Buy article online - an online subscription or single-article purchase is required to access this article.
In 2-trifluoromethyl-4-nitroaniline, C
7H
5F
3N
2O
2, (I), the molecules lie across a mirror plane in space group
Pnma. The molecules are linked by paired N-H

O hydrogen bonds to form a
C(8)[
R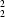
(6)] chain of rings, pairs of which are linked into a molecular ladder by a single C-H

O hydrogen bond. The isomeric 3-trifluoromethyl-4-nitroaniline, (II), has
Z' = 2 in space group
P2
1/
c. Each molecule is linked to four others by N-H

O hydrogen bonds to form sheets built from alternating
R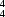
(12) and
R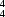
(32) rings.
Supporting information
CCDC references: 182031; 182032
Samples of compounds (I) and (II) were purchased from Aldrich. Crystals suitable
for single-crystal X-ray diffraction were grown by slow evaporation of
solutions in ethanol.
Compound (I) crystallized in the orthorhombic system. The systematic absences
permitted space groups Pna21 and Pnma; Pnma was selected, and
confirmed by the analysis. For compound (II), the space group
P21/c was uniquely assigned from the systematic absences. H
atoms were treated as riding, with C—H = 0.95 and N—H = 0.88 Å.
For both compounds, data collection: KappaCCD Server Software (Nonius, 1997); cell refinement: DENZO-SMN (Otwinowski & Minor, 1997); data reduction: DENZO-SMN; program(s) used to solve structure: SHELXS97 (Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: PLATON (Spek, 2001); software used to prepare material for publication: SHELXL97 and PRPKAPPA (Ferguson, 1999).
(I) 2-Trifluoromethyl-4-nitroaniline
top
Crystal data top
| C7H5F3N2O2 | F(000) = 416 |
| Mr = 206.13 | Dx = 1.754 Mg m−3 |
| Orthorhombic, Pnma | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2n | Cell parameters from 961 reflections |
| a = 9.0274 (4) Å | θ = 2.7–27.6° |
| b = 6.5933 (4) Å | µ = 0.18 mm−1 |
| c = 13.1170 (8) Å | T = 150 K |
| V = 780.73 (8) Å3 | Block, red |
| Z = 4 | 0.20 × 0.15 × 0.01 mm |
Data collection top
Nonius KappaCCD
diffractometer | 961 independent reflections |
| Radiation source: fine-focus sealed X-ray tube | 725 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.053 |
| ϕ scans, and ω scans with κ offsets | θmax = 27.6°, θmin = 2.7° |
Absorption correction: multi-scan
(DENZO-SMN; Otwinowski & Minor, 1997) | h = −9→11 |
| Tmin = 0.987, Tmax = 0.998 | k = −8→8 |
| 3944 measured reflections | l = −16→17 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.118 | H-atom parameters constrained |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0691P)2 + 0.053P]
where P = (Fo2 + 2Fc2)/3 |
| 961 reflections | (Δ/σ)max = 0.001 |
| 82 parameters | Δρmax = 0.31 e Å−3 |
| 0 restraints | Δρmin = −0.29 e Å−3 |
Crystal data top
| C7H5F3N2O2 | V = 780.73 (8) Å3 |
| Mr = 206.13 | Z = 4 |
| Orthorhombic, Pnma | Mo Kα radiation |
| a = 9.0274 (4) Å | µ = 0.18 mm−1 |
| b = 6.5933 (4) Å | T = 150 K |
| c = 13.1170 (8) Å | 0.20 × 0.15 × 0.01 mm |
Data collection top
Nonius KappaCCD
diffractometer | 961 independent reflections |
Absorption correction: multi-scan
(DENZO-SMN; Otwinowski & Minor, 1997) | 725 reflections with I > 2σ(I) |
| Tmin = 0.987, Tmax = 0.998 | Rint = 0.053 |
| 3944 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.041 | 0 restraints |
| wR(F2) = 0.118 | H-atom parameters constrained |
| S = 1.06 | Δρmax = 0.31 e Å−3 |
| 961 reflections | Δρmin = −0.29 e Å−3 |
| 82 parameters | |
Special details top
Experimental. The program DENZO-SMN (Otwinowski & Minor, 1997) uses a scaling algorithm
[Fox, G·C. & Holmes, K·C. (1966). Acta Cryst. 20, 886–891] which
effectively corrects for absorption effects. High-redundancy data were used in
the scaling program, hence the `multi-scan' code word was used. No
transmission coefficients are available from the program (only scale factors
for each frame). The scale factors in the experimental table are calculated
from the `size' command in the SHELXL97 input file. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N1 | −0.02222 (19) | 0.2500 | 0.01210 (14) | 0.0256 (5) | |
| C1 | 0.1272 (2) | 0.2500 | 0.01781 (15) | 0.0192 (5) | |
| C2 | 0.2192 (2) | 0.2500 | −0.07008 (15) | 0.0188 (5) | |
| C3 | 0.3712 (2) | 0.2500 | −0.06062 (15) | 0.0191 (5) | |
| C4 | 0.4351 (2) | 0.2500 | 0.03545 (15) | 0.0186 (5) | |
| C5 | 0.3483 (2) | 0.2500 | 0.12311 (15) | 0.0219 (5) | |
| C6 | 0.1968 (2) | 0.2500 | 0.11432 (16) | 0.0238 (5) | |
| C7 | 0.1532 (2) | 0.2500 | −0.17425 (15) | 0.0250 (5) | |
| F1 | 0.06499 (11) | 0.41227 (17) | −0.19085 (7) | 0.0370 (4) | |
| F3 | 0.25374 (15) | 0.2500 | −0.24767 (9) | 0.0459 (5) | |
| N2 | 0.59518 (19) | 0.2500 | 0.04274 (13) | 0.0222 (4) | |
| O1 | 0.66780 (16) | 0.2500 | −0.03648 (11) | 0.0310 (4) | |
| O2 | 0.65362 (17) | 0.2500 | 0.12830 (11) | 0.0325 (4) | |
| H12 | −0.0756 | 0.2500 | 0.0683 | 0.031* | |
| H11 | −0.0662 | 0.2500 | −0.0478 | 0.031* | |
| H3 | 0.4320 | 0.2500 | −0.1197 | 0.023* | |
| H5 | 0.3935 | 0.2500 | 0.1885 | 0.026* | |
| H6 | 0.1376 | 0.2500 | 0.1742 | 0.029* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N1 | 0.0151 (9) | 0.0348 (13) | 0.0269 (10) | 0.000 | 0.0023 (7) | 0.000 |
| C1 | 0.0168 (10) | 0.0195 (12) | 0.0214 (10) | 0.000 | 0.0017 (8) | 0.000 |
| C2 | 0.0172 (10) | 0.0233 (12) | 0.0158 (10) | 0.000 | −0.0024 (8) | 0.000 |
| C3 | 0.0184 (9) | 0.0244 (12) | 0.0144 (9) | 0.000 | 0.0020 (8) | 0.000 |
| C4 | 0.0166 (10) | 0.0217 (12) | 0.0175 (9) | 0.000 | −0.0018 (8) | 0.000 |
| C5 | 0.0247 (11) | 0.0276 (12) | 0.0134 (9) | 0.000 | −0.0025 (8) | 0.000 |
| C6 | 0.0285 (11) | 0.0293 (12) | 0.0138 (9) | 0.000 | 0.0049 (9) | 0.000 |
| C7 | 0.0194 (10) | 0.0364 (14) | 0.0193 (10) | 0.000 | −0.0053 (8) | 0.000 |
| F1 | 0.0389 (6) | 0.0414 (7) | 0.0308 (5) | 0.0069 (4) | −0.0145 (4) | 0.0058 (4) |
| F3 | 0.0280 (8) | 0.0960 (15) | 0.0136 (6) | 0.000 | −0.0010 (5) | 0.000 |
| N2 | 0.0193 (9) | 0.0249 (11) | 0.0224 (9) | 0.000 | −0.0058 (7) | 0.000 |
| O1 | 0.0154 (8) | 0.0502 (12) | 0.0274 (8) | 0.000 | 0.0031 (7) | 0.000 |
| O2 | 0.0253 (8) | 0.0487 (11) | 0.0235 (8) | 0.000 | −0.0109 (7) | 0.000 |
Geometric parameters (Å, º) top
| N1—C1 | 1.351 (3) | C4—N2 | 1.448 (3) |
| N1—H11 | 0.8800 | C5—C6 | 1.373 (3) |
| N1—H12 | 0.8800 | C5—H5 | 0.9500 |
| C1—C6 | 1.413 (3) | C6—H6 | 0.9500 |
| C1—C2 | 1.421 (3) | C7—F3 | 1.324 (2) |
| C2—C3 | 1.377 (3) | C7—F1i | 1.3511 (17) |
| C2—C7 | 1.491 (3) | C7—F1 | 1.3511 (17) |
| C3—C4 | 1.386 (3) | N2—O1 | 1.229 (2) |
| C3—H3 | 0.9500 | N2—O2 | 1.240 (2) |
| C4—C5 | 1.392 (3) | | |
| | | |
| C1—N1—H11 | 120.0 | C6—C5—C4 | 119.46 (18) |
| C1—N1—H12 | 120.0 | C6—C5—H5 | 120.3 |
| H11—N1—H12 | 120.0 | C4—C5—H5 | 120.3 |
| N1—C1—C6 | 119.56 (18) | C5—C6—C1 | 121.20 (18) |
| N1—C1—C2 | 122.60 (18) | C5—C6—H6 | 119.4 |
| C6—C1—C2 | 117.83 (19) | C1—C6—H6 | 119.4 |
| C3—C2—C1 | 120.61 (17) | F3—C7—F1i | 106.67 (12) |
| C3—C2—C7 | 118.76 (17) | F3—C7—F1 | 106.67 (12) |
| C1—C2—C7 | 120.63 (18) | F1i—C7—F1 | 104.72 (17) |
| C2—C3—C4 | 119.78 (18) | F3—C7—C2 | 113.10 (17) |
| C2—C3—H3 | 120.1 | F1i—C7—C2 | 112.54 (11) |
| C4—C3—H3 | 120.1 | F1—C7—C2 | 112.54 (11) |
| C3—C4—C5 | 121.11 (18) | O1—N2—O2 | 122.57 (16) |
| C3—C4—N2 | 118.40 (17) | O1—N2—C4 | 118.46 (15) |
| C5—C4—N2 | 120.49 (17) | O2—N2—C4 | 118.97 (16) |
| Symmetry code: (i) x, −y+1/2, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H11···O1ii | 0.88 | 2.41 | 2.870 (2) | 113 |
| N1—H12···O2ii | 0.88 | 2.57 | 3.299 (2) | 141 |
| C6—H6···O2iii | 0.95 | 2.59 | 3.398 (3) | 143 |
| Symmetry codes: (ii) x−1, y, z; (iii) x−1/2, −y+1/2, −z+1/2. |
(II) 3-Trifluoromethyl-4-nitroaniline
top
Crystal data top
| C7H5F3N2O2 | F(000) = 832 |
| Mr = 206.13 | Dx = 1.752 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3541 reflections |
| a = 7.1048 (2) Å | θ = 3.0–27.5° |
| b = 15.3832 (5) Å | µ = 0.18 mm−1 |
| c = 14.3091 (6) Å | T = 150 K |
| β = 91.9102 (12)° | Plate, yellow |
| V = 1563.04 (9) Å3 | 0.40 × 0.16 × 0.10 mm |
| Z = 8 | |
Data collection top
Nonius KappaCCD
diffractometer | 3541 independent reflections |
| Radiation source: fine-focus sealed X-ray tube | 2369 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.064 |
| ϕ scans, and ω scans with κ offsets | θmax = 27.5°, θmin = 3.0° |
Absorption correction: multi-scan
(DENZO-SMN; Otwinowski & Minor, 1997) | h = −9→9 |
| Tmin = 0.812, Tmax = 0.983 | k = −17→19 |
| 12016 measured reflections | l = −17→18 |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.044 | H-atom parameters constrained |
| wR(F2) = 0.114 | w = 1/[σ2(Fo2) + (0.0617P)2]
where P = (Fo2 + 2Fc2)/3 |
| S = 0.98 | (Δ/σ)max < 0.001 |
| 3541 reflections | Δρmax = 0.26 e Å−3 |
| 254 parameters | Δρmin = −0.32 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 1997), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0153 (19) |
Crystal data top
| C7H5F3N2O2 | V = 1563.04 (9) Å3 |
| Mr = 206.13 | Z = 8 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 7.1048 (2) Å | µ = 0.18 mm−1 |
| b = 15.3832 (5) Å | T = 150 K |
| c = 14.3091 (6) Å | 0.40 × 0.16 × 0.10 mm |
| β = 91.9102 (12)° | |
Data collection top
Nonius KappaCCD
diffractometer | 3541 independent reflections |
Absorption correction: multi-scan
(DENZO-SMN; Otwinowski & Minor, 1997) | 2369 reflections with I > 2σ(I) |
| Tmin = 0.812, Tmax = 0.983 | Rint = 0.064 |
| 12016 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.044 | 0 restraints |
| wR(F2) = 0.114 | H-atom parameters constrained |
| S = 0.98 | Δρmax = 0.26 e Å−3 |
| 3541 reflections | Δρmin = −0.32 e Å−3 |
| 254 parameters | |
Special details top
Experimental. The program DENZO-SMN (Otwinowski & Minor, 1997) uses a scaling algorithm
[Fox, G·C. & Holmes, K·C. (1966). Acta Cryst. 20, 886–891] which
effectively corrects for absorption effects. High-redundancy data were used in
the scaling program, hence the `multi-scan' code word was used. No
transmission coefficients are available from the program (only scale factors
for each frame). The scale factors in the experimental table are calculated
from the `size' command in the SHELXL97 input file. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N11 | −0.6318 (2) | 0.44448 (11) | 0.07857 (11) | 0.0299 (4) | |
| C11 | −0.5772 (2) | 0.38575 (12) | 0.14419 (12) | 0.0203 (4) | |
| C12 | −0.5337 (2) | 0.41245 (12) | 0.23643 (12) | 0.0196 (4) | |
| C13 | −0.4776 (2) | 0.35482 (11) | 0.30538 (11) | 0.0180 (4) | |
| C14 | −0.4639 (2) | 0.26582 (11) | 0.28128 (12) | 0.0190 (4) | |
| C15 | −0.5097 (2) | 0.23876 (12) | 0.19038 (12) | 0.0217 (4) | |
| C16 | −0.5656 (2) | 0.29701 (12) | 0.12290 (12) | 0.0218 (4) | |
| C17 | −0.4507 (2) | 0.39216 (12) | 0.40263 (13) | 0.0235 (4) | |
| F11 | −0.51135 (15) | 0.47424 (7) | 0.40579 (7) | 0.0308 (3) | |
| F12 | −0.27187 (14) | 0.39329 (7) | 0.43395 (7) | 0.0336 (3) | |
| F13 | −0.55087 (16) | 0.34846 (8) | 0.46541 (7) | 0.0354 (3) | |
| N12 | −0.4002 (2) | 0.19935 (10) | 0.34611 (11) | 0.0238 (4) | |
| O11 | −0.31463 (19) | 0.22002 (9) | 0.41879 (9) | 0.0336 (4) | |
| O12 | −0.4301 (2) | 0.12258 (9) | 0.32571 (10) | 0.0395 (4) | |
| N21 | −0.1232 (2) | 0.10727 (10) | 0.57317 (11) | 0.0288 (4) | |
| C21 | −0.0653 (2) | 0.16737 (12) | 0.63724 (12) | 0.0210 (4) | |
| C22 | −0.0513 (2) | 0.25579 (12) | 0.61228 (12) | 0.0212 (4) | |
| C23 | 0.0046 (2) | 0.31808 (12) | 0.67648 (12) | 0.0203 (4) | |
| C24 | 0.0481 (2) | 0.29196 (12) | 0.76917 (12) | 0.0213 (4) | |
| C25 | 0.0405 (2) | 0.20458 (12) | 0.79359 (13) | 0.0228 (4) | |
| C26 | −0.0158 (2) | 0.14327 (12) | 0.72908 (12) | 0.0231 (4) | |
| C27 | 0.0342 (2) | 0.40838 (12) | 0.63987 (13) | 0.0262 (4) | |
| F21 | 0.05603 (16) | 0.40859 (7) | 0.54741 (8) | 0.0374 (3) | |
| F22 | 0.18993 (14) | 0.44588 (7) | 0.67764 (8) | 0.0343 (3) | |
| F23 | −0.11002 (15) | 0.46208 (7) | 0.65539 (9) | 0.0398 (3) | |
| N22 | 0.0947 (2) | 0.35281 (11) | 0.84365 (11) | 0.0267 (4) | |
| O21 | 0.0298 (2) | 0.42728 (10) | 0.83940 (10) | 0.0384 (4) | |
| O22 | 0.19263 (19) | 0.32704 (10) | 0.91097 (10) | 0.0377 (4) | |
| H11 | −0.6609 | 0.4273 | 0.0212 | 0.036* | |
| H12 | −0.6383 | 0.5000 | 0.0931 | 0.036* | |
| H12A | −0.5433 | 0.4724 | 0.2516 | 0.024* | |
| H15 | −0.5020 | 0.1788 | 0.1751 | 0.026* | |
| H16 | −0.5967 | 0.2773 | 0.0614 | 0.026* | |
| H22 | −0.1301 | 0.0521 | 0.5891 | 0.035* | |
| H21 | −0.1538 | 0.1234 | 0.5157 | 0.035* | |
| H22A | −0.0813 | 0.2728 | 0.5497 | 0.025* | |
| H25 | 0.0746 | 0.1872 | 0.8556 | 0.027* | |
| H26 | −0.0213 | 0.0839 | 0.7469 | 0.028* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N11 | 0.0488 (10) | 0.0235 (9) | 0.0169 (8) | 0.0033 (7) | −0.0083 (7) | 0.0008 (7) |
| C11 | 0.0212 (9) | 0.0243 (11) | 0.0155 (9) | −0.0006 (7) | −0.0015 (7) | 0.0016 (8) |
| C12 | 0.0235 (9) | 0.0176 (9) | 0.0177 (9) | −0.0015 (7) | −0.0004 (7) | −0.0013 (8) |
| C13 | 0.0177 (8) | 0.0210 (10) | 0.0153 (9) | −0.0021 (7) | −0.0008 (6) | −0.0006 (7) |
| C14 | 0.0196 (8) | 0.0192 (10) | 0.0181 (10) | −0.0005 (7) | −0.0004 (7) | 0.0036 (8) |
| C15 | 0.0221 (9) | 0.0201 (10) | 0.0229 (10) | −0.0005 (7) | 0.0000 (7) | −0.0044 (8) |
| C16 | 0.0250 (9) | 0.0251 (11) | 0.0153 (9) | −0.0001 (7) | −0.0003 (7) | −0.0026 (8) |
| C17 | 0.0289 (10) | 0.0225 (11) | 0.0190 (10) | −0.0011 (8) | −0.0022 (7) | 0.0024 (8) |
| F11 | 0.0495 (6) | 0.0231 (6) | 0.0197 (6) | 0.0050 (5) | −0.0019 (5) | −0.0050 (5) |
| F12 | 0.0373 (6) | 0.0357 (7) | 0.0267 (6) | −0.0030 (5) | −0.0140 (5) | −0.0022 (5) |
| F13 | 0.0528 (7) | 0.0363 (7) | 0.0176 (6) | −0.0050 (5) | 0.0086 (5) | 0.0038 (5) |
| N12 | 0.0266 (8) | 0.0220 (9) | 0.0229 (9) | 0.0000 (6) | 0.0003 (6) | 0.0024 (7) |
| O11 | 0.0454 (8) | 0.0322 (9) | 0.0223 (7) | 0.0019 (6) | −0.0123 (6) | 0.0032 (6) |
| O12 | 0.0626 (10) | 0.0165 (8) | 0.0387 (9) | 0.0009 (7) | −0.0092 (7) | 0.0037 (6) |
| N21 | 0.0390 (9) | 0.0217 (9) | 0.0252 (9) | −0.0020 (7) | −0.0065 (7) | 0.0001 (7) |
| C21 | 0.0177 (8) | 0.0221 (10) | 0.0230 (10) | 0.0002 (7) | −0.0003 (7) | −0.0020 (8) |
| C22 | 0.0207 (9) | 0.0237 (10) | 0.0189 (10) | −0.0004 (7) | −0.0035 (7) | 0.0021 (8) |
| C23 | 0.0177 (8) | 0.0214 (10) | 0.0217 (10) | 0.0014 (7) | −0.0007 (7) | 0.0018 (8) |
| C24 | 0.0210 (9) | 0.0238 (10) | 0.0189 (9) | 0.0008 (7) | −0.0010 (7) | −0.0039 (8) |
| C25 | 0.0232 (9) | 0.0267 (11) | 0.0183 (9) | 0.0027 (7) | −0.0007 (7) | 0.0046 (8) |
| C26 | 0.0264 (9) | 0.0198 (10) | 0.0231 (10) | 0.0009 (7) | −0.0001 (7) | 0.0044 (8) |
| C27 | 0.0295 (10) | 0.0229 (11) | 0.0256 (11) | −0.0011 (8) | −0.0066 (8) | 0.0003 (8) |
| F21 | 0.0556 (7) | 0.0309 (7) | 0.0254 (7) | −0.0088 (5) | −0.0051 (5) | 0.0079 (5) |
| F22 | 0.0349 (6) | 0.0292 (7) | 0.0380 (7) | −0.0112 (5) | −0.0094 (5) | 0.0041 (5) |
| F23 | 0.0412 (7) | 0.0246 (7) | 0.0528 (8) | 0.0085 (5) | −0.0097 (5) | 0.0004 (6) |
| N22 | 0.0282 (8) | 0.0296 (10) | 0.0225 (9) | −0.0017 (7) | 0.0007 (7) | −0.0031 (7) |
| O21 | 0.0557 (9) | 0.0252 (9) | 0.0341 (9) | 0.0047 (7) | −0.0023 (7) | −0.0067 (7) |
| O22 | 0.0431 (8) | 0.0456 (10) | 0.0234 (8) | 0.0032 (7) | −0.0131 (6) | −0.0038 (7) |
Geometric parameters (Å, º) top
| N11—C11 | 1.351 (2) | N21—C21 | 1.356 (2) |
| N11—H11 | 0.8800 | N21—H22 | 0.8800 |
| N11—H11 | 0.8800 | N21—H21 | 0.8800 |
| C11—C16 | 1.402 (2) | C21—C26 | 1.399 (2) |
| C11—C12 | 1.407 (2) | C21—C22 | 1.411 (3) |
| C12—C13 | 1.375 (2) | C22—C23 | 1.377 (2) |
| C12—H12A | 0.9500 | C22—H22A | 0.9500 |
| C13—C14 | 1.416 (2) | C23—C24 | 1.410 (2) |
| C13—C17 | 1.512 (2) | C23—C27 | 1.502 (3) |
| C14—C15 | 1.394 (2) | C24—C25 | 1.390 (2) |
| C14—N12 | 1.443 (2) | C24—N22 | 1.448 (2) |
| C15—C16 | 1.366 (2) | C25—C26 | 1.370 (3) |
| C15—H15 | 0.9500 | C25—H25 | 0.9500 |
| C16—H16 | 0.9500 | C26—H26 | 0.9500 |
| C17—F12 | 1.333 (2) | C27—F21 | 1.337 (2) |
| C17—F11 | 1.335 (2) | C27—F23 | 1.340 (2) |
| C17—F13 | 1.345 (2) | C27—F22 | 1.345 (2) |
| N12—O11 | 1.2291 (19) | N22—O22 | 1.235 (2) |
| N12—O12 | 1.2331 (19) | N22—O21 | 1.236 (2) |
| | | |
| C11—N11—H11 | 120.0 | C21—N21—H21 | 120.0 |
| C11—N11—H12 | 120.0 | C21—N21—H22 | 120.0 |
| H11—N11—H12 | 120.0 | H21—N21—H22 | 120.0 |
| N11—C11—C16 | 121.20 (16) | N21—C21—C26 | 121.03 (17) |
| N11—C11—C12 | 120.39 (17) | N21—C21—C22 | 120.57 (16) |
| C16—C11—C12 | 118.40 (16) | C26—C21—C22 | 118.40 (16) |
| C13—C12—C11 | 122.28 (17) | C23—C22—C21 | 121.56 (16) |
| C13—C12—H12A | 118.9 | C23—C22—H22A | 119.2 |
| C11—C12—H12A | 118.9 | C21—C22—H22A | 119.2 |
| C12—C13—C14 | 117.97 (15) | C22—C23—C24 | 118.59 (17) |
| C12—C13—C17 | 116.11 (16) | C22—C23—C27 | 116.85 (16) |
| C14—C13—C17 | 125.73 (15) | C24—C23—C27 | 124.24 (16) |
| C15—C14—C13 | 120.01 (16) | C25—C24—C23 | 120.17 (16) |
| C13—C14—N12 | 123.44 (15) | C23—C24—N22 | 123.06 (16) |
| C15—C14—N12 | 116.53 (16) | C25—C24—N22 | 116.73 (16) |
| C16—C15—C14 | 121.12 (17) | C26—C25—C24 | 120.61 (17) |
| C16—C15—H15 | 119.4 | C26—C25—H25 | 119.7 |
| C14—C15—H15 | 119.4 | C24—C25—H25 | 119.7 |
| C15—C16—C11 | 120.18 (16) | C25—C26—C21 | 120.60 (17) |
| C15—C16—H16 | 119.9 | C25—C26—H26 | 119.7 |
| C11—C16—H16 | 119.9 | C21—C26—H26 | 119.7 |
| F12—C17—F11 | 106.33 (14) | F21—C27—F23 | 106.08 (14) |
| F12—C17—F13 | 107.56 (14) | F21—C27—F22 | 105.99 (14) |
| F11—C17—F13 | 105.75 (14) | F23—C27—F22 | 106.99 (15) |
| F12—C17—C13 | 113.85 (14) | F21—C27—C23 | 111.69 (15) |
| F11—C17—C13 | 111.06 (14) | F23—C27—C23 | 113.31 (15) |
| F13—C17—C13 | 111.81 (14) | F22—C27—C23 | 112.29 (14) |
| O11—N12—O12 | 121.63 (15) | O22—N22—O21 | 122.39 (16) |
| O11—N12—C14 | 119.75 (15) | O22—N22—C24 | 118.41 (16) |
| O12—N12—C14 | 118.60 (15) | O21—N22—C24 | 119.14 (15) |
| | | |
| O11—N12—C14—C13 | −16.7 (2) | O21—N22—C24—C23 | 28.1 (2) |
| O11—N12—C14—C15 | 162.0 (2) | O21—N22—C24—C25 | −149.5 (2) |
| O12—N12—C14—C13 | 164.8 (2) | O22—N22—C24—C23 | −154.6 (2) |
| O12—N12—C14—C15 | −16.4 (2) | O22—N22—C24—C25 | 27.8 (2) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N11—H11···O22i | 0.88 | 2.42 | 3.221 (2) | 152 |
| N11—H12···O12ii | 0.88 | 2.26 | 3.088 (2) | 157 |
| N21—H21···O11 | 0.88 | 2.31 | 3.088 (2) | 148 |
| N21—H22···O21iii | 0.88 | 2.28 | 3.101 (2) | 155 |
| C16—H16···F13iv | 0.95 | 2.40 | 3.180 (2) | 139 |
| C26—H26···F23iii | 0.95 | 2.50 | 3.347 (2) | 149 |
| Symmetry codes: (i) x−1, y, z−1; (ii) −x−1, y+1/2, −z+1/2; (iii) −x, y−1/2, −z+3/2; (iv) x, −y+1/2, z−1/2. |
Experimental details
| | (I) | (II) |
| Crystal data |
| Chemical formula | C7H5F3N2O2 | C7H5F3N2O2 |
| Mr | 206.13 | 206.13 |
| Crystal system, space group | Orthorhombic, Pnma | Monoclinic, P21/c |
| Temperature (K) | 150 | 150 |
| a, b, c (Å) | 9.0274 (4), 6.5933 (4), 13.1170 (8) | 7.1048 (2), 15.3832 (5), 14.3091 (6) |
| α, β, γ (°) | 90, 90, 90 | 90, 91.9102 (12), 90 |
| V (Å3) | 780.73 (8) | 1563.04 (9) |
| Z | 4 | 8 |
| Radiation type | Mo Kα | Mo Kα |
| µ (mm−1) | 0.18 | 0.18 |
| Crystal size (mm) | 0.20 × 0.15 × 0.01 | 0.40 × 0.16 × 0.10 |
| |
| Data collection |
| Diffractometer | Nonius KappaCCD
diffractometer | Nonius KappaCCD
diffractometer |
| Absorption correction | Multi-scan
(DENZO-SMN; Otwinowski & Minor, 1997) | Multi-scan
(DENZO-SMN; Otwinowski & Minor, 1997) |
| Tmin, Tmax | 0.987, 0.998 | 0.812, 0.983 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 3944, 961, 725 | 12016, 3541, 2369 |
| Rint | 0.053 | 0.064 |
| (sin θ/λ)max (Å−1) | 0.651 | 0.649 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.041, 0.118, 1.06 | 0.044, 0.114, 0.98 |
| No. of reflections | 961 | 3541 |
| No. of parameters | 82 | 254 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.31, −0.29 | 0.26, −0.32 |
Hydrogen-bond geometry (Å, º) for (I) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H11···O1i | 0.88 | 2.41 | 2.870 (2) | 113 |
| N1—H12···O2i | 0.88 | 2.57 | 3.299 (2) | 141 |
| C6—H6···O2ii | 0.95 | 2.59 | 3.398 (3) | 143 |
| Symmetry codes: (i) x−1, y, z; (ii) x−1/2, −y+1/2, −z+1/2. |
Selected bond and torsion angles (º) for (II) top| C12—C13—C17 | 116.11 (16) | C22—C23—C27 | 116.85 (16) |
| C14—C13—C17 | 125.73 (15) | C24—C23—C27 | 124.24 (16) |
| C13—C14—N12 | 123.44 (15) | C23—C24—N22 | 123.06 (16) |
| C15—C14—N12 | 116.53 (16) | C25—C24—N22 | 116.73 (16) |
| | | |
| O11—N12—C14—C13 | −16.7 (2) | O21—N22—C24—C23 | 28.1 (2) |
Hydrogen-bond geometry (Å, º) for (II) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N11—H11···O22i | 0.88 | 2.42 | 3.221 (2) | 152 |
| N11—H12···O12ii | 0.88 | 2.26 | 3.088 (2) | 157 |
| N21—H21···O11 | 0.88 | 2.31 | 3.088 (2) | 148 |
| N21—H22···O21iii | 0.88 | 2.28 | 3.101 (2) | 155 |
| Symmetry codes: (i) x−1, y, z−1; (ii) −x−1, y+1/2, −z+1/2; (iii) −x, y−1/2, −z+3/2. |

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.
 O hydrogen bonds to form a C(8)[R
O hydrogen bonds to form a C(8)[R O hydrogen bond. The isomeric 3-trifluoromethyl-4-nitroaniline, (II), has Z' = 2 in space group P21/c. Each molecule is linked to four others by N-H
O hydrogen bond. The isomeric 3-trifluoromethyl-4-nitroaniline, (II), has Z' = 2 in space group P21/c. Each molecule is linked to four others by N-H O hydrogen bonds to form sheets built from alternating R
O hydrogen bonds to form sheets built from alternating R
 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2002/02/00/sk1528/sk1528fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2002/02/00/sk1528/sk1528fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2002/02/00/sk1528/sk1528fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/c/issues/2002/02/00/sk1528/sk1528fig4thm.gif)

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry



In the structure of 4-nitroaniline (Tonogaki et al., 1993), each molecule is linked to four others by means of N—H···O hydrogen bonds, and the molecules are thereby linked into (4,4) nets (Batten & Robson, 1998). The resulting sheets, which contain a single type of R44(22) rings, are weakly linked by aromatic π···π stacking interactions. Introduction of a single methyl group in 2-methyl-4-nitroaniline gives rise to a different structure in the form of a three-dimensional framework, even though each molecule is still linked to four others by N—H···O hydrogen bonds (Ferguson et al., 2001). The structure now consists of two intersecting sets of sheets built from a single type of R88(54) ring. However, entirely different patterns of N—H···O hydrogen bonds are observed in the two polymorphs of 2-iodo-4-nitroaniline (McWilliam et al., 2001). In the triclinic polymorph, the molecules are linked into chains via a rather unusual supramolecular synthon comprising pairs of N—H···O hydrogen bonds forming an R22(6) ring, while in the orthorhombic polymorph, each molecule acts as just a single donor and single acceptor of N—H···O hydrogen bonds, with formation of simple C(8) chains. In both polymorphs, the chains are further linked into sheets by means of I···NO2 interactions.
Continuing our study of C-substituted nitroanilines (Cannon et al., 2001; Ferguson et al., 2001; Glidewell et al., 2001; McWilliam et al., 2001; Zakaria et al., 2001; Garden et al., 2001), we have now investigated the isomeric pair 2-trifluoromethyl-4-nitroaniline, (I), and 3-trifluoromethyl-4-nitroaniline, (II), which exhibit yet further modes of supramolecular aggregation. \sch
Compound (I) (Fig. 1) crystallizes in space group Pnma with Z' = 1/2, so that all the atoms, apart from two of the F atoms, lie on a mirror plane, selected as that at 1/4. The molecules are linked into chains by N—H···O hydrogen bonds (Table 1) and these chains are linked in pairs by a single C—H···O hydrogen bond. The amino atom N1 in the molecule at (x, 1/4, z) acts as a hydrogen-bond donor via H11 and H12 to O1 and O2, respectively, in the molecule at (x - 1, 1/4, z), so generating by translation a C(8)[R22(6)] chain of rings parallel to [100] (Fig. 2), where the R22(6) ring is necessarily planar. Four such chains run through each unit cell. A pair of chains related by the 21 screw axis along (x, 1/4, 1/4) are weakly linked by a soft hydrogen bond: atom C6 in the molecule at (x, 1/4, z) acts as a hydrogen-bond donor to O2 in the molecule at (x - 1/2, 1/4, 1/2 - z), while atom C6 at (x - 1/2, 1/4, 1/2 - z) in turn acts as a donor to O2 at (x - 1, 1/4, z). This combination of hard and soft hydrogen bonds thus generates a molecular ladder, in which a pair of [100] chains play the role of the uprights, while the C—H···O hydrogen bonds provide the rungs. Between the rungs, there are R32(12) rings (Fig. 2). The C(8)[R22(6)] chain of rings in (I) is thus analogous to that in triclinic 2-iodo-4-nitroaniline (McWilliam et al., 2001). In this latter compound, the linking of the chains into sheets again involves chains of like polarity, related to one another by translation. Two molecular ladders pass through each unit cell in (I), at y = 1/4 and y = 3/4, but there are no direction-specific interactions between neighbouring ladders.
Compound (II) crystallizes in space group P21/c with Z' = 2, and each molecule (Fig. 3) is linked to four others, via N—H···O hydrogen bonds (Table 3), into sheets parallel to (101). The formation of these sheets can readily be analysed in terms of three distinct one-dimensional motifs. There are two spiral chains parallel to [010], each comprising one type of molecule only, and a translational chain parallel to [101] containing both types of molecule.
Atom N11 in the type 1 molecule at (x, y, z) acts as a hydrogen-bond donor, via H12, to O12 in the type 1 molecule at (x - 1, 1/2 + y, 1/2 - z), so producing a C(8) chain parallel to [010] generated by the 21 screw axis along (-1/2, y, 1/4) (Fig. 4). Similarly, atom N21 in the type 2 molecule at (x, y, z) acts as a donor, via H22, to O21 in the type 2 molecule at (-x, y - 1/2, 3/2 - z), producing another C(8) spiral, this time generated by the 21 axis along (0, y, 3/4). Within the asymmetric unit, atom N21 acts as a donor, via H21, to O11, while atom N11 at (x, y, z) acts as a donor, via H11, to O22 at (x - 1, y, z - 1). The combination of the [101] chain with the two independent [010] chains generates a (101) sheet built of R44(12) and R44(32) rings, alternating in checkerboard fashion (Fig. 4). The interior of the large rings is occupied by the CF3 groups, the conformations of which are such that the two sets of F atoms are meshed together.
The (101) sheets in (II) are weakly linked by aromatic π···π stacking interactions. For the aromatic rings of molecules in adjacent sheets, the minimum distance between the ring centroids of a type 1 molecule at (x, y, z) and a type 2 molecule at (x - 1, 1/2 - y, z - 1/2) is 3.552 (2) Å, with the planes separated by 3.385 (3) Å.
The bond lengths in (I) and (II) present no unexpected features. In compound (II), there is evidence from the exocyclic bond angles (Table 2) for significant repulsion between the adjacent nitro and trifluoromethyl groups.