C32—C35 | 119.9 (3) | O12—S2—C12 | 106.76 (15) |
| C34—C33—C32 | 120.8 (3) | O17—S3—O18 | 113.70 (16) |
| C34—C33—H33 | 119.6 | O17—S3—O16 | 111.42 (15) |
| C32—C33—H33 | 119.6 | O18—S3—O16 | 111.19 (15) |
| C29—C34—C33 | 120.0 (4) | O17—S3—C19 | 106.86 (15) |
| C29—C34—H34 | 120.0 | O18—S3—C19 | 105.97 (16) |
| C33—C34—H34 | 120.0 | O16—S3—C19 | 107.25 (15) |
| C40—C35—C36 | 117.2 (3) | O22—S4—O23 | 110.66 (17) |
| C40—C35—C32 | 121.4 (3) | O22—S4—O24 | 113.1 (2) |
| C36—C35—C32 | 121.4 (3) | O23—S4—O24 | 111.02 (18) |
| C37—C36—C35 | 121.6 (3) | O22—S4—C26 | 108.67 (16) |
| C37—C36—H36 | 119.2 | O23—S4—C26 | 105.19 (16) |
| C35—C36—H36 | 119.2 | O24—S4—C26 | 107.80 (17) |
| | | |
| C6—C1—C2—O3 | −178.1 (3) | C29—C30—C31—C32 | 1.7 (6) |
| C7—C1—C2—O3 | 3.0 (5) | C30—C31—C32—C33 | −1.1 (6) |
| C6—C1—C2—C3 | 1.7 (5) | C30—C31—C32—C35 | 178.0 (4) |
| C7—C1—C2—C3 | −177.2 (3) | C31—C32—C33—C34 | −0.1 (6) |
| O3—C2—C3—C4 | 179.1 (3) | C35—C32—C33—C34 | −179.3 (3) |
| C1—C2—C3—C4 | −0.7 (6) | C30—C29—C34—C33 | −0.3 (6) |
| C2—C3—C4—C5 | −0.8 (5) | N1—C29—C34—C33 | 178.3 (3) |
| C3—C4—C5—C6 | 1.4 (5) | C32—C33—C34—C29 | 0.8 (6) |
| C3—C4—C5—S1 | −180.0 (3) | C31—C32—C35—C40 | 151.6 (4) |
| C4—C5—C6—C1 | −0.3 (5) | C33—C32—C35—C40 | −29.3 (5) |
| S1—C5—C6—C1 | −179.0 (3) | C31—C32—C35—C36 | −30.0 (5) |
| C2—C1—C6—C5 | −1.2 (5) | C33—C32—C35—C36 | 149.1 (4) |
| C7—C1—C6—C5 | 177.7 (3) | C40—C35—C36—C37 | 0.7 (5) |
| C6—C1—C7—O2 | 179.2 (4) | C32—C35—C36—C37 | −177.8 (3) |
| C2—C1—C7—O2 | −1.9 (6) | C35—C36—C37—C38 | −0.9 (6) |
| C6—C1—C7—O1 | −0.6 (5) | C36—C37—C38—C39 | 0.9 (5) |
| C2—C1—C7—O1 | 178.4 (3) | C36—C37—C38—N2 | 178.9 (3) |
| C13—C8—C9—O9 | −179.7 (3) | C37—C38—C39—C40 | −0.8 (6) |
| C14—C8—C9—O9 | 3.0 (5) | N2—C38—C39—C40 | −178.8 (3) |
| C13—C8—C9—C10 | 0.5 (5) | C38—C39—C40—C35 | 0.6 (6) |
| C14—C8—C9—C10 | −176.8 (3) | C36—C35—C40—C39 | −0.6 (6) |
| O9—C9—C10—C11 | −179.9 (3) | C32—C35—C40—C39 | 177.9 (3) |
| C8—C9—C10—C11 | −0.1 (5) | C46—C41—C42—C43 | 1.6 (6) |
| C9—C10—C11—C12 | −0.5 (5) | N3—C41—C42—C43 | −179.8 (3) |
| C10—C11—C12—C13 | 0.6 (5) | C41—C42—C43—C44 | −1.2 (6) |
| C10—C11—C12—S2 | 179.5 (3) | C42—C43—C44—C45 | 0.7 (6) |
| C11—C12—C13—C8 | −0.2 (5) | C42—C43—C44—C47 | −179.6 (3) |
| S2—C12—C13—C8 | −179.0 (2) | C43—C44—C45—C46 | −0.6 (6) |
| C9—C8—C13—C12 | −0.4 (5) | C47—C44—C45—C46 | 179.7 (3) |
| C14—C8—C13—C12 | 176.8 (3) | C42—C41—C46—C45 | −1.5 (6) |
| C9—C8—C14—O8 | −2.4 (5) | N3—C41—C46—C45 | 179.8 (3) |
| C13—C8—C14—O8 | −179.7 (3) | C44—C45—C46—C41 | 1.0 (6) |
| C9—C8—C14—O7 | 177.1 (3) | C43—C44—C47—C52 | −149.1 (4) |
| C13—C8—C14—O7 | −0.2 (5) | C45—C44—C47—C52 | 30.6 (5) |
| C20—C15—C16—O15 | 179.5 (3) | C43—C44—C47—C48 | 31.8 (5) |
| C21—C15—C16—O15 | −0.2 (5) | C45—C44—C47—C48 | −148.6 (4) |
| C20—C15—C16—C17 | −0.7 (5) | C52—C47—C48—C49 | 1.1 (5) |
| C21—C15—C16—C17 | 179.6 (3) | C44—C47—C48—C49 | −179.7 (3) |
| O15—C16—C17—C18 | −179.3 (3) | C47—C48—C49—C50 | −0.6 (6) |
| C15—C16—C17—C18 | 0.9 (5) | C48—C49—C50—C51 | −0.5 (6) |
| C16—C17—C18—C19 | −0.6 (5) | C48—C49—C50—N4 | −179.4 (3) |
| C17—C18—C19—C20 | 0.1 (5) | C49—C50—C51—C52 | 1.1 (6) |
| C17—C18—C19—S3 | −178.9 (3) | N4—C50—C51—C52 | −180.0 (3) |
| C18—C19—C20—C15 | 0.1 (5) | C50—C51—C52—C47 | −0.5 (6) |
| S3—C19—C20—C15 | 179.1 (3) | C48—C47—C52—C51 | −0.5 (6) |
| C16—C15—C20—C19 | 0.1 (5) | C44—C47—C52—C51 | −179.7 (3) |
| C21—C15—C20—C19 | 179.9 (3) | C6—C5—S1—O4 | −49.9 (3) |
| C20—C15—C21—O14 | 179.7 (3) | C4—C5—S1—O4 | 131.5 (3) |
| C16—C15—C21—O14 | −0.6 (5) | C6—C5—S1—O6 | −171.2 (3) |
| C20—C15—C21—O13 | −0.3 (5) | C4—C5—S1—O6 | 10.1 (3) |
| C16—C15—C21—O13 | 179.4 (3) | C6—C5—S1—O5 | 69.0 (3) |
| C27—C22—C23—O21 | −179.8 (4) | C4—C5—S1—O5 | −109.6 (3) |
| C28—C22—C23—O21 | 0.3 (6) | C13—C12—S2—O11 | −137.8 (3) |
| C27—C22—C23—C24 | 1.3 (6) | C11—C12—S2—O11 | 43.4 (3) |
| C28—C22—C23—C24 | −178.7 (4) | C13—C12—S2—O10 | −16.4 (3) |
| O21—C23—C24—C25 | −180.0 (4) | C11—C12—S2—O10 | 164.8 (3) |
| C22—C23—C24—C25 | −1.0 (6) | C13—C12—S2—O12 | 103.7 (3) |
| C23—C24—C25—C26 | 0.2 (6) | C11—C12—S2—O12 | −75.1 (3) |
| C24—C25—C26—C27 | 0.2 (6) | C20—C19—S3—O17 | 135.8 (3) |
| C24—C25—C26—S4 | −178.5 (3) | C18—C19—S3—O17 | −45.2 (3) |
| C25—C26—C27—C22 | 0.1 (5) | C20—C19—S3—O18 | 14.3 (3) |
| S4—C26—C27—C22 | 178.7 (3) | C18—C19—S3—O18 | −166.7 (3) |
| C23—C22—C27—C26 | −0.9 (5) | C20—C19—S3—O16 | −104.6 (3) |
| C28—C22—C27—C26 | 179.1 (3) | C18—C19—S3—O16 | 74.4 (3) |
| C23—C22—C28—O20 | 0.2 (6) | C27—C26—S4—O22 | 7.9 (4) |
| C27—C22—C28—O20 | −179.7 (4) | C25—C26—S4—O22 | −173.4 (3) |
| C23—C22—C28—O19 | 179.5 (3) | C27—C26—S4—O23 | −110.6 (3) |
| C27—C22—C28—O19 | −0.5 (5) | C25—C26—S4—O23 | 68.0 (3) |
| C34—C29—C30—C31 | −0.9 (6) | C27—C26—S4—O24 | 130.8 (3) |
| N1—C29—C30—C31 | −179.5 (4) | C25—C26—S4—O24 | −50.5 (3) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···O5 | 0.87 (2) | 2.05 (2) | 2.917 (4) | 175 (4) |
| N1—H1B···O24i | 0.85 (2) | 1.92 (2) | 2.756 (4) | 168 (4) |
| N1—H1C···O12 | 0.87 (2) | 2.06 (2) | 2.927 (4) | 170 (3) |
| N2—H2A···O17 | 0.87 (2) | 2.12 (2) | 2.901 (4) | 150 (3) |
| N2—H2B···O22 | 0.87 (2) | 2.17 (2) | 3.010 (5) | 162 (3) |
| N2—H2C···O12i | 0.86 (2) | 2.18 (3) | 2.918 (4) | 143 (3) |
| N2—H2C···O25ii | 0.86 (2) | 2.37 (4) | 2.868 (4) | 117 (3) |
| N3—H3A···O4 | 0.85 (2) | 1.94 (2) | 2.775 (4) | 166 (4) |
| N3—H3B···O11 | 0.87 (2) | 1.94 (2) | 2.796 (4) | 171 (4) |
| N3—H3C···O16iii | 0.87 (2) | 2.29 (3) | 3.005 (4) | 139 (3) |
| N3—H3C···O26 | 0.87 (2) | 2.38 (3) | 2.954 (5) | 124 (3) |
| N4—H4A···O16 | 0.88 (2) | 1.98 (2) | 2.858 (4) | 170 (3) |
| N4—H4B···O28iv | 0.88 (2) | 1.89 (2) | 2.755 (4) | 167 (3) |
| N4—H4C···O22 | 0.88 (2) | 2.11 (2) | 2.964 (4) | 165 (3) |
| O1—H1D···O25v | 0.83 (2) | 1.77 (2) | 2.585 (4) | 165 (5) |
| O3—H3D···O2 | 0.83 (2) | 1.87 (3) | 2.601 (4) | 147 (5) |
| O7—H7A···O5vi | 0.81 (2) | 2.04 (2) | 2.846 (3) | 169 (4) |
| O9—H9A···O8 | 0.83 (4) | 1.84 (3) | 2.580 (4) | 148 (4) |
| O13—H13A···O27vii | 0.83 (2) | 1.82 (2) | 2.639 (4) | 170 (5) |
| O15—H15A···O14 | 0.82 (4) | 1.81 (3) | 2.582 (4) | 155 (4) |
| O19—H19A···O26viii | 0.84 (2) | 1.80 (2) | 2.631 (4) | 171 (5) |
| O21—H21A···O20 | 0.81 (4) | 1.84 (3) | 2.572 (4) | 148 (5) |
| O25—H25A···O6 | 0.82 (2) | 1.97 (2) | 2.781 (4) | 177 (5) |
| O25—H25B···O10ix | 0.82 (2) | 1.91 (2) | 2.715 (4) | 168 (5) |
| O26—H26A···O23x | 0.82 (2) | 2.08 (3) | 2.822 (4) | 150 (4) |
| O26—H26B···O18iii | 0.81 (2) | 1.97 (2) | 2.782 (4) | 175 (5) |
| O27—H27A···O23 | 0.84 (2) | 2.01 (2) | 2.838 (4) | 170 (5) |
| O27—H27B···O28iv | 0.82 (2) | 2.33 (3) | 3.089 (5) | 156 (4) |
| O28—H28A···O6ii | 0.84 (2) | 2.05 (3) | 2.847 (4) | 159 (5) |
| O28—H28B···O18 | 0.84 (2) | 2.00 (2) | 2.805 (4) | 160 (5) |
| C11—H11···O4 | 0.93 | 2.45 | 3.341 (4) | 160 |
| C18—H18···O22 | 0.93 | 2.47 | 3.381 (4) | 165 |
| C48—H48···O3ii | 0.93 | 2.56 | 3.475 (5) | 168 |
| C51—H51···O23 | 0.93 | 2.52 | 3.177 (4) | 128 |
| Symmetry codes: (i) −x, −y+1, −z+1; (ii) x, −y+1/2, z+1/2; (iii) −x+1, −y+1, −z+1; (iv) −x+1, y+1/2, −z+3/2; (v) x, −y+1/2, z−1/2; (vi) −x, −y+1, −z; (vii) −x+1, −y+1, −z+2; (viii) x, y, z+1; (ix) −x, y−1/2, −z+1/2; (x) x, −y+3/2, z−1/2. |
(II) biphenyl-4,4'-diaminium bis(3-carboxy-4-hydroxybenzenesulfonate) dihydrate
top
Crystal data top
| C12H14N22+·2C7H5O6S−·2H2O | Z = 1 |
| Mr = 656.62 | F(000) = 342 |
| Triclinic, P1 | Dx = 1.518 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.6625 (14) Å | Cell parameters from 4314 reflections |
| b = 9.2851 (15) Å | θ = 2.5–28.0° |
| c = 9.7249 (16) Å | µ = 0.26 mm−1 |
| α = 76.514 (2)° | T = 298 K |
| β = 72.834 (3)° | Block, colourless |
| γ = 77.947 (3)° | 0.23 × 0.20 × 0.20 mm |
| V = 718.4 (2) Å3 | |
Data collection top
Bruker SMART APEX CCD area-detector
diffractometer | 3068 independent reflections |
| Radiation source: fine focus sealed Siemens Mo tube | 2615 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.094 |
| 0.3° wide ω exposures scans | θmax = 27.0°, θmin = 2.2° |
Absorption correction: multi-scan
(SADABS; Sheldrick, 2003) | h = −11→10 |
| Tmin = 0.932, Tmax = 0.950 | k = −11→11 |
| 7548 measured reflections | l = −12→12 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.054 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.154 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0878P)2 + 0.1335P]
where P = (Fo2 + 2Fc2)/3 |
| 3068 reflections | (Δ/σ)max < 0.001 |
| 220 parameters | Δρmax = 0.42 e Å−3 |
| 7 restraints | Δρmin = −0.41 e Å−3 |
Crystal data top
| C12H14N22+·2C7H5O6S−·2H2O | γ = 77.947 (3)° |
| Mr = 656.62 | V = 718.4 (2) Å3 |
| Triclinic, P1 | Z = 1 |
| a = 8.6625 (14) Å | Mo Kα radiation |
| b = 9.2851 (15) Å | µ = 0.26 mm−1 |
| c = 9.7249 (16) Å | T = 298 K |
| α = 76.514 (2)° | 0.23 × 0.20 × 0.20 mm |
| β = 72.834 (3)° | |
Data collection top
Bruker SMART APEX CCD area-detector
diffractometer | 3068 independent reflections |
Absorption correction: multi-scan
(SADABS; Sheldrick, 2003) | 2615 reflections with I > 2σ(I) |
| Tmin = 0.932, Tmax = 0.950 | Rint = 0.094 |
| 7548 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.054 | 7 restraints |
| wR(F2) = 0.154 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.07 | Δρmax = 0.42 e Å−3 |
| 3068 reflections | Δρmin = −0.41 e Å−3 |
| 220 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R-factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.3543 (2) | 0.6728 (2) | 0.0196 (2) | 0.0441 (4) | |
| C2 | 0.2505 (3) | 0.5798 (3) | 0.0116 (2) | 0.0530 (5) | |
| C3 | 0.1433 (3) | 0.5187 (3) | 0.1391 (3) | 0.0668 (7) | |
| H3 | 0.0761 | 0.4546 | 0.1347 | 0.080* | |
| C4 | 0.1359 (3) | 0.5521 (3) | 0.2706 (2) | 0.0585 (6) | |
| H4 | 0.0636 | 0.5107 | 0.3551 | 0.070* | |
| C5 | 0.2356 (2) | 0.6475 (2) | 0.27914 (19) | 0.0424 (4) | |
| C6 | 0.3449 (2) | 0.7071 (2) | 0.1541 (2) | 0.0432 (4) | |
| H6 | 0.4123 | 0.7703 | 0.1597 | 0.052* | |
| C7 | 0.4731 (3) | 0.7322 (2) | −0.1144 (2) | 0.0481 (5) | |
| C8 | 0.2643 (2) | 0.1713 (2) | 0.6809 (2) | 0.0426 (4) | |
| C9 | 0.2135 (4) | 0.0426 (3) | 0.6812 (2) | 0.0655 (7) | |
| H9 | 0.2473 | 0.0000 | 0.5975 | 0.079* | |
| C10 | 0.1106 (4) | −0.0249 (3) | 0.8072 (2) | 0.0643 (7) | |
| H10 | 0.0760 | −0.1130 | 0.8065 | 0.077* | |
| C11 | 0.0580 (2) | 0.0342 (2) | 0.93340 (19) | 0.0402 (4) | |
| C12 | 0.1196 (4) | 0.1609 (3) | 0.9299 (2) | 0.0706 (8) | |
| H12 | 0.0917 | 0.2014 | 1.0144 | 0.085* | |
| C13 | 0.2220 (4) | 0.2299 (3) | 0.8038 (3) | 0.0713 (8) | |
| H13 | 0.2610 | 0.3158 | 0.8041 | 0.086* | |
| N1 | 0.3671 (2) | 0.2485 (2) | 0.54603 (18) | 0.0502 (4) | |
| H1A | 0.360 (3) | 0.218 (3) | 0.471 (2) | 0.060* | |
| H1B | 0.332 (3) | 0.345 (2) | 0.542 (3) | 0.060* | |
| H1C | 0.469 (2) | 0.224 (3) | 0.551 (3) | 0.060* | |
| O1 | 0.5712 (2) | 0.8084 (2) | −0.09146 (16) | 0.0642 (5) | |
| H1 | 0.642 (3) | 0.835 (3) | −0.165 (2) | 0.077* | |
| O2 | 0.4807 (2) | 0.7094 (2) | −0.23561 (16) | 0.0609 (4) | |
| O3 | 0.2472 (3) | 0.5477 (2) | −0.11568 (19) | 0.0734 (5) | |
| H3A | 0.327 (3) | 0.577 (4) | −0.176 (3) | 0.110* | |
| O4 | 0.0499 (2) | 0.7332 (2) | 0.5150 (2) | 0.0761 (5) | |
| O5 | 0.2893 (3) | 0.55556 (18) | 0.53541 (17) | 0.0696 (5) | |
| O6 | 0.3166 (2) | 0.81066 (17) | 0.42094 (15) | 0.0548 (4) | |
| O7 | −0.2134 (3) | 0.8899 (2) | 0.6720 (2) | 0.0806 (6) | |
| H7A | −0.124 (3) | 0.852 (4) | 0.638 (4) | 0.121* | |
| H7B | −0.217 (5) | 0.978 (2) | 0.641 (4) | 0.121* | |
| S1 | 0.22125 (6) | 0.68960 (5) | 0.45057 (5) | 0.0441 (2) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1 | 0.0459 (10) | 0.0443 (10) | 0.0386 (9) | −0.0118 (8) | −0.0039 (8) | −0.0053 (7) |
| C2 | 0.0614 (13) | 0.0540 (12) | 0.0462 (10) | −0.0198 (10) | −0.0120 (9) | −0.0070 (9) |
| C3 | 0.0720 (16) | 0.0757 (16) | 0.0602 (13) | −0.0436 (13) | −0.0152 (12) | −0.0009 (12) |
| C4 | 0.0574 (13) | 0.0677 (14) | 0.0473 (11) | −0.0307 (11) | −0.0046 (9) | 0.0027 (10) |
| C5 | 0.0416 (10) | 0.0421 (10) | 0.0364 (9) | −0.0095 (8) | −0.0023 (7) | −0.0010 (7) |
| C6 | 0.0438 (10) | 0.0427 (10) | 0.0383 (9) | −0.0136 (8) | −0.0001 (7) | −0.0055 (7) |
| C7 | 0.0511 (11) | 0.0519 (11) | 0.0373 (10) | −0.0122 (9) | −0.0013 (8) | −0.0095 (8) |
| C8 | 0.0472 (10) | 0.0407 (10) | 0.0344 (9) | −0.0076 (8) | −0.0056 (7) | −0.0018 (7) |
| C9 | 0.0955 (18) | 0.0627 (14) | 0.0380 (10) | −0.0334 (13) | 0.0044 (11) | −0.0173 (10) |
| C10 | 0.0949 (18) | 0.0549 (13) | 0.0441 (11) | −0.0371 (12) | 0.0017 (11) | −0.0137 (9) |
| C11 | 0.0444 (10) | 0.0399 (9) | 0.0352 (9) | −0.0088 (8) | −0.0102 (8) | −0.0029 (7) |
| C12 | 0.0975 (19) | 0.0790 (17) | 0.0414 (11) | −0.0506 (15) | 0.0084 (11) | −0.0235 (11) |
| C13 | 0.0949 (19) | 0.0724 (16) | 0.0507 (12) | −0.0514 (15) | 0.0094 (12) | −0.0213 (11) |
| N1 | 0.0606 (11) | 0.0440 (9) | 0.0367 (8) | −0.0095 (8) | −0.0020 (8) | −0.0018 (7) |
| O1 | 0.0691 (11) | 0.0856 (12) | 0.0384 (7) | −0.0421 (9) | 0.0078 (7) | −0.0132 (7) |
| O2 | 0.0681 (10) | 0.0762 (11) | 0.0380 (7) | −0.0217 (8) | −0.0027 (7) | −0.0141 (7) |
| O3 | 0.0940 (14) | 0.0865 (13) | 0.0533 (9) | −0.0458 (11) | −0.0163 (9) | −0.0139 (9) |
| O4 | 0.0549 (10) | 0.0904 (14) | 0.0665 (11) | −0.0055 (9) | 0.0126 (8) | −0.0241 (10) |
| O5 | 0.1083 (14) | 0.0468 (9) | 0.0460 (8) | −0.0043 (9) | −0.0220 (9) | 0.0027 (7) |
| O6 | 0.0697 (10) | 0.0488 (8) | 0.0419 (7) | −0.0177 (7) | −0.0042 (7) | −0.0060 (6) |
| O7 | 0.0754 (12) | 0.0529 (10) | 0.0710 (11) | −0.0066 (9) | 0.0293 (9) | 0.0058 (8) |
| S1 | 0.0481 (3) | 0.0391 (3) | 0.0342 (3) | −0.0066 (2) | 0.00214 (19) | −0.00189 (18) |
Geometric parameters (Å, º) top
| C1—C6 | 1.393 (3) | C9—H9 | 0.9300 |
| C1—C2 | 1.398 (3) | C10—C11 | 1.377 (3) |
| C1—C7 | 1.473 (3) | C10—H10 | 0.9300 |
| C2—O3 | 1.348 (3) | C11—C12 | 1.379 (3) |
| C2—C3 | 1.391 (3) | C11—C11i | 1.485 (4) |
| C3—C4 | 1.365 (3) | C12—C13 | 1.389 (3) |
| C3—H3 | 0.9300 | C12—H12 | 0.9300 |
| C4—C5 | 1.391 (3) | C13—H13 | 0.9300 |
| C4—H4 | 0.9300 | N1—H1A | 0.861 (16) |
| C5—C6 | 1.381 (2) | N1—H1B | 0.876 (17) |
| C5—S1 | 1.763 (2) | N1—H1C | 0.879 (17) |
| C6—H6 | 0.9300 | O1—H1 | 0.819 (17) |
| C7—O2 | 1.226 (2) | O3—H3A | 0.812 (19) |
| C7—O1 | 1.308 (3) | O4—S1 | 1.4418 (18) |
| C8—C13 | 1.349 (3) | O5—S1 | 1.4418 (16) |
| C8—C9 | 1.357 (3) | O6—S1 | 1.4577 (16) |
| C8—N1 | 1.471 (2) | O7—H7A | 0.787 (19) |
| C9—C10 | 1.385 (3) | O7—H7B | 0.799 (19) |
| | | |
| C6—C1—C2 | 119.67 (18) | C11—C10—C9 | 122.1 (2) |
| C6—C1—C7 | 120.86 (18) | C11—C10—H10 | 118.9 |
| C2—C1—C7 | 119.47 (18) | C9—C10—H10 | 118.9 |
| O3—C2—C3 | 117.7 (2) | C10—C11—C12 | 116.25 (18) |
| O3—C2—C1 | 122.91 (19) | C10—C11—C11i | 122.2 (2) |
| C3—C2—C1 | 119.4 (2) | C12—C11—C11i | 121.5 (2) |
| C4—C3—C2 | 120.5 (2) | C11—C12—C13 | 122.0 (2) |
| C4—C3—H3 | 119.8 | C11—C12—H12 | 119.0 |
| C2—C3—H3 | 119.8 | C13—C12—H12 | 119.0 |
| C3—C4—C5 | 120.5 (2) | C8—C13—C12 | 119.5 (2) |
| C3—C4—H4 | 119.7 | C8—C13—H13 | 120.3 |
| C5—C4—H4 | 119.7 | C12—C13—H13 | 120.3 |
| C6—C5—C4 | 119.80 (18) | C8—N1—H1A | 109.8 (18) |
| C6—C5—S1 | 120.92 (15) | C8—N1—H1B | 106.7 (17) |
| C4—C5—S1 | 119.28 (15) | H1A—N1—H1B | 112 (2) |
| C5—C6—C1 | 120.08 (18) | C8—N1—H1C | 108.2 (17) |
| C5—C6—H6 | 120.0 | H1A—N1—H1C | 108 (2) |
| C1—C6—H6 | 120.0 | H1B—N1—H1C | 112 (2) |
| O2—C7—O1 | 123.32 (19) | C7—O1—H1 | 114 (2) |
| O2—C7—C1 | 122.82 (19) | C2—O3—H3A | 106 (3) |
| O1—C7—C1 | 113.85 (17) | H7A—O7—H7B | 107 (4) |
| C13—C8—C9 | 120.63 (19) | O5—S1—O4 | 112.12 (11) |
| C13—C8—N1 | 119.20 (19) | O5—S1—O6 | 111.65 (11) |
| C9—C8—N1 | 120.17 (18) | O4—S1—O6 | 112.81 (11) |
| C8—C9—C10 | 119.4 (2) | O5—S1—C5 | 107.10 (9) |
| C8—C9—H9 | 120.3 | O4—S1—C5 | 106.35 (10) |
| C10—C9—H9 | 120.3 | O6—S1—C5 | 106.32 (9) |
| | | |
| C6—C1—C2—O3 | 176.7 (2) | C13—C8—C9—C10 | 3.2 (4) |
| C7—C1—C2—O3 | −3.8 (3) | N1—C8—C9—C10 | −177.0 (2) |
| C6—C1—C2—C3 | −2.2 (3) | C8—C9—C10—C11 | −0.2 (4) |
| C7—C1—C2—C3 | 177.3 (2) | C9—C10—C11—C12 | −3.0 (4) |
| O3—C2—C3—C4 | −177.2 (2) | C9—C10—C11—C11i | 177.3 (3) |
| C1—C2—C3—C4 | 1.7 (4) | C10—C11—C12—C13 | 3.4 (4) |
| C2—C3—C4—C5 | −0.1 (4) | C11i—C11—C12—C13 | −176.9 (3) |
| C3—C4—C5—C6 | −1.1 (4) | C9—C8—C13—C12 | −2.8 (4) |
| C3—C4—C5—S1 | 179.4 (2) | N1—C8—C13—C12 | 177.4 (3) |
| C4—C5—C6—C1 | 0.5 (3) | C11—C12—C13—C8 | −0.6 (5) |
| S1—C5—C6—C1 | −179.91 (15) | C6—C5—S1—O5 | −109.05 (18) |
| C2—C1—C6—C5 | 1.1 (3) | C4—C5—S1—O5 | 70.5 (2) |
| C7—C1—C6—C5 | −178.41 (18) | C6—C5—S1—O4 | 130.89 (18) |
| C6—C1—C7—O2 | −177.0 (2) | C4—C5—S1—O4 | −49.5 (2) |
| C2—C1—C7—O2 | 3.4 (3) | C6—C5—S1—O6 | 10.44 (19) |
| C6—C1—C7—O1 | 4.3 (3) | C4—C5—S1—O6 | −170.00 (18) |
| C2—C1—C7—O1 | −175.2 (2) | | |
| Symmetry code: (i) −x, −y, −z+2. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···O2ii | 0.86 (2) | 2.33 (2) | 2.884 (2) | 123 (2) |
| N1—H1B···O5 | 0.88 (2) | 1.91 (2) | 2.774 (3) | 171 (2) |
| N1—H1C···O6iii | 0.88 (2) | 1.91 (2) | 2.779 (3) | 172 (2) |
| O1—H1···O7iv | 0.82 (2) | 1.75 (2) | 2.567 (2) | 177 (3) |
| O3—H3A···O2 | 0.81 (2) | 1.88 (3) | 2.601 (2) | 148 (4) |
| O7—H7A···O4 | 0.79 (2) | 1.94 (2) | 2.691 (2) | 159 (4) |
| O7—H7B···O6v | 0.80 (2) | 2.00 (2) | 2.760 (2) | 158 (4) |
| C4—H4···O4vi | 0.93 | 2.56 | 3.343 (3) | 142 |
| C13—H13···O3vii | 0.93 | 2.43 | 3.289 (3) | 155 |
| Symmetry codes: (ii) −x+1, −y+1, −z; (iii) −x+1, −y+1, −z+1; (iv) x+1, y, z−1; (v) −x, −y+2, −z+1; (vi) −x, −y+1, −z+1; (vii) x, y, z+1. |
Experimental details
| | (I) | (II) |
| Crystal data |
| Chemical formula | C12H14N22+·2C7H5O6S−·2H2O | C12H14N22+·2C7H5O6S−·2H2O |
| Mr | 656.62 | 656.62 |
| Crystal system, space group | Monoclinic, P21/c | Triclinic, P1 |
| Temperature (K) | 298 | 298 |
| a, b, c (Å) | 15.1851 (3), 18.6584 (6), 19.9114 (6) | 8.6625 (14), 9.2851 (15), 9.7249 (16) |
| α, β, γ (°) | 90, 91.300 (2), 90 | 76.514 (2), 72.834 (3), 77.947 (3) |
| V (Å3) | 5640.0 (3) | 718.4 (2) |
| Z | 8 | 1 |
| Radiation type | Mo Kα | Mo Kα |
| µ (mm−1) | 0.27 | 0.26 |
| Crystal size (mm) | 0.40 × 0.10 × 0.04 | 0.23 × 0.20 × 0.20 |
| |
| Data collection |
| Diffractometer | Bruker SMART APEX CCD area-detector
diffractometer | Bruker SMART APEX CCD area-detector
diffractometer |
| Absorption correction | Multi-scan
(SADABS; Sheldrick, 2003) | Multi-scan
(SADABS; Sheldrick, 2003) |
| Tmin, Tmax | 0.901, 0.989 | 0.932, 0.950 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 48494, 12875, 6411 | 7548, 3068, 2615 |
| Rint | 0.100 | 0.094 |
| (sin θ/λ)max (Å−1) | 0.650 | 0.639 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.063, 0.165, 0.95 | 0.054, 0.154, 1.07 |
| No. of reflections | 12875 | 3068 |
| No. of parameters | 877 | 220 |
| No. of restraints | 28 | 7 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.49, −0.27 | 0.42, −0.41 |
Hydrogen-bond geometry (Å, º) for (I) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···O5 | 0.872 (18) | 2.047 (19) | 2.917 (4) | 175 (4) |
| N1—H1B···O24i | 0.851 (18) | 1.92 (2) | 2.756 (4) | 168 (4) |
| N1—H1C···O12 | 0.874 (18) | 2.06 (2) | 2.927 (4) | 170 (3) |
| N2—H2A···O17 | 0.867 (18) | 2.12 (2) | 2.901 (4) | 150 (3) |
| N2—H2B···O22 | 0.872 (18) | 2.17 (2) | 3.010 (5) | 162 (3) |
| N2—H2C···O12i | 0.863 (18) | 2.18 (3) | 2.918 (4) | 143 (3) |
| N2—H2C···O25ii | 0.863 (18) | 2.37 (4) | 2.868 (4) | 117 (3) |
| N3—H3A···O4 | 0.854 (18) | 1.94 (2) | 2.775 (4) | 166 (4) |
| N3—H3B···O11 | 0.867 (18) | 1.936 (19) | 2.796 (4) | 171 (4) |
| N3—H3C···O16iii | 0.872 (18) | 2.29 (3) | 3.005 (4) | 139 (3) |
| N3—H3C···O26 | 0.872 (18) | 2.38 (3) | 2.954 (5) | 124 (3) |
| N4—H4A···O16 | 0.883 (18) | 1.984 (19) | 2.858 (4) | 170 (3) |
| N4—H4B···O28iv | 0.877 (18) | 1.89 (2) | 2.755 (4) | 167 (3) |
| N4—H4C···O22 | 0.876 (18) | 2.11 (2) | 2.964 (4) | 165 (3) |
| O1—H1D···O25v | 0.831 (19) | 1.77 (2) | 2.585 (4) | 165 (5) |
| O3—H3D···O2 | 0.830 (19) | 1.87 (3) | 2.601 (4) | 147 (5) |
| O7—H7A···O5vi | 0.814 (18) | 2.04 (2) | 2.846 (3) | 169 (4) |
| O9—H9A···O8 | 0.83 (4) | 1.84 (3) | 2.580 (4) | 148 (4) |
| O13—H13A···O27vii | 0.826 (19) | 1.82 (2) | 2.639 (4) | 170 (5) |
| O15—H15A···O14 | 0.82 (4) | 1.81 (3) | 2.582 (4) | 155 (4) |
| O19—H19A···O26viii | 0.840 (19) | 1.80 (2) | 2.631 (4) | 171 (5) |
| O21—H21A···O20 | 0.81 (4) | 1.84 (3) | 2.572 (4) | 148 (5) |
| O25—H25A···O6 | 0.815 (19) | 1.966 (19) | 2.781 (4) | 177 (5) |
| O25—H25B···O10ix | 0.816 (19) | 1.91 (2) | 2.715 (4) | 168 (5) |
| O26—H26A···O23x | 0.820 (19) | 2.08 (3) | 2.822 (4) | 150 (4) |
| O26—H26B···O18iii | 0.813 (19) | 1.971 (19) | 2.782 (4) | 175 (5) |
| O27—H27A···O23 | 0.836 (19) | 2.01 (2) | 2.838 (4) | 170 (5) |
| O27—H27B···O28iv | 0.817 (19) | 2.33 (3) | 3.089 (5) | 156 (4) |
| O28—H28A···O6ii | 0.842 (19) | 2.05 (3) | 2.847 (4) | 159 (5) |
| O28—H28B···O18 | 0.839 (19) | 2.00 (2) | 2.805 (4) | 160 (5) |
| C11—H11···O4 | 0.93 | 2.45 | 3.341 (4) | 160 |
| C18—H18···O22 | 0.93 | 2.47 | 3.381 (4) | 165 |
| C48—H48···O3ii | 0.93 | 2.56 | 3.475 (5) | 168 |
| C51—H51···O23 | 0.93 | 2.52 | 3.177 (4) | 128 |
| Symmetry codes: (i) −x, −y+1, −z+1; (ii) x, −y+1/2, z+1/2; (iii) −x+1, −y+1, −z+1; (iv) −x+1, y+1/2, −z+3/2; (v) x, −y+1/2, z−1/2; (vi) −x, −y+1, −z; (vii) −x+1, −y+1, −z+2; (viii) x, y, z+1; (ix) −x, y−1/2, −z+1/2; (x) x, −y+3/2, z−1/2. |
Hydrogen-bond geometry (Å, º) for (II) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···O2i | 0.861 (16) | 2.33 (2) | 2.884 (2) | 123 (2) |
| N1—H1B···O5 | 0.876 (17) | 1.905 (18) | 2.774 (3) | 171 (2) |
| N1—H1C···O6ii | 0.879 (17) | 1.905 (18) | 2.779 (3) | 172 (2) |
| O1—H1···O7iii | 0.819 (17) | 1.748 (18) | 2.567 (2) | 177 (3) |
| O3—H3A···O2 | 0.812 (19) | 1.88 (3) | 2.601 (2) | 148 (4) |
| O7—H7A···O4 | 0.787 (19) | 1.94 (2) | 2.691 (2) | 159 (4) |
| O7—H7B···O6iv | 0.799 (19) | 2.00 (2) | 2.760 (2) | 158 (4) |
| C4—H4···O4v | 0.93 | 2.56 | 3.343 (3) | 142 |
| C13—H13···O3vi | 0.93 | 2.43 | 3.289 (3) | 155 |
| Symmetry codes: (i) −x+1, −y+1, −z; (ii) −x+1, −y+1, −z+1; (iii) x+1, y, z−1; (iv) −x, −y+2, −z+1; (v) −x, −y+1, −z+1; (vi) x, y, z+1. |
 O (X = O, N and C) hydrogen bonds. In addition,
O (X = O, N and C) hydrogen bonds. In addition, ![[pi]](/logos/entities/pi_rmgif.gif) -
-![[pi]](/logos/entities/pi_rmgif.gif) interactions are observed in (I) between inversion-related benzene rings [centroid-centroid distances = 3.632 (2) and 3.627 (2) Å]. In order to simplify the complex three-dimensional networks in (I) and (II), we also give their rationalized topological analyses.
interactions are observed in (I) between inversion-related benzene rings [centroid-centroid distances = 3.632 (2) and 3.627 (2) Å]. In order to simplify the complex three-dimensional networks in (I) and (II), we also give their rationalized topological analyses.
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2010/10/00/sk3381/sk3381fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2010/10/00/sk3381/sk3381fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2010/10/00/sk3381/sk3381fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/c/issues/2010/10/00/sk3381/sk3381fig4thm.gif)
![[Figure 5]](http://journals.iucr.org/c/issues/2010/10/00/sk3381/sk3381fig5thm.gif)
![[Figure 6]](http://journals.iucr.org/c/issues/2010/10/00/sk3381/sk3381fig6thm.gif)
![[Figure 7]](http://journals.iucr.org/c/issues/2010/10/00/sk3381/sk3381fig7thm.gif)
![[Figure 8]](http://journals.iucr.org/c/issues/2010/10/00/sk3381/sk3381fig8thm.gif)




Polymorphism, the existence of more than one crystalline form of a compound, is a well known and widely studied phenomenon in the fields of materials (Girlando et al., 2010) and active pharmaceutical ingredients (APIs) (Robinson, 2010; Arenas-García et al., 2010). Benzidine (BZD) is capable of crystallizing in at least four crystalline forms, owing to rotation about its central C—C single bond (Rafilovich & Bernstein, 2006; Rafilovich et al., 2007; Dobrzycki & Wozniak, 2006). This in turn provides a key material for crystallographers to investigate the polymorph phenomenon. However, to date, many studies of polymorphism have focused on single-component compounds (Probert et al., 2010), and research into the polymorphism of multi-component compounds is comparatively rare (Stahly, 2007; Budzianowski et al., 2010). In this paper, we report two polymorphic forms of a hydrated binary organic adduct, benzidinium bis(5-sulfosalicylate) dihydrate, (I) and (II).
Both polymorphs of the title compound were unexpectedly obtained simultaneously by mixing BZD and 5-sulfosalicylic acid (5-H2SSA) in a 1:2 molar ratio. Crystals of two shapes, plates and blocks, were formed over a period of two weeks. X-ray diffraction showed the plate crystals, (I), to be monoclinic, space group P21/c, Z' = 2, with the asymmetric unit consisting of two benzidinium divalent cations, four 5-sulfonate mono-anions (5-HSSA-) and four solvent water molecules (Fig. 1). For all 5-HSSA- anions in (I), common intramolecular S(6) hydrogen-bonds were formed [see Bernstein et al. (1995) for the definition of hydrogen-bond motifs]. The sulfonic O atoms are positioned slightly differently relative to their respective benzene rings. For instance, the oxygen-to-benzene plane distances are 0.214, 1.038 and 1.326 Å, 0.353, 0.917 and 1.383 Å, 0.305, 0.947 and 1.373 Å, and 0.146, 1.037 and 1.322 Å for the S1, S2, S3 and S4 sulfonate anions, respectively, and these values are comparable with those in some analogues (Meng et al., 2007, 2008). For the two BZD2+ cations in the asymmetric unit of (I), the dihedral angles between two benzene planes are 29.5 (2) and 31.0 (2)° for the N1/N2- and N3/N4-containing cations, respectively.
In comparison, the block-shaped crystals of (II) crystallize in the triclinic space group P1 with Z' = 0.5, i.e. its asymmetric unit is composed of half a benzidinium cation, one 5-HSSA- anion and one water molecule (Fig. 2). An inversion centre is located at the mid-point of the central C—C single bond of the BZD2+ cation and a dihedral angle of exactly 0° is formed between the two benzene rings. The three sulfonic O atoms, O4, O5 and O6 [Please check order of added atoms], are 0.278, 1.073 and 1.275 Å, respectively, away from the benzene plane. These conformational diversities of both cations and anions may favour the formation of even more polymorphs.
In the packing structures of both (I) and (II), the component ions are linked into complex three-dimensional frameworks by a combination of X—H···O (X = O, N, C) hydrogen bonds. In (I), the supramolecular structure can be relatively easily analysed in terms of the three aspects mentioned below.
Firstly, ammonium atoms N1 act as hydrogen-bond donors via atoms H1A, H1B and H1C, and are hydrogen-bonded to three independent sulfonate O atoms, O5, O24(-x, 1 - y, 1 - z) and O12, respectively. In the same way, atom N2 is also hydrogen-bonded to three other independent sulfonate O atoms, O12, O17 and O12(-x, 1 - y, 1 - z). In addition, atom N2 is also hydrogen-bonded to water molecule O25 at (x, 1/2 - y, 1/2 + z). Thus, the BZD2+ cation, as a whole, is linked to six symmetry-independent 5-HSSA- anions and one water molecule. Similarly, the other benzidinium N atoms, N3 and N4, acting as hydrogen-bond donors via six H atoms, H3A–H3C and H4A–H4C, are linked to seven independent O atoms, of which five are from sulfonate anions and the remaining two from water O atoms.
Secondly, carboxylic atoms O1, O7, O13 and O19 in the four sulfonate anions act as hydrogen-bond donors and link to sulfonate atom O5 at (x, y, 1 + z) and three water atoms, O25 at (x, 1/2 - y, z - 1/2), O26 at (x, y, 1 + z) and O27 at (1 - x, 1 - y, 2 - z), respectively.
Thirdly, as far as the four solvent water molecules are concerned, atom O27 acts as a hydrogen-bond donor to sulfonate atom O23 and water atom O28. The other three water molecules are all hydrogen-bonded to two sulfonate O atoms, i.e. atoms O6 and O10(- x, y - 1/2, 1/2 - z) for water O25, atoms O23(x, 3/2 - y, z - 1/2) and O18(1 - x, 1 - y, 1 - z) for water O26, and atoms O6 and O18(x, 1/2 - y, 1/2 + z) for water O28. These extensive hydrogen bonds are sufficient to link the BZD2+ cations, 5-HSSA- anions and H2O molecules into a complex three-dimensional network in (I) (Table 1 and Fig. 3).
Aiming to understand the network better, we attempted to give it a standard topological simplification (Baburin & Blatov, 2007), i.e. all the ion centroids were regarded as different nodes in a new network. According to the numbers of intermolecular hydrogen bonds originating from one individual ion as both hydrogen-bond donor and acceptor, the two benzidinium cations can be regarded as seven-connected nodes, the S1-, S3- and S4-containing 5-HSSA- anions as six-connected nodes, the S2-containing 5-HSSA- anion as a five-connected node, water molecules O25, O26 and O28 as four-connected nodes and water molecule O27 as a three-connected node. The final 3,4,5,6,7-connected three-dimensionally topological network with stoichiometry (3 - c):(4 - c)3:(5 - c):(6 - c)3:(7 - c)2 was then rationalized with a Schlafli symbol of (3.4.54)(3.4.55.67.7)(3.411.55.64)(3.43.55.65.7)(3.48.55.67)(32.4.52.6)(32.44.52.62)(4.54.6)(42.6)(45.53.65.72) (Fig. 4).
Although it may seem meaningless to simplify the three-dimensional hydrogen-bonding network in (I), it could indeed help us to analyse the topology of the hydrogen-bond nets and molecular packing conveniently. For instance, from a topological viewpoint it can be easily seen that the BZD2+ cations and 5-HSSA- anions in (I) are linked together into a two-dimensional layer running parallel to the (010) plane (Fig. 5). These adjacent (010) layers are further joined together by water molecules, resulting in the final three-dimensional network with hydrogen-bonds as the edges.
In addition to the above-mentioned hydrogen bonds, four C—H···O hydrogen bonds (Table 1) and two π–π stacking interactions are observed in (I). For the two π–π interactions, one exists between inversion-related C8–C13 benzene rings [Cg1···Cg1 = 3.632 (2) Å; symmetry code (-x, 1 - y, z; Cg1 is the centroid of the C8–C13 ring) and the other between inversion-related C15–C20 benzene rings [Cg2···Cg2 = 3.627 (2) Å; symmetry code (1 - x, 1 - y, 2 - z); Cg2 is the centroid of the C15–C20 ring].
In (II), benzidinium atom N1 in the asymmetric unit acts as a hydrogen-bond donor, via atoms H1A, H1B and H1C, to carboxylate atom O2 at (1 - x, 1 - y, - z) and sulfonate atoms O5 and O6 at (1 - x, 1 - y, 1 - z), respectively (Table 2). Thus, each BZD2+ cation should be linked to six independent 5-HSSA- anions, of which four are N—H···Osulfonate and two are N—H···Ocarboxylate hydrogen bonds, forming a two-dimensional layer structure running parallel to the (110) plane (Fig. 6). In one layer, three types of cyclic hydrogen-bond motifs of R44(12), R44(20) and R44(38) are observed. Carboxylate atom O1 is hydrogen-bonded to water atom O7 at (1 + x, y, z - 1), which thus acts as a hydrogen-bond donor. Water atom O7 is hydrogen-bonded to sulfonate atoms O4 at (x + 1, y, z - 1) and O6 at (x - 1, 2 - y, - z) through atoms H7A and H7B, forming an R44(12) hydrogen motif. With the aid of water molecules, these adjacent (110) layers are linked together, forming the final complex three-dimensional network (Fig. 7).
In order to simplify the network in (II), the BZD2+ cation and 5-HSSA- anion can both be regarded as six-connected nodes and water atom O7 as a three-connected node. The whole structure can be rationalized as a 3,6-connected topological three-dimensional network with stoichiometry (3 - c)2:(6 - c)3 and a Schlafli symbol of (42.6)2(45.65.85)2(46.66.83) (Fig. 8).
Further analysis with PLATON (Spek, 2009) shows that no C—H···π or π–π interactions exist in (II). In comparison, two-dimensional layers are formed in both (I) and (II) by hydrogen bonds between BZD2+ cations and 5-HSSA- anions, but their packing patterns are different. This may, to a large extent, be attributed to the differently oriented N—H···O hydrogen bonds, because of their different BZD2+ and 5-HSSA- configurations.
In short, two polymorphs of benzidinium bis(5-sulfosalicylate) dihydrate were obtained simultaneously at room temperature, and this is largely attributed to the conformational diversities of the BZD2+ and 5-HSSA- ions. In order to simplify the hydrogen-bonded network, we have given their standard topological analysis. If the crystallization conditions were to be varied, more polymorphs may be obtained. Further research on this is ongoing in our laboratory.