
In the title compounds, 4-aminopyridinium 4-aminobenzoate dihydrate, C
7H
6NO
2-·C
5H
7N
2+·2H
2O, (I), and 4-aminopyridinium nicotinate, C
5H
7N
2+·C
6H
4NO
2-, (II), the aromatic N atoms of the 4-aminopyridinium cations are protonated. In (I), the asymmetric unit is composed of two 4-aminopyridinium cations, two 4-aminobenzoate anions and four water molecules, and the compound crystallizes in a noncentrosymmetric space group. The two sets of independent molecules of (I) are related by a centre of symmetry which is not part of the space group. In (I), the protonated pyridinium ring H atoms are involved in bifurcated hydrogen bonding with carboxylate O atoms to form an
R12(4) ring motif. The water molecules link the ions to form a two-dimensional network along the (10
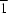
) plane. In (II), an intramolecular bifurcated hydrogen bond generates an
R12(4) ring motif and inter-ion hydrogen bonding generates an
R42(16) ring motif. The packing of adduct (II) is consolidated
via N-H

O and N-H

N hydrogen bonds to form a two-dimensional network along the (10
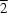
) plane.
Supporting information
CCDC references: 718876; 724593
Compound (I) was prepared by dissolving 4-aminopyridine (0.094 g, 1 mmol) in water (10 ml) and 4-aminobenzoic acid (0.169 g, 1 mmol) in ethanol (10 ml). [The solutions were mixed and the mixture] was stirred well for 3 h. Colourless crystals of (I) were obtained by slow evaporation of the solution over a period of one month. Compound (II) was prepared by dissolving 4-aminopyridine (0.094 g, 1 mmol) in ethanol and nicotinic acid (0.123 g, 1 mmol) in water. The nicotinic acid solution was added dropwise to the 4-aminopyridine. The clear solution obtained was allowed to evaporate slowly. Colourless crystals of (II) suitable for X-ray diffraction were obtained after two weeks.
In (I) and (II), H atoms bonded to C and N atoms were positioned geometrically and treated as riding, with C—H = 0.95 Å and N—H = 0.88 Å, and with Uiso(H) = 1.2Ueq(C,N). H atoms bonded to water O atoms were located in a difference Fourier map and constraints were applied to refine them with Uiso(H) = 1.5Ueq(O). The number of Friedel pairs is 2236 in molecule (I).
For both compounds, data collection: CAD-4 EXPRESS (Enraf–Nonius, 1994); cell refinement: CAD-4 EXPRESS (Enraf–Nonius, 1994); data reduction: CORINC (Draeger & Gattow, 1971); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008); molecular graphics: SHELXTL (Sheldrick, 2008); software used to prepare material for publication: SHELXTL (Sheldrick, 2008) and PLATON (Spek, 2009).
(I) 4-aminopyridinium 4-aminobenzoate dihydrate
top
Crystal data top
| C5H7N2+·C7H6NO2−·2H2O | F(000) = 1136 |
| Mr = 267.29 | Dx = 1.321 Mg m−3 |
| Monoclinic, Cc | Cu Kα radiation, λ = 1.54178 Å |
| Hall symbol: C -2yc | Cell parameters from 25 reflections |
| a = 18.9692 (18) Å | θ = 60–69° |
| b = 7.8092 (4) Å | µ = 0.84 mm−1 |
| c = 19.5944 (19) Å | T = 193 K |
| β = 112.213 (4)° | Block, colourless |
| V = 2687.2 (4) Å3 | 0.38 × 0.32 × 0.16 mm |
| Z = 8 | |
Data collection top
Enraf–Nonius CAD-4
diffractometer | 2334 reflections with I > 2σ(I) |
| Radiation source: rotating anode | Rint = 0.045 |
| Graphite monochromator | θmax = 70.0°, θmin = 4.9° |
| ω/2θ scans | h = −22→22 |
Absorption correction: ψ scan
(CORINC; Draeger & Gattow, 1971) | k = 0→9 |
| Tmin = 0.741, Tmax = 0.877 | l = −23→23 |
| 4793 measured reflections | 3 standard reflections every 60 min |
| 2557 independent reflections | intensity decay: 2% |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.055 | H-atom parameters constrained |
| wR(F2) = 0.156 | w = 1/[σ2(Fo2) + (0.1162P)2 + 0.3843P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max < 0.002 |
| 2557 reflections | Δρmax = 0.28 e Å−3 |
| 344 parameters | Δρmin = −0.37 e Å−3 |
| 14 restraints | Absolute structure: Flack (1983), with 2236 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Absolute structure parameter: 0.2 (3) |
Crystal data top
| C5H7N2+·C7H6NO2−·2H2O | V = 2687.2 (4) Å3 |
| Mr = 267.29 | Z = 8 |
| Monoclinic, Cc | Cu Kα radiation |
| a = 18.9692 (18) Å | µ = 0.84 mm−1 |
| b = 7.8092 (4) Å | T = 193 K |
| c = 19.5944 (19) Å | 0.38 × 0.32 × 0.16 mm |
| β = 112.213 (4)° | |
Data collection top
Enraf–Nonius CAD-4
diffractometer | 2334 reflections with I > 2σ(I) |
Absorption correction: ψ scan
(CORINC; Draeger & Gattow, 1971) | Rint = 0.045 |
| Tmin = 0.741, Tmax = 0.877 | 3 standard reflections every 60 min |
| 4793 measured reflections | intensity decay: 2% |
| 2557 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.055 | H-atom parameters constrained |
| wR(F2) = 0.156 | Δρmax = 0.28 e Å−3 |
| S = 1.05 | Δρmin = −0.37 e Å−3 |
| 2557 reflections | Absolute structure: Flack (1983), with 2236 Friedel pairs |
| 344 parameters | Absolute structure parameter: 0.2 (3) |
| 14 restraints | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1A | 0.4483 (2) | 0.0745 (5) | 0.3941 (2) | 0.0428 (8) | |
| C2A | 0.47909 (18) | 0.0212 (5) | 0.46685 (19) | 0.0420 (8) | |
| H2A | 0.5292 | −0.0244 | 0.4864 | 0.050* | |
| C3A | 0.43750 (19) | 0.0338 (5) | 0.51106 (19) | 0.0412 (8) | |
| H3A | 0.4592 | −0.0031 | 0.5609 | 0.049* | |
| C4A | 0.36214 (16) | 0.1017 (4) | 0.48236 (17) | 0.0361 (7) | |
| C5A | 0.33385 (17) | 0.1549 (5) | 0.41012 (18) | 0.0367 (7) | |
| H5A | 0.2834 | 0.1988 | 0.3900 | 0.044* | |
| C6A | 0.37500 (18) | 0.1474 (5) | 0.36545 (19) | 0.0412 (8) | |
| H6A | 0.3544 | 0.1904 | 0.3165 | 0.049* | |
| C7A | 0.3182 (2) | 0.1135 (4) | 0.53087 (19) | 0.0431 (7) | |
| O8A | 0.24875 (15) | 0.1565 (3) | 0.50276 (15) | 0.0504 (6) | |
| O9A | 0.34981 (17) | 0.0812 (3) | 0.59912 (14) | 0.0522 (7) | |
| N10A | 0.49082 (19) | 0.0594 (6) | 0.35071 (19) | 0.0632 (11) | |
| H10A | 0.5371 | 0.0162 | 0.3693 | 0.076* | |
| H10B | 0.4717 | 0.0931 | 0.3044 | 0.076* | |
| C1B | 0.12976 (19) | 0.8248 (4) | 0.21892 (18) | 0.0370 (7) | |
| C2B | 0.20204 (19) | 0.9014 (5) | 0.2432 (2) | 0.0454 (9) | |
| H2B | 0.2238 | 0.9434 | 0.2922 | 0.054* | |
| C3B | 0.2416 (2) | 0.9171 (6) | 0.1987 (3) | 0.0602 (12) | |
| H3B | 0.2897 | 0.9729 | 0.2160 | 0.072* | |
| N4B | 0.2116 (2) | 0.8516 (4) | 0.1285 (2) | 0.0596 (9) | |
| H4B | 0.2376 | 0.8594 | 0.0997 | 0.072* | |
| C5B | 0.1452 (3) | 0.7783 (5) | 0.1042 (2) | 0.0543 (10) | |
| H5B | 0.1260 | 0.7341 | 0.0554 | 0.065* | |
| C6B | 0.1025 (2) | 0.7621 (5) | 0.14495 (19) | 0.0478 (9) | |
| H6B | 0.0540 | 0.7088 | 0.1246 | 0.057* | |
| N7B | 0.09003 (17) | 0.8107 (4) | 0.26088 (18) | 0.0470 (7) | |
| H7B | 0.1085 | 0.8502 | 0.3063 | 0.056* | |
| H7C | 0.0449 | 0.7618 | 0.2436 | 0.056* | |
| C1C | 0.12642 (19) | 0.3243 (4) | 0.22138 (18) | 0.0375 (7) | |
| C2C | 0.0975 (2) | 0.2690 (5) | 0.14609 (19) | 0.0465 (9) | |
| H2C | 0.0479 | 0.2209 | 0.1251 | 0.056* | |
| C3C | 0.1404 (2) | 0.2846 (5) | 0.10423 (19) | 0.0444 (9) | |
| H3C | 0.1191 | 0.2490 | 0.0542 | 0.053* | |
| C4C | 0.21200 (19) | 0.3486 (4) | 0.13078 (17) | 0.0376 (7) | |
| C5C | 0.24278 (19) | 0.4046 (5) | 0.20511 (18) | 0.0419 (8) | |
| H5C | 0.2927 | 0.4514 | 0.2251 | 0.050* | |
| C6C | 0.20032 (17) | 0.3911 (5) | 0.24801 (18) | 0.0388 (8) | |
| H6C | 0.2219 | 0.4286 | 0.2978 | 0.047* | |
| C7C | 0.2584 (2) | 0.3631 (4) | 0.08407 (18) | 0.0420 (6) | |
| O8C | 0.22755 (17) | 0.3273 (3) | 0.01618 (14) | 0.0514 (7) | |
| O9C | 0.32715 (16) | 0.4120 (3) | 0.11228 (15) | 0.0510 (6) | |
| N10C | 0.08559 (19) | 0.3106 (5) | 0.26362 (19) | 0.0544 (9) | |
| H10C | 0.1050 | 0.3442 | 0.3099 | 0.065* | |
| H10D | 0.0392 | 0.2680 | 0.2452 | 0.065* | |
| C1D | 0.44629 (19) | 0.5743 (5) | 0.39756 (18) | 0.0398 (8) | |
| C2D | 0.4741 (2) | 0.5146 (5) | 0.4696 (2) | 0.0464 (9) | |
| H2D | 0.5230 | 0.4630 | 0.4898 | 0.056* | |
| C3D | 0.4310 (3) | 0.5302 (6) | 0.5113 (2) | 0.0566 (11) | |
| H3D | 0.4496 | 0.4889 | 0.5606 | 0.068* | |
| N4D | 0.35979 (19) | 0.6066 (5) | 0.4815 (2) | 0.0596 (9) | |
| H4D | 0.3317 | 0.6161 | 0.5083 | 0.072* | |
| C5D | 0.3342 (2) | 0.6645 (5) | 0.4139 (3) | 0.0524 (10) | |
| H5D | 0.2851 | 0.7157 | 0.3952 | 0.063* | |
| C6D | 0.37390 (19) | 0.6554 (5) | 0.3690 (2) | 0.0447 (8) | |
| H6D | 0.3541 | 0.7015 | 0.3206 | 0.054* | |
| N7D | 0.48796 (18) | 0.5564 (5) | 0.35534 (18) | 0.0532 (8) | |
| H7D | 0.5329 | 0.5067 | 0.3735 | 0.064* | |
| H7E | 0.4702 | 0.5944 | 0.3097 | 0.064* | |
| O1W | 0.41925 (15) | 0.6648 (4) | 0.20049 (14) | 0.0551 (8) | |
| H1W | 0.4027 | 0.7709 | 0.1742 | 0.083* | |
| H2W | 0.3819 | 0.5799 | 0.1750 | 0.083* | |
| O2W | 0.14559 (17) | 0.9180 (4) | 0.41753 (14) | 0.0556 (7) | |
| H3W | 0.1832 | 1.0052 | 0.4424 | 0.083* | |
| H4W | 0.1605 | 0.8195 | 0.4502 | 0.083* | |
| O3W | 0.15272 (18) | 0.4099 (4) | 0.42329 (16) | 0.0664 (9) | |
| H5W | 0.1731 | 0.5028 | 0.4574 | 0.100* | |
| H6W | 0.1831 | 0.3106 | 0.4442 | 0.100* | |
| O4W | 0.43348 (15) | 0.1709 (4) | 0.19169 (14) | 0.0517 (7) | |
| H7W | 0.4061 | 0.2770 | 0.1736 | 0.078* | |
| H8W | 0.4037 | 0.0835 | 0.1587 | 0.078* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1A | 0.0345 (17) | 0.052 (2) | 0.0415 (19) | −0.0015 (15) | 0.0142 (14) | −0.0063 (16) |
| C2A | 0.0282 (15) | 0.0472 (18) | 0.0452 (19) | 0.0069 (13) | 0.0080 (14) | −0.0021 (15) |
| C3A | 0.0316 (16) | 0.0497 (19) | 0.0330 (16) | 0.0020 (12) | 0.0014 (13) | −0.0036 (14) |
| C4A | 0.0236 (14) | 0.0436 (17) | 0.0336 (15) | 0.0008 (12) | 0.0023 (11) | −0.0064 (13) |
| C5A | 0.0282 (16) | 0.0391 (17) | 0.0378 (16) | 0.0002 (12) | 0.0069 (12) | −0.0013 (13) |
| C6A | 0.0356 (17) | 0.049 (2) | 0.0324 (16) | −0.0028 (13) | 0.0058 (13) | 0.0022 (14) |
| C7A | 0.0469 (17) | 0.0398 (15) | 0.0404 (16) | −0.0023 (12) | 0.0141 (13) | −0.0047 (11) |
| O8A | 0.0442 (14) | 0.0561 (14) | 0.0487 (14) | 0.0052 (10) | 0.0149 (10) | −0.0009 (10) |
| O9A | 0.0579 (16) | 0.0574 (13) | 0.0348 (12) | 0.0053 (11) | 0.0102 (10) | −0.0004 (9) |
| N10A | 0.0445 (18) | 0.101 (3) | 0.0486 (18) | 0.0131 (18) | 0.0225 (15) | 0.0039 (19) |
| C1B | 0.0333 (16) | 0.0335 (15) | 0.0361 (17) | 0.0024 (13) | 0.0039 (13) | 0.0000 (14) |
| C2B | 0.0341 (18) | 0.049 (2) | 0.0421 (19) | −0.0025 (14) | 0.0018 (15) | 0.0001 (15) |
| C3B | 0.041 (2) | 0.056 (2) | 0.078 (3) | −0.0020 (17) | 0.0157 (19) | 0.012 (2) |
| N4B | 0.081 (3) | 0.0481 (18) | 0.064 (2) | 0.0145 (18) | 0.0434 (18) | 0.0118 (16) |
| C5B | 0.068 (3) | 0.055 (2) | 0.042 (2) | 0.0009 (18) | 0.0221 (19) | −0.0066 (17) |
| C6B | 0.046 (2) | 0.052 (2) | 0.0365 (19) | −0.0047 (15) | 0.0060 (15) | −0.0080 (15) |
| N7B | 0.0402 (15) | 0.0585 (19) | 0.0409 (16) | −0.0105 (13) | 0.0136 (13) | −0.0083 (14) |
| C1C | 0.0352 (17) | 0.0345 (15) | 0.0348 (17) | 0.0011 (13) | 0.0040 (12) | −0.0005 (14) |
| C2C | 0.0361 (17) | 0.057 (2) | 0.0355 (18) | −0.0100 (15) | 0.0010 (14) | −0.0093 (15) |
| C3C | 0.0426 (19) | 0.053 (2) | 0.0296 (16) | −0.0058 (14) | 0.0040 (14) | −0.0110 (14) |
| C4C | 0.0460 (19) | 0.0314 (15) | 0.0299 (15) | 0.0051 (13) | 0.0082 (13) | 0.0011 (12) |
| C5C | 0.0328 (17) | 0.050 (2) | 0.0307 (15) | −0.0029 (14) | −0.0016 (12) | 0.0024 (14) |
| C6C | 0.0341 (18) | 0.0448 (18) | 0.0279 (15) | −0.0039 (13) | 0.0008 (13) | −0.0012 (13) |
| C7C | 0.0444 (15) | 0.0404 (14) | 0.0368 (15) | 0.0009 (11) | 0.0105 (12) | 0.0046 (11) |
| O8C | 0.0610 (16) | 0.0537 (13) | 0.0355 (11) | −0.0012 (11) | 0.0138 (10) | −0.0015 (9) |
| O9C | 0.0448 (13) | 0.0596 (14) | 0.0463 (13) | −0.0057 (10) | 0.0147 (10) | −0.0029 (10) |
| N10C | 0.0454 (18) | 0.075 (2) | 0.0430 (17) | −0.0171 (16) | 0.0166 (14) | −0.0126 (16) |
| C1D | 0.0331 (16) | 0.052 (2) | 0.0349 (17) | −0.0042 (14) | 0.0131 (13) | −0.0030 (15) |
| C2D | 0.0453 (19) | 0.0461 (18) | 0.044 (2) | 0.0025 (15) | 0.0131 (16) | −0.0029 (15) |
| C3D | 0.077 (3) | 0.054 (2) | 0.044 (2) | −0.0085 (18) | 0.028 (2) | −0.0078 (17) |
| N4D | 0.0560 (18) | 0.065 (2) | 0.075 (2) | −0.0101 (16) | 0.0441 (16) | −0.0187 (18) |
| C5D | 0.0384 (19) | 0.046 (2) | 0.077 (3) | −0.0056 (14) | 0.0268 (18) | −0.0127 (18) |
| C6D | 0.0361 (18) | 0.0454 (19) | 0.0454 (19) | −0.0038 (14) | 0.0075 (15) | −0.0034 (15) |
| N7D | 0.0407 (15) | 0.078 (2) | 0.0440 (17) | 0.0000 (15) | 0.0196 (13) | 0.0059 (16) |
| O1W | 0.0524 (16) | 0.0566 (15) | 0.0396 (13) | −0.0102 (11) | −0.0017 (10) | 0.0056 (11) |
| O2W | 0.0509 (14) | 0.0623 (16) | 0.0432 (14) | −0.0037 (12) | 0.0059 (11) | 0.0019 (12) |
| O3W | 0.0573 (16) | 0.0682 (18) | 0.0502 (16) | 0.0116 (13) | −0.0063 (12) | −0.0056 (13) |
| O4W | 0.0390 (13) | 0.0574 (14) | 0.0456 (13) | −0.0033 (10) | 0.0012 (10) | −0.0063 (12) |
Geometric parameters (Å, º) top
| C1A—N10A | 1.381 (5) | C2C—H2C | 0.9500 |
| C1A—C2A | 1.384 (5) | C3C—C4C | 1.353 (5) |
| C1A—C6A | 1.409 (5) | C3C—H3C | 0.9500 |
| C2A—C3A | 1.378 (5) | C4C—C5C | 1.418 (5) |
| C2A—H2A | 0.9500 | C4C—C7C | 1.495 (5) |
| C3A—C4A | 1.426 (4) | C5C—C6C | 1.371 (5) |
| C3A—H3A | 0.9500 | C5C—H5C | 0.9500 |
| C4A—C5A | 1.375 (5) | C6C—H6C | 0.9500 |
| C4A—C7A | 1.485 (5) | C7C—O8C | 1.265 (4) |
| C5A—C6A | 1.377 (5) | C7C—O9C | 1.268 (4) |
| C5A—H5A | 0.9500 | N10C—H10C | 0.8800 |
| C6A—H6A | 0.9500 | N10C—H10D | 0.8800 |
| C7A—O8A | 1.266 (4) | C1D—N7D | 1.350 (4) |
| C7A—O9A | 1.267 (4) | C1D—C2D | 1.387 (5) |
| N10A—H10A | 0.8800 | C1D—C6D | 1.421 (5) |
| N10A—H10B | 0.8800 | C2D—C3D | 1.364 (5) |
| C1B—N7B | 1.313 (5) | C2D—H2D | 0.9500 |
| C1B—C2B | 1.404 (5) | C3D—N4D | 1.387 (6) |
| C1B—C6B | 1.429 (5) | C3D—H3D | 0.9500 |
| C2B—C3B | 1.353 (7) | N4D—C5D | 1.307 (6) |
| C2B—H2B | 0.9500 | N4D—H4D | 0.8800 |
| C3B—N4B | 1.374 (6) | C5D—C6D | 1.360 (6) |
| C3B—H3B | 0.9500 | C5D—H5D | 0.9500 |
| N4B—C5B | 1.300 (6) | C6D—H6D | 0.9500 |
| N4B—H4B | 0.8800 | N7D—H7D | 0.8800 |
| C5B—C6B | 1.341 (6) | N7D—H7E | 0.8800 |
| C5B—H5B | 0.9500 | O1W—H1W | 0.9635 |
| C6B—H6B | 0.9500 | O1W—H2W | 0.9611 |
| N7B—H7B | 0.8800 | O2W—H3W | 0.9734 |
| N7B—H7C | 0.8800 | O2W—H4W | 0.9723 |
| C1C—N10C | 1.335 (5) | O3W—H5W | 0.9623 |
| C1C—C6C | 1.399 (5) | O3W—H6W | 0.9604 |
| C1C—C2C | 1.432 (5) | O4W—H7W | 0.9715 |
| C2C—C3C | 1.361 (5) | O4W—H8W | 0.9630 |
| | | |
| N10A—C1A—C2A | 119.5 (3) | C6C—C1C—C2C | 115.9 (3) |
| N10A—C1A—C6A | 120.5 (4) | C3C—C2C—C1C | 120.7 (3) |
| C2A—C1A—C6A | 120.1 (3) | C3C—C2C—H2C | 119.7 |
| C3A—C2A—C1A | 120.5 (3) | C1C—C2C—H2C | 119.7 |
| C3A—C2A—H2A | 119.8 | C4C—C3C—C2C | 122.9 (3) |
| C1A—C2A—H2A | 119.8 | C4C—C3C—H3C | 118.6 |
| C2A—C3A—C4A | 120.4 (3) | C2C—C3C—H3C | 118.6 |
| C2A—C3A—H3A | 119.8 | C3C—C4C—C5C | 118.2 (3) |
| C4A—C3A—H3A | 119.8 | C3C—C4C—C7C | 122.0 (3) |
| C5A—C4A—C3A | 117.5 (3) | C5C—C4C—C7C | 119.8 (3) |
| C5A—C4A—C7A | 123.0 (3) | C6C—C5C—C4C | 119.8 (3) |
| C3A—C4A—C7A | 119.5 (3) | C6C—C5C—H5C | 120.1 |
| C4A—C5A—C6A | 123.2 (3) | C4C—C5C—H5C | 120.1 |
| C4A—C5A—H5A | 118.4 | C5C—C6C—C1C | 122.6 (3) |
| C6A—C5A—H5A | 118.4 | C5C—C6C—H6C | 118.7 |
| C5A—C6A—C1A | 118.4 (3) | C1C—C6C—H6C | 118.7 |
| C5A—C6A—H6A | 120.8 | O8C—C7C—O9C | 121.0 (3) |
| C1A—C6A—H6A | 120.8 | O8C—C7C—C4C | 118.8 (3) |
| O8A—C7A—O9A | 120.8 (3) | O9C—C7C—C4C | 120.2 (3) |
| O8A—C7A—C4A | 118.8 (3) | C1C—N10C—H10C | 120.0 |
| O9A—C7A—C4A | 120.5 (3) | C1C—N10C—H10D | 120.0 |
| C1A—N10A—H10A | 120.0 | H10C—N10C—H10D | 120.0 |
| C1A—N10A—H10B | 120.0 | N7D—C1D—C2D | 120.2 (4) |
| H10A—N10A—H10B | 120.0 | N7D—C1D—C6D | 120.5 (4) |
| N7B—C1B—C2B | 122.8 (3) | C2D—C1D—C6D | 119.3 (3) |
| N7B—C1B—C6B | 122.2 (3) | C3D—C2D—C1D | 119.7 (4) |
| C2B—C1B—C6B | 114.9 (3) | C3D—C2D—H2D | 120.1 |
| C3B—C2B—C1B | 122.0 (4) | C1D—C2D—H2D | 120.1 |
| C3B—C2B—H2B | 119.0 | C2D—C3D—N4D | 119.7 (4) |
| C1B—C2B—H2B | 119.0 | C2D—C3D—H3D | 120.2 |
| C2B—C3B—N4B | 119.4 (4) | N4D—C3D—H3D | 120.2 |
| C2B—C3B—H3B | 120.3 | C5D—N4D—C3D | 120.4 (3) |
| N4B—C3B—H3B | 120.3 | C5D—N4D—H4D | 119.8 |
| C5B—N4B—C3B | 120.5 (4) | C3D—N4D—H4D | 119.8 |
| C5B—N4B—H4B | 119.7 | N4D—C5D—C6D | 123.6 (4) |
| C3B—N4B—H4B | 119.7 | N4D—C5D—H5D | 118.2 |
| N4B—C5B—C6B | 122.9 (4) | C6D—C5D—H5D | 118.2 |
| N4B—C5B—H5B | 118.5 | C5D—C6D—C1D | 117.3 (4) |
| C6B—C5B—H5B | 118.5 | C5D—C6D—H6D | 121.4 |
| C5B—C6B—C1B | 120.2 (4) | C1D—C6D—H6D | 121.4 |
| C5B—C6B—H6B | 119.9 | C1D—N7D—H7D | 120.0 |
| C1B—C6B—H6B | 119.9 | C1D—N7D—H7E | 120.0 |
| C1B—N7B—H7B | 120.0 | H7D—N7D—H7E | 120.0 |
| C1B—N7B—H7C | 120.0 | H1W—O1W—H2W | 106.8 |
| H7B—N7B—H7C | 120.0 | H3W—O2W—H4W | 104.4 |
| N10C—C1C—C6C | 122.2 (3) | H5W—O3W—H6W | 107.1 |
| N10C—C1C—C2C | 121.9 (3) | H7W—O4W—H8W | 105.2 |
| | | |
| N10A—C1A—C2A—C3A | −179.5 (4) | N10C—C1C—C2C—C3C | 179.9 (4) |
| C6A—C1A—C2A—C3A | 2.1 (6) | C6C—C1C—C2C—C3C | −0.9 (6) |
| C1A—C2A—C3A—C4A | 0.1 (6) | C1C—C2C—C3C—C4C | 1.4 (6) |
| C2A—C3A—C4A—C5A | −0.7 (5) | C2C—C3C—C4C—C5C | −1.2 (6) |
| C2A—C3A—C4A—C7A | 179.7 (3) | C2C—C3C—C4C—C7C | 179.4 (4) |
| C3A—C4A—C5A—C6A | −0.9 (5) | C3C—C4C—C5C—C6C | 0.7 (5) |
| C7A—C4A—C5A—C6A | 178.6 (4) | C7C—C4C—C5C—C6C | −179.9 (3) |
| C4A—C5A—C6A—C1A | 3.1 (6) | C4C—C5C—C6C—C1C | −0.4 (5) |
| N10A—C1A—C6A—C5A | 178.0 (4) | N10C—C1C—C6C—C5C | 179.6 (4) |
| C2A—C1A—C6A—C5A | −3.6 (6) | C2C—C1C—C6C—C5C | 0.5 (5) |
| C5A—C4A—C7A—O8A | 8.0 (5) | C3C—C4C—C7C—O8C | 5.8 (5) |
| C3A—C4A—C7A—O8A | −172.5 (3) | C5C—C4C—C7C—O8C | −173.6 (3) |
| C5A—C4A—C7A—O9A | −171.9 (3) | C3C—C4C—C7C—O9C | −174.8 (3) |
| C3A—C4A—C7A—O9A | 7.7 (5) | C5C—C4C—C7C—O9C | 5.8 (4) |
| N7B—C1B—C2B—C3B | −179.0 (4) | N7D—C1D—C2D—C3D | −178.7 (4) |
| C6B—C1B—C2B—C3B | 1.4 (5) | C6D—C1D—C2D—C3D | 1.8 (6) |
| C1B—C2B—C3B—N4B | −2.1 (6) | C1D—C2D—C3D—N4D | −0.4 (6) |
| C2B—C3B—N4B—C5B | 1.4 (6) | C2D—C3D—N4D—C5D | −0.4 (6) |
| C3B—N4B—C5B—C6B | 0.1 (7) | C3D—N4D—C5D—C6D | −0.3 (6) |
| N4B—C5B—C6B—C1B | −0.8 (7) | N4D—C5D—C6D—C1D | 1.6 (6) |
| N7B—C1B—C6B—C5B | −179.6 (4) | N7D—C1D—C6D—C5D | 178.1 (4) |
| C2B—C1B—C6B—C5B | 0.1 (6) | C2D—C1D—C6D—C5D | −2.4 (6) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1W—H1W···O9Ai | 0.96 | 1.84 | 2.763 (4) | 159 |
| O1W—H2W···O9C | 0.96 | 1.83 | 2.767 (4) | 165 |
| O2W—H3W···O8Aii | 0.97 | 1.80 | 2.760 (4) | 170 |
| N4B—H4B···O8Ai | 0.88 | 1.99 | 2.808 (5) | 153 |
| N4B—H4B···O9Ai | 0.88 | 2.18 | 2.936 (5) | 143 |
| N4D—H4D···O8Ciii | 0.88 | 2.09 | 2.883 (5) | 150 |
| N4D—H4D···O9Ciii | 0.88 | 2.08 | 2.859 (5) | 146 |
| O2W—H4W···O8Ciii | 0.97 | 1.83 | 2.748 (4) | 156 |
| O3W—H5W···O8Ciii | 0.96 | 1.80 | 2.754 (4) | 169 |
| O3W—H6W···O8A | 0.96 | 1.80 | 2.741 (4) | 166 |
| N7B—H7B···O2W | 0.88 | 2.09 | 2.965 (4) | 173 |
| N7B—H7C···O4Wiv | 0.88 | 2.09 | 2.967 (5) | 172 |
| N7D—H7D···O2Wv | 0.88 | 2.10 | 2.973 (5) | 173 |
| N7D—H7E···O1W | 0.88 | 2.06 | 2.938 (4) | 174 |
| O4W—H7W···O9C | 0.97 | 1.85 | 2.768 (4) | 156 |
| O4W—H8W···O9Avi | 0.96 | 1.78 | 2.740 (4) | 179 |
| N10A—H10A···O3Wv | 0.88 | 2.21 | 3.085 (5) | 176 |
| N10A—H10B···O4W | 0.88 | 2.14 | 3.015 (4) | 176 |
| N10C—H10C···O3W | 0.88 | 2.12 | 3.000 (5) | 177 |
| N10C—H10D···O1Wvii | 0.88 | 2.26 | 3.136 (5) | 178 |
| Symmetry codes: (i) x, −y+1, z−1/2; (ii) x, y+1, z; (iii) x, −y+1, z+1/2; (iv) x−1/2, y+1/2, z; (v) x+1/2, y−1/2, z; (vi) x, −y, z−1/2; (vii) x−1/2, y−1/2, z. |
(II) 4-aminopyridinium nicotinate
top
Crystal data top
| C5H7N2+·C6H4NO2− | F(000) = 456 |
| Mr = 217.23 | Dx = 1.391 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation, λ = 1.54178 Å |
| Hall symbol: -P 2ybc | Cell parameters from 25 reflections |
| a = 11.9645 (17) Å | θ = 65–69° |
| b = 8.2635 (5) Å | µ = 0.82 mm−1 |
| c = 11.305 (3) Å | T = 193 K |
| β = 111.854 (7)° | Block, colourless |
| V = 1037.4 (3) Å3 | 0.26 × 0.19 × 0.13 mm |
| Z = 4 | |
Data collection top
Enraf–Nonius CAD-4
diffractometer | 1837 reflections with I > 2σ(I) |
| Radiation source: rotating anode | Rint = 0.022 |
| Graphite monochromator | θmax = 70.2°, θmin = 4.0° |
| ω/2θ scans | h = −14→13 |
Absorption correction: ψ scan
(CORINC; Draeger & Gattow, 1971) | k = 0→10 |
| Tmin = 0.815, Tmax = 0.901 | l = 0→13 |
| 2068 measured reflections | 3 standard reflections every 60 min |
| 1960 independent reflections | intensity decay: 2% |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.062 | H-atom parameters constrained |
| wR(F2) = 0.220 | w = 1/[σ2(Fo2) + (0.1456P)2 + 0.5559P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.13 | (Δ/σ)max < 0.001 |
| 1960 reflections | Δρmax = 0.35 e Å−3 |
| 146 parameters | Δρmin = −0.38 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.016 (3) |
Crystal data top
| C5H7N2+·C6H4NO2− | V = 1037.4 (3) Å3 |
| Mr = 217.23 | Z = 4 |
| Monoclinic, P21/c | Cu Kα radiation |
| a = 11.9645 (17) Å | µ = 0.82 mm−1 |
| b = 8.2635 (5) Å | T = 193 K |
| c = 11.305 (3) Å | 0.26 × 0.19 × 0.13 mm |
| β = 111.854 (7)° | |
Data collection top
Enraf–Nonius CAD-4
diffractometer | 1837 reflections with I > 2σ(I) |
Absorption correction: ψ scan
(CORINC; Draeger & Gattow, 1971) | Rint = 0.022 |
| Tmin = 0.815, Tmax = 0.901 | 3 standard reflections every 60 min |
| 2068 measured reflections | intensity decay: 2% |
| 1960 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.062 | 0 restraints |
| wR(F2) = 0.220 | H-atom parameters constrained |
| S = 1.13 | Δρmax = 0.35 e Å−3 |
| 1960 reflections | Δρmin = −0.38 e Å−3 |
| 146 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N1 | 0.45422 (16) | 0.2987 (2) | 0.44351 (18) | 0.0329 (5) | |
| H1 | 0.5078 | 0.2289 | 0.5008 | 0.039* | |
| C2 | 0.47405 (19) | 0.3494 (3) | 0.3405 (2) | 0.0331 (6) | |
| H2 | 0.5457 | 0.3168 | 0.3295 | 0.040* | |
| C3 | 0.39413 (18) | 0.4466 (3) | 0.2510 (2) | 0.0306 (5) | |
| H3 | 0.4094 | 0.4795 | 0.1779 | 0.037* | |
| C4 | 0.28780 (17) | 0.4982 (2) | 0.2681 (2) | 0.0285 (5) | |
| C5 | 0.27057 (18) | 0.4429 (3) | 0.3777 (2) | 0.0327 (6) | |
| H5 | 0.2005 | 0.4739 | 0.3926 | 0.039* | |
| C6 | 0.3541 (2) | 0.3449 (3) | 0.4629 (2) | 0.0336 (6) | |
| H6 | 0.3416 | 0.3088 | 0.5368 | 0.040* | |
| N7 | 0.20697 (16) | 0.5928 (2) | 0.18238 (18) | 0.0339 (5) | |
| H7A | 0.1417 | 0.6262 | 0.2090 | 0.051* | |
| H7B | 0.2280 | 0.6310 | 0.1203 | 0.051* | |
| C8 | 0.79754 (17) | −0.0010 (2) | 0.6998 (2) | 0.0279 (5) | |
| C9 | 0.89015 (19) | −0.0577 (3) | 0.6641 (2) | 0.0322 (5) | |
| H9 | 0.8880 | −0.0307 | 0.5816 | 0.039* | |
| N10 | 0.98191 (16) | −0.1482 (2) | 0.7393 (2) | 0.0382 (6) | |
| C11 | 0.9832 (2) | −0.1853 (3) | 0.8545 (2) | 0.0390 (6) | |
| H11 | 1.0483 | −0.2481 | 0.9095 | 0.047* | |
| C12 | 0.8944 (2) | −0.1371 (3) | 0.8982 (2) | 0.0408 (6) | |
| H12 | 0.8981 | −0.1675 | 0.9806 | 0.049* | |
| C13 | 0.7999 (2) | −0.0433 (3) | 0.8188 (2) | 0.0373 (6) | |
| H13 | 0.7376 | −0.0087 | 0.8460 | 0.045* | |
| C14 | 0.69835 (18) | 0.1037 (3) | 0.60888 (19) | 0.0285 (5) | |
| O15 | 0.71755 (14) | 0.1689 (2) | 0.51889 (15) | 0.0372 (5) | |
| O16 | 0.60445 (13) | 0.1179 (2) | 0.63279 (16) | 0.0386 (5) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N1 | 0.0228 (9) | 0.0315 (9) | 0.0417 (10) | 0.0024 (7) | 0.0090 (8) | 0.0014 (7) |
| C2 | 0.0213 (10) | 0.0336 (11) | 0.0461 (13) | 0.0005 (8) | 0.0143 (9) | −0.0030 (9) |
| C3 | 0.0247 (10) | 0.0336 (11) | 0.0359 (11) | −0.0019 (8) | 0.0143 (8) | −0.0019 (8) |
| C4 | 0.0200 (10) | 0.0271 (10) | 0.0374 (11) | −0.0033 (7) | 0.0093 (8) | −0.0044 (8) |
| C5 | 0.0232 (10) | 0.0344 (11) | 0.0440 (12) | 0.0021 (8) | 0.0166 (9) | 0.0033 (9) |
| C6 | 0.0304 (11) | 0.0354 (11) | 0.0369 (11) | 0.0005 (9) | 0.0148 (9) | 0.0002 (8) |
| N7 | 0.0236 (9) | 0.0384 (11) | 0.0408 (10) | 0.0032 (7) | 0.0132 (8) | 0.0051 (8) |
| C8 | 0.0201 (10) | 0.0259 (10) | 0.0381 (11) | −0.0020 (8) | 0.0115 (8) | −0.0011 (8) |
| C9 | 0.0251 (10) | 0.0312 (11) | 0.0432 (12) | 0.0009 (8) | 0.0161 (9) | 0.0020 (9) |
| N10 | 0.0227 (10) | 0.0340 (10) | 0.0591 (13) | 0.0046 (7) | 0.0167 (9) | 0.0038 (9) |
| C11 | 0.0279 (11) | 0.0346 (12) | 0.0509 (14) | 0.0053 (9) | 0.0105 (10) | 0.0064 (9) |
| C12 | 0.0406 (13) | 0.0444 (13) | 0.0397 (13) | 0.0090 (10) | 0.0174 (10) | 0.0097 (10) |
| C13 | 0.0299 (11) | 0.0399 (13) | 0.0472 (13) | 0.0075 (9) | 0.0202 (10) | 0.0059 (10) |
| C14 | 0.0227 (10) | 0.0291 (11) | 0.0341 (11) | −0.0002 (8) | 0.0110 (8) | −0.0040 (8) |
| O15 | 0.0327 (9) | 0.0447 (10) | 0.0374 (9) | 0.0089 (7) | 0.0165 (7) | 0.0053 (7) |
| O16 | 0.0227 (8) | 0.0474 (10) | 0.0483 (10) | 0.0083 (6) | 0.0163 (7) | 0.0093 (7) |
Geometric parameters (Å, º) top
| N1—C2 | 1.339 (3) | C8—C13 | 1.380 (3) |
| N1—C6 | 1.350 (3) | C8—C9 | 1.394 (3) |
| N1—H1 | 0.9237 | C8—C14 | 1.518 (3) |
| C2—C3 | 1.365 (3) | C9—N10 | 1.339 (3) |
| C2—H2 | 0.9500 | C9—H9 | 0.9500 |
| C3—C4 | 1.422 (3) | N10—C11 | 1.332 (3) |
| C3—H3 | 0.9500 | C11—C12 | 1.387 (3) |
| C4—N7 | 1.337 (3) | C11—H11 | 0.9500 |
| C4—C5 | 1.406 (3) | C12—C13 | 1.389 (3) |
| C5—C6 | 1.366 (3) | C12—H12 | 0.9500 |
| C5—H5 | 0.9500 | C13—H13 | 0.9500 |
| C6—H6 | 0.9500 | C14—O15 | 1.246 (3) |
| N7—H7A | 0.9757 | C14—O16 | 1.255 (2) |
| N7—H7B | 0.8870 | | |
| | | |
| C2—N1—C6 | 120.52 (19) | H7A—N7—H7B | 129.7 |
| C2—N1—H1 | 120.2 | C13—C8—C9 | 117.84 (19) |
| C6—N1—H1 | 119.2 | C13—C8—C14 | 122.09 (18) |
| N1—C2—C3 | 121.79 (19) | C9—C8—C14 | 120.08 (19) |
| N1—C2—H2 | 119.1 | N10—C9—C8 | 123.8 (2) |
| C3—C2—H2 | 119.1 | N10—C9—H9 | 118.1 |
| C2—C3—C4 | 119.4 (2) | C8—C9—H9 | 118.1 |
| C2—C3—H3 | 120.3 | C11—N10—C9 | 117.27 (19) |
| C4—C3—H3 | 120.3 | N10—C11—C12 | 123.4 (2) |
| N7—C4—C5 | 121.67 (18) | N10—C11—H11 | 118.3 |
| N7—C4—C3 | 121.27 (19) | C12—C11—H11 | 118.3 |
| C5—C4—C3 | 117.05 (19) | C11—C12—C13 | 118.5 (2) |
| C6—C5—C4 | 120.34 (19) | C11—C12—H12 | 120.8 |
| C6—C5—H5 | 119.8 | C13—C12—H12 | 120.8 |
| C4—C5—H5 | 119.8 | C8—C13—C12 | 119.2 (2) |
| N1—C6—C5 | 120.9 (2) | C8—C13—H13 | 120.4 |
| N1—C6—H6 | 119.5 | C12—C13—H13 | 120.4 |
| C5—C6—H6 | 119.5 | O15—C14—O16 | 125.99 (19) |
| C4—N7—H7A | 112.8 | O15—C14—C8 | 117.64 (17) |
| C4—N7—H7B | 116.1 | O16—C14—C8 | 116.37 (18) |
| | | |
| C6—N1—C2—C3 | −1.0 (3) | C8—C9—N10—C11 | 0.2 (3) |
| N1—C2—C3—C4 | 1.1 (3) | C9—N10—C11—C12 | 0.9 (4) |
| C2—C3—C4—N7 | −179.67 (19) | N10—C11—C12—C13 | −0.9 (4) |
| C2—C3—C4—C5 | −0.8 (3) | C9—C8—C13—C12 | 1.2 (3) |
| N7—C4—C5—C6 | 179.3 (2) | C14—C8—C13—C12 | −178.9 (2) |
| C3—C4—C5—C6 | 0.4 (3) | C11—C12—C13—C8 | −0.2 (4) |
| C2—N1—C6—C5 | 0.6 (3) | C13—C8—C14—O15 | 162.6 (2) |
| C4—C5—C6—N1 | −0.3 (3) | C9—C8—C14—O15 | −17.5 (3) |
| C13—C8—C9—N10 | −1.2 (3) | C13—C8—C14—O16 | −16.6 (3) |
| C14—C8—C9—N10 | 178.81 (19) | C9—C8—C14—O16 | 163.32 (19) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O16 | 0.92 | 1.77 | 2.680 (2) | 169 |
| N1—H1···O15 | 0.92 | 2.49 | 3.131 (2) | 127 |
| N7—H7A···N10i | 0.98 | 2.07 | 3.027 (3) | 166 |
| N7—H7B···O15ii | 0.89 | 1.94 | 2.814 (2) | 168 |
| Symmetry codes: (i) x−1, −y+1/2, z−1/2; (ii) −x+1, y+1/2, −z+1/2. |
Experimental details
| | (I) | (II) |
| Crystal data |
| Chemical formula | C5H7N2+·C7H6NO2−·2H2O | C5H7N2+·C6H4NO2− |
| Mr | 267.29 | 217.23 |
| Crystal system, space group | Monoclinic, Cc | Monoclinic, P21/c |
| Temperature (K) | 193 | 193 |
| a, b, c (Å) | 18.9692 (18), 7.8092 (4), 19.5944 (19) | 11.9645 (17), 8.2635 (5), 11.305 (3) |
| β (°) | 112.213 (4) | 111.854 (7) |
| V (Å3) | 2687.2 (4) | 1037.4 (3) |
| Z | 8 | 4 |
| Radiation type | Cu Kα | Cu Kα |
| µ (mm−1) | 0.84 | 0.82 |
| Crystal size (mm) | 0.38 × 0.32 × 0.16 | 0.26 × 0.19 × 0.13 |
| |
| Data collection |
| Diffractometer | Enraf–Nonius CAD-4
diffractometer | Enraf–Nonius CAD-4
diffractometer |
| Absorption correction | ψ scan
(CORINC; Draeger & Gattow, 1971) | ψ scan
(CORINC; Draeger & Gattow, 1971) |
| Tmin, Tmax | 0.741, 0.877 | 0.815, 0.901 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 4793, 2557, 2334 | 2068, 1960, 1837 |
| Rint | 0.045 | 0.022 |
| (sin θ/λ)max (Å−1) | 0.609 | 0.610 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.055, 0.156, 1.05 | 0.062, 0.220, 1.13 |
| No. of reflections | 2557 | 1960 |
| No. of parameters | 344 | 146 |
| No. of restraints | 14 | 0 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.28, −0.37 | 0.35, −0.38 |
| Absolute structure | Flack (1983), with 2236 Friedel pairs | ? |
| Absolute structure parameter | 0.2 (3) | ? |
Selected bond angles (º) for (I) top| C5B—N4B—C3B | 120.5 (4) | C5D—N4D—C3D | 120.4 (3) |
Hydrogen-bond geometry (Å, º) for (I) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1W—H1W···O9Ai | 0.96 | 1.84 | 2.763 (4) | 159 |
| O1W—H2W···O9C | 0.96 | 1.83 | 2.767 (4) | 165 |
| O2W—H3W···O8Aii | 0.97 | 1.80 | 2.760 (4) | 170 |
| N4B—H4B···O8Ai | 0.88 | 1.99 | 2.808 (5) | 153 |
| N4B—H4B···O9Ai | 0.88 | 2.18 | 2.936 (5) | 143 |
| N4D—H4D···O8Ciii | 0.88 | 2.09 | 2.883 (5) | 150 |
| N4D—H4D···O9Ciii | 0.88 | 2.08 | 2.859 (5) | 146 |
| O2W—H4W···O8Ciii | 0.97 | 1.83 | 2.748 (4) | 156 |
| O3W—H5W···O8Ciii | 0.96 | 1.80 | 2.754 (4) | 169 |
| O3W—H6W···O8A | 0.96 | 1.80 | 2.741 (4) | 166 |
| N7B—H7B···O2W | 0.88 | 2.09 | 2.965 (4) | 173 |
| N7B—H7C···O4Wiv | 0.88 | 2.09 | 2.967 (5) | 172 |
| N7D—H7D···O2Wv | 0.88 | 2.10 | 2.973 (5) | 173 |
| N7D—H7E···O1W | 0.88 | 2.06 | 2.938 (4) | 174 |
| O4W—H7W···O9C | 0.97 | 1.85 | 2.768 (4) | 156 |
| O4W—H8W···O9Avi | 0.96 | 1.78 | 2.740 (4) | 179 |
| N10A—H10A···O3Wv | 0.88 | 2.21 | 3.085 (5) | 176 |
| N10A—H10B···O4W | 0.88 | 2.14 | 3.015 (4) | 176 |
| N10C—H10C···O3W | 0.88 | 2.12 | 3.000 (5) | 177 |
| N10C—H10D···O1Wvii | 0.88 | 2.26 | 3.136 (5) | 178 |
| Symmetry codes: (i) x, −y+1, z−1/2; (ii) x, y+1, z; (iii) x, −y+1, z+1/2; (iv) x−1/2, y+1/2, z; (v) x+1/2, y−1/2, z; (vi) x, −y, z−1/2; (vii) x−1/2, y−1/2, z. |
Selected geometric parameters (Å, º) for (II) top| N1—C2 | 1.339 (3) | C14—O15 | 1.246 (3) |
| N1—C6 | 1.350 (3) | C14—O16 | 1.255 (2) |
| | | |
| C2—N1—C6 | 120.52 (19) | O16—C14—C8 | 116.37 (18) |
| O15—C14—C8 | 117.64 (17) | | |
Hydrogen-bond geometry (Å, º) for (II) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O16 | 0.92 | 1.77 | 2.680 (2) | 169 |
| N1—H1···O15 | 0.92 | 2.49 | 3.131 (2) | 127 |
| N7—H7A···N10i | 0.98 | 2.07 | 3.027 (3) | 166 |
| N7—H7B···O15ii | 0.89 | 1.94 | 2.814 (2) | 168 |
| Symmetry codes: (i) x−1, −y+1/2, z−1/2; (ii) −x+1, y+1/2, −z+1/2. |
 O and N-H
O and N-H N hydrogen bonds to form a two-dimensional network along the (10
N hydrogen bonds to form a two-dimensional network along the (10
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2009/07/00/sk3319/sk3319fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2009/07/00/sk3319/sk3319fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2009/07/00/sk3319/sk3319fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/c/issues/2009/07/00/sk3319/sk3319fig4thm.gif)




4-aminopyridine (fampridine) is used clinically in Lambert-Eaton myasthenic syndrome and multiple sclerosis because by blocking potassium channels it prolongs action potentials, thereby increasing transmitter release at the neuromuscular junction (Judge & Bever, 2006; Schwid et al., 1997; Strupp et al., 2004). The structure of 4-aminopyridine was first reported by Chao & Schempp (1977) and a redetermination was reported by Anderson et al. (2005). 4-Aminobenzoic acid acts as a bacterial cofactor involved in the biosynthesis of folic acid, which is a constituent of the vitamin B complex and is found in animal and plant tissues (Robinson, 1966; Zoroddu et al., 1996). Two different polymorphs of 4-aminobenzoic acid have been re-investigated recently, the α-form (Athimoolam & Natarajan, 2007) and the β-form (Gracin & Fischer, 2005). We have already reported the crystal structure of the salt of 4-aminobenzoic acid-nicotinic acid (Jebas & Balasubramanian, 2006). Nicotinic acid (vitamin B3) is a 3-pyridine carboxylic acid, known as niacin. It is a lipid-lowering agent widely used to treat hypertriglyceridemia by the inhibition of lipolysis in adipose tissues (Athimoolam & Rajaram, 2005). The crystal structure of nicotinic acid was first determined by photographic methods (Wright & King, 1953), and a redetermination was reported by Kutoglu & Scheringer (1983). The crystal structures of isonicotinic acid (Takusagawa & Shimada, 1976), 2-aminonicotinic acid (Dobson & Gerkin, 1997), 6-aminonicotinic acid hydrochloride (Giantsidis & Turnbull, 2000) and 2-(methylsulfanyl)nicotinic acid (Basavoju et al., 2005) have been reported. The nicotinic acid complex 5-methylpyrazine-2-carboxylic acid 4-oxide is a commonly used drug for the treatment of hypercholesterolemia (Lorenzen et al., 2001). Coordination complexes of nicotinic acid with metals such as Sn possess antitumour activity greater than that of the well known cis-platin or doxorubicin (Gielen et al., 1992). The enzyme nicotinic acid mononucleotide adenyltransferase is essential for the synthesis of nicotinamide adenine dinucleotide in all living cells and is a potential target for antibiotics (Kim et al., 2004). As an extension of our systematic study of hydrogen-bonding patterns of 4-aminopyridine with carboxylic acids, the title compounds, (I) and (II), have been synthesized and their crystal structures are presented here.
The asymmetric unit of (I) (Fig. 1), consists of two 4-aminopyridinium cations protonated at atoms N4B and N4D, two 4-aminobenzoate anions with the carboxyl groups deprotonated, and four water molecules, while that of (II) contains one 4-aminopyridinium cation protonated at atom N1 and one nicotinic acetate anion with the carboxylate group deprotonated (Fig. 2). Compound (I) crystallizes in a noncentrosymmetric space group. The bond lengths and angles of the protonated 4-aminopyridinium in (I) and (II) are comparable with the values reported earlier for 4-aminopyridine in its protonated form (Fun et al., 2009a,b). There is a significant decrease in the length of the C1B—N7B bond, which is 1.315 (4) Å in (I) compared with 1.3597 (18) Å in the neutral 4-aminopyridine molecule (Anderson et al., 2005). The bond lengths and angles in 4-aminobenzoic acid and nicotinic acid are comparable with the values reported earlier (Gracin & Fischer, 2005; Jebas et al., 2006; Kutoglu & Scheringer, 1983; Jebas & Balasubramanian, 2006). As expected, all the aromatic rings in the asymmetric units of (I) and (II) are essentially planar, with the maximum deviations of the ring atoms from the least-squares plane being 0.018 (4) Å for atom C6A and 0.006 (4) Å for atom C3C in 4-aminobenzoate, and 0.010 (5) Å for atom C3B and 0.011 (4) Å for atoms C1D and C6D in 4-aminopyridine. In (II), the mean-plane deviation is 0.005 (3) Å for atom C2 and −0.008 Å for atom C8. 4-Aminopyridine is protonated in molecules (I) and (II) to form a salt, with the H atom from the carboxyl group of 4-aminobenzoic acid in (I) transferred to atoms N4B and N4D of the 4-aminopyridine in (I), and the H atom from the carboxyl group of nicotinic acid transferred to atom N1 of the 4-aminopyridine in (II). The protonation is evidenced by the widening of the internal angles (C3B—N4B—C5B and C3D—N4D—C5D) of the pyridine rings to 120.5 (4)° in molecule B and 120.4 (3)° in molecule D of (I), and of the C2—N1—C6 angle to 120.51° in molecule (II), compared with 115.25 (13)° in unprotonated 4-aminopyridine (Anderson et al., 2005; Chao & Schempp, 1977). Similar protonation is observed in various 4-aminopyridine–acid complexes, such as 4-aminopyridinium hydrogen succinate (Fun et al., 2009a) and bis(4-aminopyridinium)bis(hydrogen oxalate)monohydrate (Fun et al., 2009b).
In (I), the dihedral angles formed by the carboxylate groups (O8A—C7A—O9A—C4A and O8C—C7C—O9C—C4C) of the aminobenzoate anions with the attached benzene rings (C1A–C6A and C1C–C6C) are 8.21 (18) and 5.92 (18)°, respectively, indicating that they are twisted from the mean planes of the benzene rings. The two pyridinium rings (C1B–C3B/N4B/C5B/C6B and C1D–C3D/N4D/C5D/C6D) in (I) are twisted from away each other, forming a dihedral angle of 55.10 (17)°, and the two 4-aminobenzoate anions are twisted away from each other with a dihedral angle of 47.78 (18)°. Pyramidalization can be observed in the sums of the three angles at N of 356.78° in molecule B and 359.97° in molecule D of (I), and 358.62° in (II). The nicotinate and 4-aminopyridinium cations are oriented at an angle of 9.04 (7)° in (II).
The hydrogen-bonding pattern of (I) is shown in Fig. 3. Twenty intermolecular hydrogen bonds are observed in the structure of (I). The carboxylate O atoms of the 4-aminobenzoate anion act as acceptors of bifurcated N—H···O hydrogen bonds with the protonated 4-aminopyridinium cation (Table 2), forming a ring with the graph-set notation R12(4) (Bernstein et al., 1995). In addition to the bifurcated hydrogen bonding linking the cation with the anion, the water O atoms play a crucial role in linking the cations with the anions. In the crystal packing of (I) (Fig. 3), the molecules are linked by N—H···O and O—H···O hydrogen bonds to form a two-dimensional network along the (101) plane. In (II), the carboxylate O atoms of the nicotinate anion act as acceptors of bifurcated N—H···O hydrogen bonds with the protonated 4-aminopyridinium cation (Table 4), forming a ring with the graph-set notation R12(4). The amino H atoms are involved in forming N—H···N and N—H···O hydrogen bonds with the inversion-related molecules to complete a loop of graph-set motif R44(16) (Fig. 4). The packing of adduct (II) is consolidated via N—H···O and N—H···N hydrogen bonds to form a two-dimensional network along the (102) plane.