In the adduct 1,2-bis(4-pyridyl)ethane-1,1,1-tris(4-hydroxyphenyl)ethane (1/2), C
12H
12N
2·2C
20H
18O
3, the bipyridyl component lies across an inversion centre in
P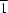
. The tris-phenol molecules [systematic name: 4,4',4''-(ethane-1,1,1-triyl)triphenol] are linked by O-H

O hydrogen bonds to form sheets built from
R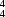
(38) rings, and symmetry-related pairs of sheets are linked by the bipyridyl molecules
via O-H

N hydrogen bonds to form open bilayers. Each bilayer is interwoven with two adjacent bilayers, forming a continuous three-dimensional structure. In the adduct 1,2-bis(4-pyridyl)ethene-1,1,1-tris(4-hydroxyphenyl)ethane-methanol (1/1/1), C
12H
10N
2·C
20H
18O
3·CH
4O, the molecules are linked by O-H

O and O-H

N hydrogen bonds into three interwoven three-dimensional frameworks, generated by single spiral chains along [010] and [001] and a triple-helical spiral along [100].
Supporting information
CCDC references: 180149; 180150
Equimolar quantities of the 1,1,1-tris(4-hydroxyphenyl)ethane and the
appropriate bipyridyl were separately dissolved in methanol. For each
compound, solutions of the tris-phenol and the appropriate bipyridyl were
mixed and set aside to crystallize, providing (I) and (II), respectively.
Analyses, (I): found C 77.8, H 6.2, N 3.5%; C50H48N2O6 requires C
77.7, H 6.3, N 3.6%; (II): found C 76.0, H 6.7, N 5.4%; C33H32N2O4
requires C 76.1, H 6.2, N 5.4%. Crystals suitable for single-crystal X-ray
diffraction were selected directly from the analytical samples.
Compound (I) crystallized in the triclinic system; space group P1 was
assumed and subsequently confirmed by the analysis. For compound (II), space
group P212121 was uniquely assigned from the systematic absences.
The refined value, 2(3), of the Flack (1983) parameterwas inconclusive (Flack
& Bernardinelli, 2000), hence Friedel equivalents were merged prior to the
final refinements. All H atoms were treated as riding atoms, with C—H
distances in the range 0.95–0.99 Å and O—H distances of 0.84 Å.
For both compounds, data collection: KappaCCD Server Software (Nonius, 1997); cell refinement: DENZO-SMN (Otwinowski & Minor, 1997); data reduction: DENZO-SMN; program(s) used to solve structure: SHELXS97 (Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: PLATON (Spek, 2001). Software used to prepare material for publication: SHELXL97 and PRPKAPPA (Ferguson, 1999) for (I); SHELXL97 (Sheldrick, 1997) and PRPKAPPA (Ferguson, 1999) for (II).
(I) 1,2-Bis(4'-pyridyl)ethane–1,1,1-tris(4-hydroxyphenyl)ethane (1/2)
top
Crystal data top
| C12H12N2·2C20H18O3 | Z = 1 |
| Mr = 796.92 | F(000) = 422 |
| Triclinic, P1 | Dx = 1.255 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.2076 (2) Å | Cell parameters from 4446 reflections |
| b = 10.6559 (2) Å | θ = 3.1–27.5° |
| c = 10.9341 (2) Å | µ = 0.08 mm−1 |
| α = 100.9920 (16)° | T = 150 K |
| β = 109.4530 (14)° | Block, colourless |
| γ = 101.6700 (11)° | 0.30 × 0.20 × 0.18 mm |
| V = 1054.49 (3) Å3 | |
Data collection top
Kappa-CCD
diffractometer | 4797 independent reflections |
| Radiation source: fine-focus sealed X-ray tube | 3886 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.031 |
| ϕ scans, and ω scans with κ offsets | θmax = 27.5°, θmin = 3.1° |
Absorption correction: multi-scan
(DENZO-SMN; Otwinowski & Minor, 1997) | h = 0→13 |
| Tmin = 0.976, Tmax = 0.985 | k = −13→13 |
| 13679 measured reflections | l = −14→12 |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.044 | H-atom parameters constrained |
| wR(F2) = 0.123 | w = 1/[σ2(Fo2) + (0.052P)2 + 0.196P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.09 | (Δ/σ)max = 0.001 |
| 4797 reflections | Δρmax = 0.22 e Å−3 |
| 276 parameters | Δρmin = −0.28 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.074 (8) |
Crystal data top
| C12H12N2·2C20H18O3 | γ = 101.6700 (11)° |
| Mr = 796.92 | V = 1054.49 (3) Å3 |
| Triclinic, P1 | Z = 1 |
| a = 10.2076 (2) Å | Mo Kα radiation |
| b = 10.6559 (2) Å | µ = 0.08 mm−1 |
| c = 10.9341 (2) Å | T = 150 K |
| α = 100.9920 (16)° | 0.30 × 0.20 × 0.18 mm |
| β = 109.4530 (14)° | |
Data collection top
Kappa-CCD
diffractometer | 4797 independent reflections |
Absorption correction: multi-scan
(DENZO-SMN; Otwinowski & Minor, 1997) | 3886 reflections with I > 2σ(I) |
| Tmin = 0.976, Tmax = 0.985 | Rint = 0.031 |
| 13679 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.044 | 0 restraints |
| wR(F2) = 0.123 | H-atom parameters constrained |
| S = 1.09 | Δρmax = 0.22 e Å−3 |
| 4797 reflections | Δρmin = −0.28 e Å−3 |
| 276 parameters | |
Special details top
Experimental. The program DENZO-SMN (Otwinowski & Minor, 1997) uses a scaling algorithm
[Fox, G·C. & Holmes, K·C. (1966). Acta Cryst. 20, 886–891] which
effectively corrects for absorption effects. High redundancy data were used in
the scaling program hence the 'multi-scan' code word was used. No transmission
coefficients are available from the program (only scale factors for each
frame). The scale factors in the experimental table are calculated from the
'size' command in the SHELXL97 input file. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 1.07976 (11) | 0.19588 (11) | 0.54229 (9) | 0.0364 (3) | |
| O2 | 1.24756 (12) | 0.34767 (10) | −0.20730 (9) | 0.0363 (2) | |
| O3 | 1.07237 (12) | −0.50019 (10) | −0.14460 (11) | 0.0426 (3) | |
| C1 | 1.34508 (13) | 0.04092 (12) | 0.17400 (12) | 0.0237 (3) | |
| C2 | 1.51061 (13) | 0.07340 (14) | 0.25400 (13) | 0.0299 (3) | |
| C11 | 1.27299 (13) | 0.08509 (12) | 0.27198 (12) | 0.0241 (3) | |
| C12 | 1.31224 (17) | 0.06065 (16) | 0.39778 (14) | 0.0392 (4) | |
| C13 | 1.24684 (18) | 0.09705 (18) | 0.48618 (14) | 0.0443 (4) | |
| C14 | 1.13922 (14) | 0.15989 (13) | 0.45060 (12) | 0.0280 (3) | |
| C15 | 1.09683 (13) | 0.18381 (13) | 0.32569 (12) | 0.0259 (3) | |
| C16 | 1.16273 (13) | 0.14611 (13) | 0.23781 (12) | 0.0262 (3) | |
| C21 | 1.32052 (13) | 0.11895 (12) | 0.06746 (12) | 0.0234 (3) | |
| C22 | 1.38429 (14) | 0.25688 (13) | 0.10620 (12) | 0.0281 (3) | |
| C23 | 1.35965 (15) | 0.33185 (13) | 0.01504 (13) | 0.0300 (3) | |
| C24 | 1.27008 (14) | 0.26951 (13) | −0.11934 (12) | 0.0276 (3) | |
| C25 | 1.20579 (14) | 0.13329 (13) | −0.16115 (12) | 0.0284 (3) | |
| C26 | 1.23157 (13) | 0.05955 (12) | −0.06776 (12) | 0.0261 (3) | |
| C31 | 1.27828 (13) | −0.10920 (12) | 0.10285 (12) | 0.0241 (3) | |
| C32 | 1.35497 (14) | −0.18380 (13) | 0.05045 (13) | 0.0307 (3) | |
| C33 | 1.28890 (15) | −0.31416 (13) | −0.03031 (14) | 0.0336 (3) | |
| C34 | 1.14415 (15) | −0.37398 (13) | −0.06062 (13) | 0.0307 (3) | |
| C35 | 1.06642 (14) | −0.30349 (13) | −0.00677 (13) | 0.0310 (3) | |
| C36 | 1.13350 (13) | −0.17336 (13) | 0.07377 (13) | 0.0281 (3) | |
| N41 | 0.86824 (12) | 0.30647 (12) | 0.49551 (11) | 0.0327 (3) | |
| C42 | 0.81396 (16) | 0.29772 (16) | 0.58974 (15) | 0.0395 (3) | |
| C43 | 0.71869 (17) | 0.36662 (18) | 0.61082 (15) | 0.0457 (4) | |
| C44 | 0.67745 (15) | 0.45044 (15) | 0.53263 (16) | 0.0406 (4) | |
| C45 | 0.73375 (16) | 0.45926 (15) | 0.43425 (17) | 0.0408 (3) | |
| C46 | 0.82787 (15) | 0.38664 (14) | 0.41886 (15) | 0.0360 (3) | |
| C47 | 0.57473 (17) | 0.52853 (18) | 0.55278 (19) | 0.0561 (5) | |
| H1 | 1.0110 | 0.2263 | 0.5072 | 0.055* | |
| H2 | 1.1993 | 0.2986 | −0.2865 | 0.054* | |
| H3 | 1.1316 | −0.5353 | −0.1652 | 0.064* | |
| H2A | 1.5291 | 0.0120 | 0.3097 | 0.045* | |
| H2B | 1.5481 | 0.1654 | 0.3122 | 0.045* | |
| H2C | 1.5592 | 0.0634 | 0.1906 | 0.045* | |
| H12 | 1.3860 | 0.0178 | 0.4239 | 0.047* | |
| H13 | 1.2759 | 0.0788 | 0.5714 | 0.053* | |
| H15 | 1.0225 | 0.2261 | 0.2997 | 0.031* | |
| H16 | 1.1316 | 0.1625 | 0.1518 | 0.031* | |
| H22 | 1.4464 | 0.3004 | 0.1979 | 0.034* | |
| H23 | 1.4038 | 0.4255 | 0.0443 | 0.036* | |
| H25 | 1.1443 | 0.0902 | −0.2531 | 0.034* | |
| H26 | 1.1870 | −0.0341 | −0.0975 | 0.031* | |
| H32 | 1.4546 | −0.1443 | 0.0706 | 0.037* | |
| H33 | 1.3434 | −0.3627 | −0.0650 | 0.040* | |
| H35 | 0.9676 | −0.3444 | −0.0251 | 0.037* | |
| H36 | 1.0793 | −0.1262 | 0.1104 | 0.034* | |
| H42 | 0.8421 | 0.2414 | 0.6452 | 0.047* | |
| H43 | 0.6818 | 0.3564 | 0.6786 | 0.055* | |
| H45 | 0.7077 | 0.5151 | 0.3776 | 0.049* | |
| H46 | 0.8654 | 0.3940 | 0.3510 | 0.043* | |
| H47A | 0.5706 | 0.5311 | 0.6424 | 0.067* | |
| H47B | 0.6133 | 0.6215 | 0.5526 | 0.067* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0413 (6) | 0.0546 (7) | 0.0281 (5) | 0.0307 (5) | 0.0193 (4) | 0.0159 (4) |
| O2 | 0.0528 (6) | 0.0311 (5) | 0.0297 (5) | 0.0161 (5) | 0.0168 (5) | 0.0128 (4) |
| O3 | 0.0437 (6) | 0.0261 (5) | 0.0517 (6) | 0.0065 (4) | 0.0179 (5) | 0.0011 (4) |
| C1 | 0.0223 (6) | 0.0273 (6) | 0.0220 (5) | 0.0093 (5) | 0.0088 (5) | 0.0057 (5) |
| C2 | 0.0236 (6) | 0.0348 (7) | 0.0297 (6) | 0.0106 (5) | 0.0079 (5) | 0.0072 (5) |
| C11 | 0.0249 (6) | 0.0250 (6) | 0.0228 (6) | 0.0091 (5) | 0.0092 (5) | 0.0054 (5) |
| C12 | 0.0479 (8) | 0.0579 (9) | 0.0317 (7) | 0.0386 (8) | 0.0205 (6) | 0.0225 (7) |
| C13 | 0.0559 (9) | 0.0717 (11) | 0.0298 (7) | 0.0451 (9) | 0.0235 (7) | 0.0274 (7) |
| C14 | 0.0304 (6) | 0.0337 (7) | 0.0241 (6) | 0.0147 (5) | 0.0130 (5) | 0.0073 (5) |
| C15 | 0.0230 (6) | 0.0299 (6) | 0.0254 (6) | 0.0117 (5) | 0.0077 (5) | 0.0076 (5) |
| C16 | 0.0247 (6) | 0.0327 (7) | 0.0215 (5) | 0.0112 (5) | 0.0074 (5) | 0.0082 (5) |
| C21 | 0.0224 (6) | 0.0268 (6) | 0.0233 (6) | 0.0091 (5) | 0.0112 (5) | 0.0060 (5) |
| C22 | 0.0297 (6) | 0.0276 (7) | 0.0236 (6) | 0.0064 (5) | 0.0097 (5) | 0.0028 (5) |
| C23 | 0.0360 (7) | 0.0230 (6) | 0.0312 (6) | 0.0072 (5) | 0.0152 (6) | 0.0050 (5) |
| C24 | 0.0330 (7) | 0.0292 (7) | 0.0273 (6) | 0.0135 (5) | 0.0157 (5) | 0.0106 (5) |
| C25 | 0.0307 (6) | 0.0301 (7) | 0.0227 (6) | 0.0083 (5) | 0.0094 (5) | 0.0056 (5) |
| C26 | 0.0267 (6) | 0.0247 (6) | 0.0247 (6) | 0.0060 (5) | 0.0091 (5) | 0.0047 (5) |
| C31 | 0.0252 (6) | 0.0256 (6) | 0.0238 (6) | 0.0100 (5) | 0.0099 (5) | 0.0084 (5) |
| C32 | 0.0265 (6) | 0.0297 (7) | 0.0370 (7) | 0.0097 (5) | 0.0145 (6) | 0.0063 (5) |
| C33 | 0.0344 (7) | 0.0293 (7) | 0.0404 (7) | 0.0135 (6) | 0.0184 (6) | 0.0055 (6) |
| C34 | 0.0353 (7) | 0.0241 (6) | 0.0321 (6) | 0.0083 (5) | 0.0125 (6) | 0.0074 (5) |
| C35 | 0.0269 (6) | 0.0301 (7) | 0.0369 (7) | 0.0072 (5) | 0.0136 (6) | 0.0101 (5) |
| C36 | 0.0269 (6) | 0.0309 (7) | 0.0311 (6) | 0.0117 (5) | 0.0141 (5) | 0.0099 (5) |
| N41 | 0.0282 (6) | 0.0360 (6) | 0.0338 (6) | 0.0125 (5) | 0.0119 (5) | 0.0063 (5) |
| C42 | 0.0362 (8) | 0.0511 (9) | 0.0348 (7) | 0.0183 (7) | 0.0146 (6) | 0.0122 (6) |
| C43 | 0.0345 (8) | 0.0660 (11) | 0.0368 (8) | 0.0192 (8) | 0.0168 (6) | 0.0040 (7) |
| C44 | 0.0243 (7) | 0.0355 (8) | 0.0472 (8) | 0.0079 (6) | 0.0075 (6) | −0.0091 (6) |
| C45 | 0.0315 (7) | 0.0302 (7) | 0.0567 (9) | 0.0099 (6) | 0.0123 (7) | 0.0110 (7) |
| C46 | 0.0302 (7) | 0.0376 (8) | 0.0428 (8) | 0.0110 (6) | 0.0164 (6) | 0.0119 (6) |
| C47 | 0.0320 (8) | 0.0480 (10) | 0.0690 (11) | 0.0165 (7) | 0.0107 (8) | −0.0162 (8) |
Geometric parameters (Å, º) top
| O1—C14 | 1.3688 (14) | C24—C25 | 1.3814 (18) |
| O1—H1 | 0.84 | C25—C26 | 1.3928 (17) |
| O2—C24 | 1.3810 (14) | C25—H25 | 0.95 |
| O2—H2 | 0.84 | C26—H26 | 0.95 |
| O3—C34 | 1.3678 (16) | C31—C36 | 1.3957 (17) |
| O3—H3 | 0.84 | C31—C32 | 1.3963 (17) |
| C1—C31 | 1.5362 (17) | C32—C33 | 1.3860 (19) |
| C1—C21 | 1.5396 (16) | C32—H32 | 0.95 |
| C1—C11 | 1.5439 (15) | C33—C34 | 1.3826 (19) |
| C1—C2 | 1.5503 (17) | C33—H33 | 0.95 |
| C2—H2A | 0.98 | C34—C35 | 1.3872 (18) |
| C2—H2B | 0.98 | C35—C36 | 1.3841 (19) |
| C2—H2C | 0.98 | C35—H35 | 0.95 |
| C11—C12 | 1.3900 (17) | C36—H36 | 0.95 |
| C11—C16 | 1.3920 (16) | N41—C42 | 1.3316 (18) |
| C12—C13 | 1.3864 (18) | N41—C46 | 1.3369 (17) |
| C12—H12 | 0.95 | C42—C43 | 1.383 (2) |
| C13—C14 | 1.3852 (18) | C42—H42 | 0.95 |
| C13—H13 | 0.95 | C43—C44 | 1.382 (2) |
| C14—C15 | 1.3789 (17) | C43—H43 | 0.95 |
| C15—C16 | 1.3893 (16) | C44—C45 | 1.387 (2) |
| C15—H15 | 0.95 | C44—C47 | 1.5091 (19) |
| C16—H16 | 0.95 | C45—C46 | 1.3817 (19) |
| C21—C26 | 1.3884 (17) | C45—H45 | 0.95 |
| C21—C22 | 1.3983 (18) | C46—H46 | 0.95 |
| C22—C23 | 1.3831 (17) | C47—C47i | 1.488 (3) |
| C22—H22 | 0.95 | C47—H47A | 0.99 |
| C23—C24 | 1.3869 (18) | C47—H47B | 0.99 |
| C23—H23 | 0.95 | | |
| | | |
| C14—O1—H1 | 109.5 | C24—C25—H25 | 120.2 |
| C24—O2—H2 | 109.5 | C26—C25—H25 | 120.2 |
| C34—O3—H3 | 109.5 | C21—C26—C25 | 121.90 (11) |
| C31—C1—C21 | 109.27 (9) | C21—C26—H26 | 119.0 |
| C31—C1—C11 | 109.93 (9) | C25—C26—H26 | 119.0 |
| C21—C1—C11 | 109.30 (9) | C36—C31—C32 | 116.93 (12) |
| C31—C1—C2 | 110.58 (10) | C36—C31—C1 | 121.58 (10) |
| C21—C1—C2 | 108.43 (10) | C32—C31—C1 | 121.08 (11) |
| C11—C1—C2 | 109.29 (10) | C33—C32—C31 | 121.45 (12) |
| C1—C2—H2A | 109.5 | C33—C32—H32 | 119.3 |
| C1—C2—H2B | 109.5 | C31—C32—H32 | 119.3 |
| H2A—C2—H2B | 109.5 | C34—C33—C32 | 120.36 (12) |
| C1—C2—H2C | 109.5 | C34—C33—H33 | 119.8 |
| H2A—C2—H2C | 109.5 | C32—C33—H33 | 119.8 |
| H2B—C2—H2C | 109.5 | O3—C34—C33 | 122.59 (12) |
| C12—C11—C16 | 116.72 (11) | O3—C34—C35 | 117.99 (12) |
| C12—C11—C1 | 120.93 (10) | C33—C34—C35 | 119.41 (12) |
| C16—C11—C1 | 122.31 (10) | C36—C35—C34 | 119.72 (12) |
| C13—C12—C11 | 121.87 (12) | C36—C35—H35 | 120.1 |
| C13—C12—H12 | 119.1 | C34—C35—H35 | 120.1 |
| C11—C12—H12 | 119.1 | C35—C36—C31 | 122.09 (11) |
| C14—C13—C12 | 120.21 (12) | C35—C36—H36 | 119.0 |
| C14—C13—H13 | 119.9 | C31—C36—H36 | 119.0 |
| C12—C13—H13 | 119.9 | C42—N41—C46 | 117.53 (12) |
| O1—C14—C15 | 122.87 (11) | N41—C42—C43 | 123.19 (14) |
| O1—C14—C13 | 117.98 (11) | N41—C42—H42 | 118.4 |
| C15—C14—C13 | 119.15 (11) | C43—C42—H42 | 118.4 |
| C14—C15—C16 | 120.03 (11) | C44—C43—C42 | 119.55 (14) |
| C14—C15—H15 | 120.0 | C44—C43—H43 | 120.2 |
| C16—C15—H15 | 120.0 | C42—C43—H43 | 120.2 |
| C15—C16—C11 | 122.01 (11) | C43—C44—C45 | 117.24 (12) |
| C15—C16—H16 | 119.0 | C43—C44—C47 | 121.42 (15) |
| C11—C16—H16 | 119.0 | C45—C44—C47 | 121.35 (16) |
| C26—C21—C22 | 116.96 (11) | C46—C45—C44 | 119.80 (14) |
| C26—C21—C1 | 122.94 (11) | C46—C45—H45 | 120.1 |
| C22—C21—C1 | 120.04 (10) | C44—C45—H45 | 120.1 |
| C23—C22—C21 | 121.98 (11) | N41—C46—C45 | 122.70 (13) |
| C23—C22—H22 | 119.0 | N41—C46—H46 | 118.7 |
| C21—C22—H22 | 119.0 | C45—C46—H46 | 118.7 |
| C22—C23—C24 | 119.64 (12) | C47i—C47—C44 | 113.21 (16) |
| C22—C23—H23 | 120.2 | C47i—C47—H47A | 108.9 |
| C24—C23—H23 | 120.2 | C44—C47—H47A | 108.9 |
| O2—C24—C25 | 122.11 (11) | C47i—C47—H47B | 108.9 |
| O2—C24—C23 | 118.01 (11) | C44—C47—H47B | 108.9 |
| C25—C24—C23 | 119.88 (11) | H47A—C47—H47B | 107.7 |
| C24—C25—C26 | 119.63 (11) | | |
| | | |
| C31—C1—C11—C12 | 78.67 (15) | C22—C21—C26—C25 | −0.33 (17) |
| C21—C1—C11—C12 | −161.39 (13) | C1—C21—C26—C25 | 177.02 (10) |
| C2—C1—C11—C12 | −42.87 (17) | C24—C25—C26—C21 | 0.09 (19) |
| C31—C1—C11—C16 | −98.83 (13) | C21—C1—C31—C36 | −88.13 (13) |
| C21—C1—C11—C16 | 21.11 (16) | C11—C1—C31—C36 | 31.82 (15) |
| C2—C1—C11—C16 | 139.63 (12) | C2—C1—C31—C36 | 152.59 (11) |
| C16—C11—C12—C13 | −1.0 (2) | C21—C1—C31—C32 | 84.24 (13) |
| C1—C11—C12—C13 | −178.68 (14) | C11—C1—C31—C32 | −155.81 (11) |
| C11—C12—C13—C14 | −0.1 (3) | C2—C1—C31—C32 | −35.04 (15) |
| C12—C13—C14—O1 | −178.90 (15) | C36—C31—C32—C33 | 2.03 (19) |
| C12—C13—C14—C15 | 0.9 (2) | C1—C31—C32—C33 | −170.68 (12) |
| O1—C14—C15—C16 | 179.26 (12) | C31—C32—C33—C34 | −0.3 (2) |
| C13—C14—C15—C16 | −0.6 (2) | C32—C33—C34—O3 | 177.04 (12) |
| C14—C15—C16—C11 | −0.6 (2) | C32—C33—C34—C35 | −1.6 (2) |
| C12—C11—C16—C15 | 1.4 (2) | O3—C34—C35—C36 | −177.11 (12) |
| C1—C11—C16—C15 | 179.02 (12) | C33—C34—C35—C36 | 1.6 (2) |
| C31—C1—C21—C26 | 9.19 (15) | C34—C35—C36—C31 | 0.28 (19) |
| C11—C1—C21—C26 | −111.15 (12) | C32—C31—C36—C35 | −2.04 (18) |
| C2—C1—C21—C26 | 129.80 (12) | C1—C31—C36—C35 | 170.63 (11) |
| C31—C1—C21—C22 | −173.54 (10) | C46—N41—C42—C43 | 0.3 (2) |
| C11—C1—C21—C22 | 66.12 (13) | N41—C42—C43—C44 | −0.8 (2) |
| C2—C1—C21—C22 | −52.93 (14) | C42—C43—C44—C45 | 0.9 (2) |
| C26—C21—C22—C23 | 0.53 (17) | C42—C43—C44—C47 | −179.48 (14) |
| C1—C21—C22—C23 | −176.90 (11) | C43—C44—C45—C46 | −0.5 (2) |
| C21—C22—C23—C24 | −0.48 (19) | C47—C44—C45—C46 | 179.81 (14) |
| C22—C23—C24—O2 | 179.74 (11) | C42—N41—C46—C45 | 0.1 (2) |
| C22—C23—C24—C25 | 0.21 (19) | C44—C45—C46—N41 | 0.1 (2) |
| O2—C24—C25—C26 | −179.53 (11) | C43—C44—C47—C47i | −104.0 (3) |
| C23—C24—C25—C26 | −0.02 (18) | C45—C44—C47—C47i | 75.7 (3) |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N41 | 0.84 | 1.81 | 2.6145 (18) | 159 |
| O2—H2···O1ii | 0.84 | 1.83 | 2.6671 (13) | 175 |
| O3—H3···O2iii | 0.84 | 1.98 | 2.8039 (17) | 168 |
| Symmetry codes: (ii) x, y, z−1; (iii) x, y−1, z. |
(II) 1,2-Bis(4'-pyridyl)ethene–1,1,1-tris(4-hydroxyphenyl)ethane–methanol (1/1/1)
top
Crystal data top
| C12H10N2·C20H18O3·CH4O | F(000) = 1104 |
| Mr = 520.61 | Dx = 1.223 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 2663 reflections |
| a = 11.8264 (9) Å | θ = 2.6–25.0° |
| b = 14.2158 (9) Å | µ = 0.08 mm−1 |
| c = 16.8109 (12) Å | T = 150 K |
| V = 2826.3 (3) Å3 | Block, colourless |
| Z = 4 | 0.30 × 0.26 × 0.16 mm |
Data collection top
KappaCCD
diffractometer | 2795 independent reflections |
| Radiation source: fine-focus sealed X-ray tube | 1859 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.082 |
| ϕ scans, and ω scans with κ offsets | θmax = 25.0°, θmin = 2.6° |
Absorption correction: multi-scan
(DENZO-SMN; Otwinowski & Minor, 1997) | h = 0→14 |
| Tmin = 0.976, Tmax = 0.987 | k = 0→16 |
| 11922 measured reflections | l = 0→20 |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.057 | H-atom parameters constrained |
| wR(F2) = 0.149 | w = 1/[σ2(Fo2) + (0.0458P)2 + 1.525P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.08 | (Δ/σ)max < 0.001 |
| 2795 reflections | Δρmax = 0.22 e Å−3 |
| 359 parameters | Δρmin = −0.19 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0144 (17) |
Crystal data top
| C12H10N2·C20H18O3·CH4O | V = 2826.3 (3) Å3 |
| Mr = 520.61 | Z = 4 |
| Orthorhombic, P212121 | Mo Kα radiation |
| a = 11.8264 (9) Å | µ = 0.08 mm−1 |
| b = 14.2158 (9) Å | T = 150 K |
| c = 16.8109 (12) Å | 0.30 × 0.26 × 0.16 mm |
Data collection top
KappaCCD
diffractometer | 2795 independent reflections |
Absorption correction: multi-scan
(DENZO-SMN; Otwinowski & Minor, 1997) | 1859 reflections with I > 2σ(I) |
| Tmin = 0.976, Tmax = 0.987 | Rint = 0.082 |
| 11922 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.057 | 0 restraints |
| wR(F2) = 0.149 | H-atom parameters constrained |
| S = 1.08 | Δρmax = 0.22 e Å−3 |
| 2795 reflections | Δρmin = −0.19 e Å−3 |
| 359 parameters | |
Special details top
Experimental. The program DENZO-SMN (Otwinowski & Minor, 1997) uses a scaling algorithm
[Fox, G·C. & Holmes, K·C. (1966). Acta Cryst. 20, 886–891] which
effectively corrects for absorption effects. High redundancy data were used in
the scaling program hence the 'multi-scan' code word was used. No transmission
coefficients are available from the program (only scale factors for each
frame). The scale factors in the experimental table are calculated from the
'size' command in the SHELXL97 input file. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.2045 (3) | −0.0490 (3) | 0.2848 (2) | 0.0561 (11) | |
| O2 | 0.1312 (4) | 0.2606 (3) | 0.7819 (2) | 0.0501 (10) | |
| O3 | 0.4144 (3) | 0.5744 (3) | 0.3525 (2) | 0.0551 (11) | |
| C1 | 0.0895 (4) | 0.2946 (4) | 0.4439 (3) | 0.0400 (13) | |
| C2 | −0.0310 (4) | 0.3264 (4) | 0.4233 (3) | 0.0458 (14) | |
| C11 | 0.1193 (4) | 0.2023 (4) | 0.4002 (3) | 0.0394 (13) | |
| C12 | 0.0349 (5) | 0.1374 (4) | 0.3789 (3) | 0.0450 (14) | |
| C13 | 0.0612 (5) | 0.0548 (4) | 0.3409 (3) | 0.0487 (15) | |
| C14 | 0.1726 (5) | 0.0327 (4) | 0.3232 (3) | 0.0436 (14) | |
| C15 | 0.2576 (5) | 0.0944 (4) | 0.3439 (3) | 0.0495 (15) | |
| C16 | 0.2294 (4) | 0.1778 (4) | 0.3820 (3) | 0.0447 (14) | |
| C21 | 0.0967 (4) | 0.2817 (4) | 0.5350 (3) | 0.0396 (13) | |
| C22 | 0.0716 (5) | 0.3574 (4) | 0.5850 (3) | 0.0447 (14) | |
| C23 | 0.0820 (5) | 0.3528 (4) | 0.6658 (3) | 0.0434 (14) | |
| C24 | 0.1185 (4) | 0.2701 (4) | 0.7016 (3) | 0.0396 (13) | |
| C25 | 0.1439 (5) | 0.1944 (4) | 0.6539 (3) | 0.0470 (14) | |
| C26 | 0.1333 (4) | 0.1997 (4) | 0.5711 (3) | 0.0437 (14) | |
| C31 | 0.1761 (4) | 0.3720 (4) | 0.4204 (3) | 0.0378 (13) | |
| C32 | 0.1576 (5) | 0.4327 (4) | 0.3564 (3) | 0.0449 (14) | |
| C33 | 0.2388 (5) | 0.5006 (4) | 0.3337 (3) | 0.0461 (14) | |
| C34 | 0.3398 (4) | 0.5070 (4) | 0.3749 (3) | 0.0417 (13) | |
| C35 | 0.3607 (4) | 0.4452 (4) | 0.4376 (3) | 0.0443 (14) | |
| C36 | 0.2783 (4) | 0.3800 (4) | 0.4603 (3) | 0.0425 (14) | |
| O4 | 0.0484 (3) | −0.1821 (3) | 0.2977 (3) | 0.0622 (12) | |
| C3 | 0.0843 (6) | −0.2770 (4) | 0.3092 (4) | 0.0665 (19) | |
| N41 | 0.6098 (4) | 0.5851 (3) | 0.4268 (3) | 0.0473 (12) | |
| C42 | 0.6364 (5) | 0.6162 (5) | 0.4993 (3) | 0.0541 (16) | |
| C43 | 0.7478 (5) | 0.6248 (5) | 0.5260 (3) | 0.0551 (16) | |
| C44 | 0.8363 (4) | 0.5987 (4) | 0.4764 (3) | 0.0418 (13) | |
| C45 | 0.8089 (5) | 0.5678 (4) | 0.4012 (3) | 0.0492 (15) | |
| C46 | 0.6964 (5) | 0.5615 (4) | 0.3787 (3) | 0.0487 (14) | |
| C47 | 0.9559 (4) | 0.6054 (4) | 0.5015 (3) | 0.0439 (14) | |
| N51 | 1.3371 (4) | 0.6634 (4) | 0.6443 (3) | 0.0606 (14) | |
| C52 | 1.3093 (5) | 0.6232 (5) | 0.5749 (4) | 0.069 (2) | |
| C53 | 1.1983 (5) | 0.6113 (5) | 0.5487 (4) | 0.0608 (18) | |
| C54 | 1.1125 (5) | 0.6423 (4) | 0.5952 (3) | 0.0470 (15) | |
| C55 | 1.1392 (5) | 0.6828 (5) | 0.6674 (4) | 0.0625 (18) | |
| C56 | 1.2518 (6) | 0.6925 (5) | 0.6894 (4) | 0.0626 (18) | |
| C57 | 0.9919 (5) | 0.6339 (4) | 0.5706 (3) | 0.0489 (15) | |
| H1 | 0.1477 | −0.0734 | 0.2630 | 0.084* | |
| H2 | 0.1109 | 0.3105 | 0.8045 | 0.075* | |
| H3 | 0.4719 | 0.5718 | 0.3818 | 0.083* | |
| H2A | −0.0856 | 0.2851 | 0.4497 | 0.069* | |
| H2B | −0.0420 | 0.3233 | 0.3656 | 0.069* | |
| H2C | −0.0423 | 0.3913 | 0.4415 | 0.069* | |
| H12 | −0.0418 | 0.1509 | 0.3910 | 0.054* | |
| H13 | 0.0025 | 0.0124 | 0.3265 | 0.058* | |
| H15 | 0.3343 | 0.0801 | 0.3322 | 0.059* | |
| H16 | 0.2884 | 0.2199 | 0.3962 | 0.054* | |
| H22 | 0.0463 | 0.4145 | 0.5618 | 0.054* | |
| H23 | 0.0643 | 0.4061 | 0.6975 | 0.052* | |
| H25 | 0.1690 | 0.1374 | 0.6776 | 0.056* | |
| H26 | 0.1515 | 0.1465 | 0.5394 | 0.052* | |
| H32 | 0.0889 | 0.4282 | 0.3274 | 0.054* | |
| H33 | 0.2240 | 0.5417 | 0.2903 | 0.055* | |
| H35 | 0.4309 | 0.4474 | 0.4650 | 0.053* | |
| H36 | 0.2927 | 0.3400 | 0.5045 | 0.051* | |
| H4 | −0.0118 | −0.1728 | 0.3229 | 0.093* | |
| H3A | 0.1501 | −0.2898 | 0.2754 | 0.100* | |
| H3B | 0.0226 | −0.3198 | 0.2949 | 0.100* | |
| H3C | 0.1048 | −0.2864 | 0.3651 | 0.100* | |
| H42 | 0.5769 | 0.6333 | 0.5343 | 0.065* | |
| H43 | 0.7629 | 0.6484 | 0.5777 | 0.066* | |
| H45 | 0.8669 | 0.5509 | 0.3649 | 0.059* | |
| H46 | 0.6795 | 0.5393 | 0.3268 | 0.058* | |
| H47 | 1.0116 | 0.5873 | 0.4638 | 0.053* | |
| H52 | 1.3687 | 0.6016 | 0.5416 | 0.083* | |
| H53 | 1.1831 | 0.5820 | 0.4991 | 0.073* | |
| H55 | 1.0808 | 0.7040 | 0.7018 | 0.075* | |
| H56 | 1.2686 | 0.7212 | 0.7391 | 0.075* | |
| H57 | 0.9362 | 0.6508 | 0.6088 | 0.059* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.065 (3) | 0.047 (3) | 0.056 (2) | −0.002 (2) | 0.013 (2) | −0.012 (2) |
| O2 | 0.064 (3) | 0.048 (3) | 0.038 (2) | −0.003 (2) | −0.0025 (19) | 0.0007 (18) |
| O3 | 0.047 (2) | 0.062 (3) | 0.056 (2) | −0.019 (2) | −0.0120 (19) | 0.018 (2) |
| C1 | 0.040 (3) | 0.042 (3) | 0.038 (3) | −0.003 (3) | −0.001 (2) | 0.004 (2) |
| C2 | 0.045 (3) | 0.048 (4) | 0.045 (3) | −0.002 (3) | −0.003 (3) | 0.003 (3) |
| C11 | 0.040 (3) | 0.041 (3) | 0.037 (3) | −0.005 (3) | 0.002 (2) | 0.001 (2) |
| C12 | 0.038 (3) | 0.052 (4) | 0.045 (3) | 0.001 (3) | 0.003 (3) | −0.002 (3) |
| C13 | 0.048 (4) | 0.046 (4) | 0.052 (3) | −0.011 (3) | 0.001 (3) | −0.002 (3) |
| C14 | 0.053 (4) | 0.039 (3) | 0.039 (3) | 0.001 (3) | 0.014 (3) | −0.005 (3) |
| C15 | 0.044 (3) | 0.052 (4) | 0.053 (3) | −0.004 (3) | 0.007 (3) | −0.008 (3) |
| C16 | 0.038 (3) | 0.048 (4) | 0.048 (3) | −0.004 (3) | 0.002 (3) | −0.001 (3) |
| C21 | 0.041 (3) | 0.041 (3) | 0.036 (3) | −0.001 (3) | 0.000 (2) | 0.002 (3) |
| C22 | 0.054 (3) | 0.038 (3) | 0.042 (3) | 0.004 (3) | 0.003 (3) | 0.004 (3) |
| C23 | 0.053 (3) | 0.037 (3) | 0.040 (3) | 0.003 (3) | 0.005 (3) | −0.006 (3) |
| C24 | 0.037 (3) | 0.047 (3) | 0.035 (3) | −0.002 (3) | 0.001 (2) | 0.001 (3) |
| C25 | 0.051 (3) | 0.044 (3) | 0.045 (3) | 0.002 (3) | −0.001 (3) | 0.008 (3) |
| C26 | 0.046 (3) | 0.039 (3) | 0.046 (3) | 0.000 (3) | 0.001 (3) | 0.001 (3) |
| C31 | 0.037 (3) | 0.041 (3) | 0.035 (3) | −0.002 (3) | −0.002 (2) | 0.001 (3) |
| C32 | 0.043 (3) | 0.047 (3) | 0.044 (3) | 0.001 (3) | −0.007 (3) | 0.004 (3) |
| C33 | 0.047 (3) | 0.049 (4) | 0.042 (3) | −0.001 (3) | −0.009 (3) | 0.011 (3) |
| C34 | 0.037 (3) | 0.047 (3) | 0.041 (3) | −0.006 (3) | 0.002 (3) | 0.005 (3) |
| C35 | 0.037 (3) | 0.055 (4) | 0.041 (3) | 0.001 (3) | −0.006 (2) | 0.006 (3) |
| C36 | 0.043 (3) | 0.045 (3) | 0.039 (3) | 0.002 (3) | 0.001 (2) | 0.011 (3) |
| O4 | 0.049 (3) | 0.051 (3) | 0.086 (3) | −0.007 (2) | 0.018 (2) | −0.005 (2) |
| C3 | 0.057 (4) | 0.047 (4) | 0.096 (5) | 0.000 (3) | 0.014 (4) | −0.001 (4) |
| N41 | 0.043 (3) | 0.049 (3) | 0.050 (3) | −0.007 (2) | −0.002 (2) | 0.004 (2) |
| C42 | 0.041 (3) | 0.069 (4) | 0.053 (4) | 0.004 (3) | 0.003 (3) | −0.003 (3) |
| C43 | 0.050 (4) | 0.070 (4) | 0.045 (3) | −0.003 (3) | −0.001 (3) | −0.005 (3) |
| C44 | 0.045 (3) | 0.042 (3) | 0.038 (3) | 0.000 (3) | 0.003 (3) | 0.006 (3) |
| C45 | 0.045 (3) | 0.058 (4) | 0.045 (3) | 0.004 (3) | 0.003 (3) | 0.002 (3) |
| C46 | 0.055 (4) | 0.041 (3) | 0.050 (3) | −0.004 (3) | −0.006 (3) | 0.003 (3) |
| C47 | 0.038 (3) | 0.048 (4) | 0.046 (3) | −0.002 (3) | 0.004 (3) | −0.004 (3) |
| N51 | 0.051 (3) | 0.063 (3) | 0.068 (3) | 0.006 (3) | −0.012 (3) | −0.011 (3) |
| C52 | 0.040 (4) | 0.094 (5) | 0.074 (5) | −0.002 (4) | −0.010 (3) | −0.033 (4) |
| C53 | 0.033 (3) | 0.090 (5) | 0.059 (4) | −0.001 (3) | 0.004 (3) | −0.026 (4) |
| C54 | 0.045 (3) | 0.045 (3) | 0.051 (3) | 0.002 (3) | −0.010 (3) | 0.000 (3) |
| C55 | 0.061 (4) | 0.068 (4) | 0.059 (4) | 0.015 (4) | −0.012 (3) | −0.014 (3) |
| C56 | 0.055 (4) | 0.068 (4) | 0.065 (4) | 0.011 (4) | −0.019 (3) | −0.012 (4) |
| C57 | 0.043 (3) | 0.052 (4) | 0.052 (3) | 0.007 (3) | 0.000 (3) | −0.002 (3) |
Geometric parameters (Å, º) top
| O1—C14 | 1.381 (6) | C33—C34 | 1.385 (8) |
| O1—H1 | 0.84 | C33—H33 | 0.95 |
| O2—C24 | 1.364 (6) | C34—C35 | 1.394 (7) |
| O2—H2 | 0.84 | C35—C36 | 1.397 (7) |
| O3—C34 | 1.355 (6) | C35—H35 | 0.95 |
| O3—H3 | 0.84 | C36—H36 | 0.95 |
| C1—C2 | 1.535 (7) | O4—C3 | 1.427 (7) |
| C1—C21 | 1.545 (7) | O4—H4 | 0.84 |
| C1—C11 | 1.545 (8) | C3—H3A | 0.98 |
| C1—C31 | 1.554 (7) | C3—H3B | 0.98 |
| C2—H2A | 0.98 | C3—H3C | 0.98 |
| C2—H2B | 0.98 | N41—C42 | 1.333 (7) |
| C2—H2C | 0.98 | N41—C46 | 1.347 (7) |
| C11—C16 | 1.382 (7) | C42—C43 | 1.397 (8) |
| C11—C12 | 1.406 (7) | C42—H42 | 0.95 |
| C12—C13 | 1.371 (8) | C43—C44 | 1.388 (7) |
| C12—H12 | 0.95 | C43—H43 | 0.95 |
| C13—C14 | 1.387 (8) | C44—C45 | 1.377 (7) |
| C13—H13 | 0.95 | C44—C47 | 1.480 (7) |
| C14—C15 | 1.379 (8) | C45—C46 | 1.386 (7) |
| C15—C16 | 1.388 (7) | C45—H45 | 0.95 |
| C15—H15 | 0.95 | C46—H46 | 0.95 |
| C16—H16 | 0.95 | C47—C57 | 1.301 (7) |
| C21—C26 | 1.384 (7) | C47—H47 | 0.95 |
| C21—C22 | 1.397 (7) | N51—C56 | 1.328 (7) |
| C22—C23 | 1.365 (7) | N51—C52 | 1.339 (7) |
| C22—H22 | 0.95 | C52—C53 | 1.395 (8) |
| C23—C24 | 1.390 (7) | C52—H52 | 0.95 |
| C23—H23 | 0.95 | C53—C54 | 1.354 (7) |
| C24—C25 | 1.376 (7) | C53—H53 | 0.95 |
| C25—C26 | 1.398 (7) | C54—C55 | 1.379 (8) |
| C25—H25 | 0.95 | C54—C57 | 1.490 (7) |
| C26—H26 | 0.95 | C55—C56 | 1.390 (8) |
| C31—C36 | 1.387 (7) | C55—H55 | 0.95 |
| C31—C32 | 1.396 (7) | C56—H56 | 0.95 |
| C32—C33 | 1.414 (8) | C57—H57 | 0.95 |
| C32—H32 | 0.95 | | |
| | | |
| C14—O1—H1 | 109.5 | C34—C33—C32 | 119.7 (5) |
| C24—O2—H2 | 109.5 | C34—C33—H33 | 120.2 |
| C34—O3—H3 | 109.5 | C32—C33—H33 | 120.2 |
| C2—C1—C21 | 108.0 (4) | O3—C34—C33 | 118.0 (5) |
| C2—C1—C11 | 110.8 (4) | O3—C34—C35 | 122.7 (5) |
| C21—C1—C11 | 111.0 (4) | C33—C34—C35 | 119.3 (5) |
| C2—C1—C31 | 110.3 (4) | C34—C35—C36 | 120.1 (5) |
| C21—C1—C31 | 107.5 (4) | C34—C35—H35 | 120.0 |
| C11—C1—C31 | 109.3 (4) | C36—C35—H35 | 120.0 |
| C1—C2—H2A | 109.5 | C31—C36—C35 | 122.0 (5) |
| C1—C2—H2B | 109.5 | C31—C36—H36 | 119.0 |
| H2A—C2—H2B | 109.5 | C35—C36—H36 | 119.0 |
| C1—C2—H2C | 109.5 | C3—O4—H4 | 109.5 |
| H2A—C2—H2C | 109.5 | O4—C3—H3A | 109.5 |
| H2B—C2—H2C | 109.5 | O4—C3—H3B | 109.5 |
| C16—C11—C12 | 116.5 (5) | H3A—C3—H3B | 109.5 |
| C16—C11—C1 | 122.3 (5) | O4—C3—H3C | 109.5 |
| C12—C11—C1 | 121.2 (5) | H3A—C3—H3C | 109.5 |
| C13—C12—C11 | 121.3 (5) | H3B—C3—H3C | 109.5 |
| C13—C12—H12 | 119.3 | C42—N41—C46 | 116.8 (5) |
| C11—C12—H12 | 119.3 | N41—C42—C43 | 123.0 (5) |
| C12—C13—C14 | 120.6 (5) | N41—C42—H42 | 118.5 |
| C12—C13—H13 | 119.7 | C43—C42—H42 | 118.5 |
| C14—C13—H13 | 119.7 | C44—C43—C42 | 119.6 (5) |
| C15—C14—O1 | 117.0 (5) | C44—C43—H43 | 120.2 |
| C15—C14—C13 | 119.6 (5) | C42—C43—H43 | 120.2 |
| O1—C14—C13 | 123.4 (5) | C45—C44—C43 | 117.3 (5) |
| C14—C15—C16 | 119.0 (5) | C45—C44—C47 | 120.5 (5) |
| C14—C15—H15 | 120.5 | C43—C44—C47 | 122.2 (5) |
| C16—C15—H15 | 120.5 | C44—C45—C46 | 119.8 (5) |
| C11—C16—C15 | 123.0 (5) | C44—C45—H45 | 120.1 |
| C11—C16—H16 | 118.5 | C46—C45—H45 | 120.1 |
| C15—C16—H16 | 118.5 | N41—C46—C45 | 123.3 (5) |
| C26—C21—C22 | 116.8 (5) | N41—C46—H46 | 118.3 |
| C26—C21—C1 | 123.6 (5) | C45—C46—H46 | 118.3 |
| C22—C21—C1 | 119.5 (5) | C57—C47—C44 | 125.9 (5) |
| C23—C22—C21 | 122.9 (5) | C57—C47—H47 | 117.0 |
| C23—C22—H22 | 118.6 | C44—C47—H47 | 117.0 |
| C21—C22—H22 | 118.6 | C56—N51—C52 | 116.4 (5) |
| C22—C23—C24 | 120.0 (5) | N51—C52—C53 | 123.8 (6) |
| C22—C23—H23 | 120.0 | N51—C52—H52 | 118.1 |
| C24—C23—H23 | 120.0 | C53—C52—H52 | 118.1 |
| O2—C24—C25 | 118.4 (5) | C54—C53—C52 | 118.9 (6) |
| O2—C24—C23 | 123.1 (5) | C54—C53—H53 | 120.5 |
| C25—C24—C23 | 118.5 (4) | C52—C53—H53 | 120.5 |
| C24—C25—C26 | 121.3 (5) | C53—C54—C55 | 118.1 (5) |
| C24—C25—H25 | 119.4 | C53—C54—C57 | 122.1 (5) |
| C26—C25—H25 | 119.4 | C55—C54—C57 | 119.8 (5) |
| C21—C26—C25 | 120.6 (5) | C54—C55—C56 | 119.7 (6) |
| C21—C26—H26 | 119.7 | C54—C55—H55 | 120.2 |
| C25—C26—H26 | 119.7 | C56—C55—H55 | 120.2 |
| C36—C31—C32 | 117.3 (5) | N51—C56—C55 | 123.0 (6) |
| C36—C31—C1 | 120.6 (4) | N51—C56—H56 | 118.5 |
| C32—C31—C1 | 122.0 (4) | C55—C56—H56 | 118.5 |
| C31—C32—C33 | 121.6 (5) | C47—C57—C54 | 125.8 (5) |
| C31—C32—H32 | 119.2 | C47—C57—H57 | 117.1 |
| C33—C32—H32 | 119.2 | C54—C57—H57 | 117.1 |
| | | |
| C2—C1—C11—C16 | 153.9 (5) | C2—C1—C31—C32 | −30.6 (7) |
| C21—C1—C11—C16 | −86.1 (6) | C21—C1—C31—C32 | −148.1 (5) |
| C31—C1—C11—C16 | 32.2 (6) | C11—C1—C31—C32 | 91.4 (6) |
| C2—C1—C11—C12 | −28.0 (6) | C36—C31—C32—C33 | −0.9 (8) |
| C21—C1—C11—C12 | 92.0 (6) | C1—C31—C32—C33 | −177.7 (5) |
| C31—C1—C11—C12 | −149.6 (5) | C31—C32—C33—C34 | 0.8 (8) |
| C16—C11—C12—C13 | −1.0 (8) | C32—C33—C34—O3 | −178.5 (5) |
| C1—C11—C12—C13 | −179.2 (5) | C32—C33—C34—C35 | 0.9 (8) |
| C11—C12—C13—C14 | 0.7 (8) | O3—C34—C35—C36 | 177.0 (5) |
| C12—C13—C14—C15 | −0.2 (9) | C33—C34—C35—C36 | −2.4 (8) |
| C12—C13—C14—O1 | −179.7 (5) | C32—C31—C36—C35 | −0.6 (8) |
| O1—C14—C15—C16 | 179.5 (5) | C1—C31—C36—C35 | 176.2 (5) |
| C13—C14—C15—C16 | −0.1 (8) | C34—C35—C36—C31 | 2.3 (8) |
| C12—C11—C16—C15 | 0.7 (8) | C46—N41—C42—C43 | 0.1 (9) |
| C1—C11—C16—C15 | 179.0 (5) | N41—C42—C43—C44 | −1.2 (10) |
| C14—C15—C16—C11 | −0.2 (8) | C42—C43—C44—C45 | 2.0 (9) |
| C2—C1—C21—C26 | 126.1 (5) | C42—C43—C44—C47 | −179.4 (6) |
| C11—C1—C21—C26 | 4.5 (7) | C43—C44—C45—C46 | −1.7 (9) |
| C31—C1—C21—C26 | −114.9 (6) | C47—C44—C45—C46 | 179.7 (5) |
| C2—C1—C21—C22 | −57.8 (6) | C42—N41—C46—C45 | 0.2 (9) |
| C11—C1—C21—C22 | −179.4 (5) | C44—C45—C46—N41 | 0.7 (9) |
| C31—C1—C21—C22 | 61.1 (6) | C45—C44—C47—C57 | 179.7 (6) |
| C26—C21—C22—C23 | 0.0 (8) | C43—C44—C47—C57 | 1.2 (9) |
| C1—C21—C22—C23 | −176.3 (5) | C56—N51—C52—C53 | 0.2 (11) |
| C21—C22—C23—C24 | −0.3 (9) | N51—C52—C53—C54 | 0.5 (12) |
| C22—C23—C24—O2 | 179.8 (5) | C52—C53—C54—C55 | −1.2 (10) |
| C22—C23—C24—C25 | 0.3 (8) | C52—C53—C54—C57 | 178.7 (7) |
| O2—C24—C25—C26 | −179.6 (5) | C53—C54—C55—C56 | 1.3 (10) |
| C23—C24—C25—C26 | −0.1 (8) | C57—C54—C55—C56 | −178.6 (6) |
| C22—C21—C26—C25 | 0.2 (8) | C52—N51—C56—C55 | −0.1 (10) |
| C1—C21—C26—C25 | 176.4 (5) | C54—C55—C56—N51 | −0.7 (10) |
| C24—C25—C26—C21 | −0.1 (9) | C44—C47—C57—C54 | −178.7 (6) |
| C2—C1—C31—C36 | 152.7 (5) | C53—C54—C57—C47 | −6.8 (10) |
| C21—C1—C31—C36 | 35.2 (7) | C55—C54—C57—C47 | 173.2 (6) |
| C11—C1—C31—C36 | −85.3 (6) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O4 | 0.84 | 2.03 | 2.652 (6) | 131 |
| O2—H2···O3i | 0.84 | 1.85 | 2.684 (6) | 172 |
| O3—H3···N41 | 0.84 | 1.81 | 2.631 (6) | 166 |
| O4—H4···N51ii | 0.84 | 1.87 | 2.696 (6) | 166 |
| Symmetry codes: (i) −x+1/2, −y+1, z+1/2; (ii) x−3/2, −y+1/2, −z+1. |
Experimental details
| | (I) | (II) |
| Crystal data |
| Chemical formula | C12H12N2·2C20H18O3 | C12H10N2·C20H18O3·CH4O |
| Mr | 796.92 | 520.61 |
| Crystal system, space group | Triclinic, P1 | Orthorhombic, P212121 |
| Temperature (K) | 150 | 150 |
| a, b, c (Å) | 10.2076 (2), 10.6559 (2), 10.9341 (2) | 11.8264 (9), 14.2158 (9), 16.8109 (12) |
| α, β, γ (°) | 100.9920 (16), 109.4530 (14), 101.6700 (11) | 90, 90, 90 |
| V (Å3) | 1054.49 (3) | 2826.3 (3) |
| Z | 1 | 4 |
| Radiation type | Mo Kα | Mo Kα |
| µ (mm−1) | 0.08 | 0.08 |
| Crystal size (mm) | 0.30 × 0.20 × 0.18 | 0.30 × 0.26 × 0.16 |
| |
| Data collection |
| Diffractometer | Kappa-CCD
diffractometer | KappaCCD
diffractometer |
| Absorption correction | Multi-scan
(DENZO-SMN; Otwinowski & Minor, 1997) | Multi-scan
(DENZO-SMN; Otwinowski & Minor, 1997) |
| Tmin, Tmax | 0.976, 0.985 | 0.976, 0.987 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 13679, 4797, 3886 | 11922, 2795, 1859 |
| Rint | 0.031 | 0.082 |
| (sin θ/λ)max (Å−1) | 0.650 | 0.595 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.044, 0.123, 1.09 | 0.057, 0.149, 1.08 |
| No. of reflections | 4797 | 2795 |
| No. of parameters | 276 | 359 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.22, −0.28 | 0.22, −0.19 |
Selected torsion angles (º) for (I) top| C2—C1—C11—C12 | −42.87 (17) | C2—C1—C31—C32 | −35.04 (15) |
| C2—C1—C21—C22 | −52.93 (14) | C43—C44—C47—C47i | −104.0 (3) |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) for (I) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N41 | 0.84 | 1.81 | 2.6145 (18) | 159 |
| O2—H2···O1ii | 0.84 | 1.83 | 2.6671 (13) | 175 |
| O3—H3···O2iii | 0.84 | 1.98 | 2.8039 (17) | 168 |
| Symmetry codes: (ii) x, y, z−1; (iii) x, y−1, z. |
Selected torsion angles (º) for (II) top| C2—C1—C11—C12 | −28.0 (6) | C43—C44—C47—C57 | 1.2 (9) |
| C2—C1—C21—C22 | −57.8 (6) | C53—C54—C57—C47 | −6.8 (10) |
| C2—C1—C31—C32 | −30.6 (7) | | |
Hydrogen-bond geometry (Å, º) for (II) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O4 | 0.84 | 2.03 | 2.652 (6) | 131 |
| O2—H2···O3i | 0.84 | 1.85 | 2.684 (6) | 172 |
| O3—H3···N41 | 0.84 | 1.81 | 2.631 (6) | 166 |
| O4—H4···N51ii | 0.84 | 1.87 | 2.696 (6) | 166 |
| Symmetry codes: (i) −x+1/2, −y+1, z+1/2; (ii) x−3/2, −y+1/2, −z+1. |
 O hydrogen bonds to form sheets built from R
O hydrogen bonds to form sheets built from R N hydrogen bonds to form open bilayers. Each bilayer is interwoven with two adjacent bilayers, forming a continuous three-dimensional structure. In the adduct 1,2-bis(4-pyridyl)ethene-1,1,1-tris(4-hydroxyphenyl)ethane-methanol (1/1/1), C12H10N2·C20H18O3·CH4O, the molecules are linked by O-H
N hydrogen bonds to form open bilayers. Each bilayer is interwoven with two adjacent bilayers, forming a continuous three-dimensional structure. In the adduct 1,2-bis(4-pyridyl)ethene-1,1,1-tris(4-hydroxyphenyl)ethane-methanol (1/1/1), C12H10N2·C20H18O3·CH4O, the molecules are linked by O-H O and O-H
O and O-H N hydrogen bonds into three interwoven three-dimensional frameworks, generated by single spiral chains along [010] and [001] and a triple-helical spiral along [100].
N hydrogen bonds into three interwoven three-dimensional frameworks, generated by single spiral chains along [010] and [001] and a triple-helical spiral along [100].
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2002/01/00/sk1516/sk1516fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2002/01/00/sk1516/sk1516fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2002/01/00/sk1516/sk1516fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/c/issues/2002/01/00/sk1516/sk1516fig4thm.gif)
![[Figure 5]](http://journals.iucr.org/c/issues/2002/01/00/sk1516/sk1516fig5thm.gif)
![[Figure 6]](http://journals.iucr.org/c/issues/2002/01/00/sk1516/sk1516fig6thm.gif)
![[Figure 7]](http://journals.iucr.org/c/issues/2002/01/00/sk1516/sk1516fig7thm.gif)




When cocrystallized with 4,4'-bipyridyl the threefold symmetric tris-phenol 1,1,1-tris(4-hydroxyphenyl)ethane [systematic name: 4,4',4''-(ethane-1,1,1-triyl)triphenol], CH3C(C6H4OH)3, forms a hydrogen-bonded adduct of 3:2 stoichiometry, 3C10H8N2·2C20H18O3, in which all the hydrogen bonds are of the O—H···N type and whose supramolecular structure consists of tenfold interwoven nets built from R1212(126) rings (Bényei et al., 1998). In a similar way, 4,4'-bipyridyl forms 3:2 adducts both with 1,3,5-benzenetricarboxylic acid (Sharma & Zaworotko, 1996) and with 2,3,5,6-tetrahydroxybenzoquinone (Cowan et al., 2001); in the 1,3,5-benzenetricarboxylic acid adduct, the components are linked into threefold interwoven nets built from R1212(102) rings, while the 2,3,5,6-tetrahydroxybenzoquinone adduct has a three-dimensional supramolecular structure consisting of three interpenetrating frameworks. On the basis of the two-dimensional examples above, there seems to be no obvious impediment to the formation of nets containing yet larger rings and, accordingly, we have now investigated the cocrystallization of 1,1,1-tris(4-hydroxyphenyl)ethane with the extended bipyridyl species 1,2-bis(4-pyridyl)ethane and 1,2-bis(4-pyridyl)ethene in the hope, and expectation, of generating nets built from R1212(138) rings. In the event, no 3:2 adducts could be found; instead, 1,2-bis(4-pyridyl)ethane yields a 1:2 adduct, C12H12N2·2C20H18O3, (I), while 1,2-bis(4-pyridyl)ethene yields a methanol-solvated 1:1 adduct, C12H10N2·C20H18O3·CH4O, (II).
In compound (I), the bis(4-pyridyl)ethane molecule lies across a centre of inversion in space group P1, chosen for the sake of convenience as that at (1/2, 1/2, 1/2), and the tris-phenol component lies in a general position (Fig. 1). The supramolecular structure consists of continuously interwoven bilayers, giving a three-dimensional structure overall; the constitution of the bilayers, and thence of the overall structure, is most simply analysed in terms of the substructure built from just the tris-phenol component.
Atom O2 in the tris-phenol at (x, y, z) acts as a hydrogen-bond donor to O1 in the molecule at (x, y, -1 + z), so generating by translation a C(12) chain running parallel to the [001] direction. Atom O3 at (x, y, z) similarly acts as donor to O2 at (x, -1 + y, z), producing another C(12) chain, this time by translation along the [010] direction. The combination of these two simple chain motifs generates a sheet parallel to (100) built from a single type of R44(38) ring (Fig. 2). This reference sheet lies in the domain 0.96 < x < 1.56, and there is a symmetry-related sheet, generated by the inversion centres at (1/2, y, z), which lies in the domain -0.56 < x < 0.04. These two sheets are linked into a hydrogen-bonded bilayer by the amines; atoms of type O1 at (x, y, z) and (1 - x, 1 - y, 1 - z) in the domains 0.96 < x < 1.56 and -0.56 < x < 0.04, respectively, act as hydrogen-bond donors to the N1 atoms at (x, y, z) and (1 - x, 1 - y, 1 - z), respectively, which both lie in the reference amine centred at (1/2, 1/2, 1/2). Hence, these two (100) sheets are multiply linked by the series of amines centred at (1/2, m+0.5, n+0.5), where m, n = zero or integer.
Each bilayer is continuously interwoven with the two adjacent bilayers, as the bis(4-pyridyl)ethane linkers in one bilayer pass through the R44(38) rings in the adjacent sheet. In the (100) sheet containing the tris-phenol at (x, y, z), the centroids of the rings are at approximately (1.30, 0.70, 0.45) and similarly by translation along [010] and [001], and there are bis(4-pyridyl)ethane units centred close to these points, at (1.5, m+0.5, n+0.5), where m, n = zero or integer. In the other component of the reference bilayer, there is a ring centroid at approximately (-0.30, 0.30, 0.55), close to the centre of the bis(4-pyridyl)ethane molecule at (-0.5, 1/2, 1/2), and so on. Thus, the bilayer centred at x = 0.5 is interwoven with the two bilayers centred at x = 1.5 and x = -0.5; with each bilayer interwoven with its two neighbouring bilayers, the overall supramolecular structure is three-dimensional (Fig. 3).
The (100) sheets of tris-phenol molecules are generated by translation and hence are not puckered; nor are they interwoven. It is of interest to compare these sheets, built from a single tris-phenol molecule, with the analogous sheets of R44(38) rings in the 1:2 adduct C2H8N2·2C20H18O3, (III), formed with 1,2-diaminoethane (Ferguson et al., 1998). Here the sheets are built from two independent tris-phenol molecules and they are so deeply interwoven as to be pairwise interwoven; nonetheless the individual ring structures containing seven O atoms, but only four O—H···O hydrogen bonds, are identical in compounds (I) and (III).
The interweaving of the two-dimensional hydrogen-bonded bilayers in (I) to form an overall three-dimensional structure may be compared with the interweaving of hydrogen-bonded molecular ladders to form sheets in the 2:1 adduct of 4,4'-sulfonyldiphenol and pyrazine, (IV) (Ferguson et al., 1999), where the ladders have R66(50) rings between the rungs and each ladder is interwoven with two adjacent ladders. We have previously noted (Ferguson et al., 1999) that for systems of this type based on substituted aryl components and containing interwoven substructures, ring types such as R44(32) and R44(38) can permit just one strand of an interwoven substructure to pass through each ring, while larger types such as R66(48) can accommodate up to three strands; the number apparently depends upon the extent to which the rings are puckered. Consistent with this, in compounds (I) and (III), the R44(38) rings can accommodate either part of another tris-phenol molecule, as in (III), or part of a bipyridyl, as in (I), but these choices are mutually exclusive. The larger R66(50) rings in the interwoven ladders of (IV) can readily accommodate two strands from adjacent ladders.
In the methanol solvate (II), atoms O1 and O3 in the tris-phenol component act as hydrogen-bond donors to the methanol O4 and bipyridyl N41 atoms, respectively, within the asymmetric unit (Fig. 4). The two remaining hydrogen bonds (Table 4) serve to link the molecules in three dimensions; the hydrogen bonds having O2 and O4 as donors independently generate spiral chains along [001] and [100], respectively, while their combined action generates a spiral chain long [010].
The tris-phenol O2 atom at (x, y, z) acts as hydrogen-bond donor to O3 at (0.5 - x, 1 - y, 0.5 + z), while O2 at (0.5 - x, 1 - y, 0.5 + z) acts as donor to O3 at (x, y, 1 + z), so producing a C(12) chain running parallel to [001] and generated by the 21 screw axis along (1/4, 1/2, z) (Fig. 5). The methanol O4 atom at (x, y, z) acts as donor to N51 at (-1.5 + x, 0.5 - y, 1 - z), while O4 at (-1.5 + x, 0.5 - y, 1 - z),in turn, acts as donor to N51 at (-3 + x, y, z). The resulting C33(25) chain parallel to [100] (Fig. 6) is generated by the 21 axis along (x, 1/4, 1/2) and it has a repeat vector along [100] of three unit-cell lengths; there are thus three such spiral chains, related by translation, forming a triple helix along [100]. Each of these chains along [001] and [100] involves just one hydrogen bond outside the asymmetric unit; the combination of these two spiral chains generates a third spiral, along [010], involving both of the hydrogen bonds outside the asymmetric unit,
Just as O4 at (x, y, z) acts as donor to N51 at (-1.5 + x, 0.5 - y, 1 - z) to generate the [100] chain, so O4 at (0.5 - x, 1 - y, 0.5 + z) in the [001] chain acts as donor to N51 at (2 - x, 0.5 + y, 1.5 - z); O2 at (2 - x, 0.5 + y, 1.5 - z), in turn, acts as donor to O3 at (1.5 + x, 1.5 - y, 1 - z) and O4 at (1.5 + x, 1.5 - y, 1 - z) acts as donor to N51 at (x, 1 + y, z). In this way, a C44(35) chain is generated parallel to [010] (Fig. 7), which links the [100] and [001] spirals, so forming a three-dimensional framework. Since there are three unconnected chains in the triple helix along [100], there are three interwoven frameworks in the structure.
In both (I) and (II), the tris-phenol component shows significant deviations from the idealized C3 molecular symmetry (Tables 1 and 2). While the bipyridyl component in (II) is almost flat, in (I), the bipyridyl skeleton is very far from being planar. The bond lengths and angles present no unusual features.