
The title compound, 3,5,7-triaza-1-azoniatricyclo[3.3.1.1
3,7]decane 2,4-dinitrophenolate monohydrate, C
6H
13N
4+·C
6H
3N
2O
5-·H
2O, the 1:1 hydrate adduct of hexamethylenetetramine (HMT) and 2,4-dinitrophenol, undergoes a temperature phase transition. In the room-temperature phase, the adduct crystallizes in the monoclinic
P2
1/
m space group, whereas in the low-temperature phase, the adduct crystallizes in the triclinic
P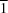
space group. This phase transition is reversible, with the transition temperature at 273 K, and the phase transition is governed by hydrogen bonds and weak interactions. In both these temperature-dependent polymorphs, the crystal structure is alternately layered with sheets of hexamethylenetetramine and sheets of dinitrophenol stacked along the
c axis. The hexamethylenetetramine and dinitrophenol moieties are linked by intermolecular hydrogen bonds. The water molecule in the adduct plays an important role, forming O-H

O hydrogen bonds which, together with C-H

O hydrogen bonds, bridge the adducts into molecular ribbons. Extra hydrogen bonds and weak interactions exist for the low-temperature polymorph and these interconnect the molecular ribbons into a three-dimensional packing structure. Also in these two temperature-dependent polymorphs, dinitrophenol acts as a hydrogen-bond acceptor and HMT acts as a hydrogen-bond donor.
Supporting information
CCDC references: 179279; 179280
HMT (1.4 g, 10 mmol) and 2,4-dinitrophenol (1.8 g, 10 mmol) were mixed and
dissolved in acetone (30 ml) together with a few drops of water. The resulting
mixture was heated until a clear solution was obtained. The solution was then
filtered and the filtrate was left to evaporate slowly in air. Yellow single
crystals suitable for X-ray diffraction studies were obtained after a few
days.
For the two polymorphs, all the H atoms were located from the difference map and
refined isotropically [C—H = 0.87 (3)–1.03 (2) Å].
For both compounds, data collection: SMART (Siemens, 1996); cell refinement: SAINT (Siemens, 1996); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 1997); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL, PARST (Nardelli, 1995) and PLATON (Spek, 1990).
(Iat300K) 3,5,7-triaza-1-azoniatricyclo[3.3.1.1
3,7]decane 2,4-dinitrophenolate
monohydrate
top
Crystal data top
| C6H13N4+·C6H3N2O5−·H2O | F(000) = 360 |
| Mr = 342.32 | Dx = 1.524 Mg m−3 |
| Monoclinic, P21/m | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.8610 (5) Å | Cell parameters from 3560 reflections |
| b = 6.5980 (4) Å | θ = 1.4–29.6° |
| c = 14.4339 (9) Å | µ = 0.12 mm−1 |
| β = 94.915 (1)° | T = 300 K |
| V = 745.89 (8) Å3 | Block, yellow |
| Z = 2 | 0.42 × 0.24 × 0.20 mm |
Data collection top
Siemens SMART CCD area-detector
diffractometer | 1900 independent reflections |
| Radiation source: fine-focus sealed tube | 1288 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.069 |
| Detector resolution: 8.33 pixels mm-1 | θmax = 28.0°, θmin = 2.6° |
| ω scans | h = −10→9 |
Absorption correction: empirical (using intensity measurements)
(SADABS; Sheldrick, 1996) | k = −8→7 |
| Tmin = 0.950, Tmax = 0.976 | l = −19→18 |
| 5088 measured reflections | |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.059 | All H-atom parameters refined |
| wR(F2) = 0.144 | w = 1/[σ2(Fo2) + (0.0622P)2]
where P = (Fo2 + 2Fc2)/3 |
| S = 0.95 | (Δ/σ)max < 0.001 |
| 1900 reflections | Δρmax = 0.33 e Å−3 |
| 178 parameters | Δρmin = −0.31 e Å−3 |
| 0 restraints | Extinction correction: SHELXTL, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.145 (17) |
Crystal data top
| C6H13N4+·C6H3N2O5−·H2O | V = 745.89 (8) Å3 |
| Mr = 342.32 | Z = 2 |
| Monoclinic, P21/m | Mo Kα radiation |
| a = 7.8610 (5) Å | µ = 0.12 mm−1 |
| b = 6.5980 (4) Å | T = 300 K |
| c = 14.4339 (9) Å | 0.42 × 0.24 × 0.20 mm |
| β = 94.915 (1)° | |
Data collection top
Siemens SMART CCD area-detector
diffractometer | 1900 independent reflections |
Absorption correction: empirical (using intensity measurements)
(SADABS; Sheldrick, 1996) | 1288 reflections with I > 2σ(I) |
| Tmin = 0.950, Tmax = 0.976 | Rint = 0.069 |
| 5088 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.059 | 0 restraints |
| wR(F2) = 0.144 | All H-atom parameters refined |
| S = 0.95 | Δρmax = 0.33 e Å−3 |
| 1900 reflections | Δρmin = −0.31 e Å−3 |
| 178 parameters | |
Special details top
Experimental. The data collection covered over a hemisphere of reciprocal space by a
combination of three sets of exposures; each set had a different ϕ angle (0,
88 and 180°) for the crystal and each exposure of 30 s covered 0.3° in ω.
The crystal-to-detector distance was 4 cm and the detector swing angle was
-35°. Crystal decay was monitored by repeating fifty initial frames at the
end of data collection and analysing the intensity of duplicate reflections,
and was found to be negligible. |
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. # loop_ # _publ_manuscript_incl_extra_item # # '_geom_extra_tableA_col_1' #
'_geom_extra_tableA_col_2' # '_geom_extra_table_head_A' #
'_geom_table_footnote_A' # # _geom_extra_table_head_A #; # Average C—N bond
distances (Å) along with the puckering parameters (Å, °) # of the
six-membered C—N—C—N—C—N rings of the HMT #; # # loop_ #
_geom_extra_tableA_col_1 # _geom_extra_tableA_col_2 # 'Scheme 2 here' ? # ? ?
# 'T = 300K' 'T = 143K' # ? ? # 'Average C—N = 1.474' 'Average C—N =
1.480 (3)' # 'Q2 = 0.014 (2)' 'Q2 = 0.009 (3)' # 'Q3 = 0.592 (2)' 'Q3 =
0.596 (3)' # 'QT = 0.593 (2)' 'QT = 0.596 (3)' # 'θ = 0.4 (2)' 'θ = 0.0 (3)'
# ? ? # 'Average C—N = 1.474 (3)' 'Average C—N = 1.480 (3)' # 'Q2 =
0.014 (2)' 'Q2 = 0.017 (3)' # 'Q3 = 0.592 (2)' 'Q3 = 0.600 (3)' # 'QT =
0.593 (2)' 'QT = 0.600 (3)' # 'θ = 0.4 (2)' 'θ = 1.8 (3)' # ? ? # 'Average
C—N = 1.471 (3)' 'Average C—N = 1.479 (3)' # 'Q2 = 0.008 (2)' 'Q2 =
0.007 (2)' # 'Q3 = 0.598 (2)' 'Q3 = 0.597 (2)' # 'QT = 0.598 (2)' 'QT =
0.597 (3)' # 'θ = 1.6 (2)' 'θ = 2.1 (3)' # ? ? # 'Average C—N = 1.469 (3)'
'Average C—N = 1.473 (3)' # 'Q2 = 0.004 (2)' 'Q2 = 0.009 (3)' # 'Q3 =
0.584 (2)' 'Q3 = 0.578 (3)' # 'QT = 0.585 (2)' 'QT = 0.578 (3)' # 'θ =
1.1 (2)' 'θ = 0.0 (3)' # ? ? # # _geom_table_footnote_A #; # Note: All the
rings adopt a chair conformation. #; # |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on all data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N1 | 0.3354 (3) | 0.2500 | 0.24704 (13) | 0.0402 (5) | |
| N2 | 0.0558 (2) | 0.2500 | 0.53083 (13) | 0.0432 (6) | |
| N3 | 0.7478 (2) | 0.7500 | 0.20852 (12) | 0.0325 (5) | |
| H1N3 | 0.727 (4) | 0.7500 | 0.265 (2) | 0.047* | |
| N4 | 0.88545 (17) | 0.5660 (2) | 0.09159 (9) | 0.0337 (4) | |
| N5 | 0.6216 (2) | 0.7500 | 0.04920 (13) | 0.0351 (5) | |
| O1 | 0.4692 (3) | 0.2500 | 0.20695 (11) | 0.0526 (6) | |
| O2 | 0.1928 (3) | 0.2500 | 0.20416 (12) | 0.0621 (6) | |
| O3 | −0.0775 (2) | 0.2500 | 0.48100 (14) | 0.0754 (8) | |
| O4 | 0.0538 (2) | 0.2500 | 0.61478 (13) | 0.0811 (9) | |
| O5 | 0.3979 (2) | 0.2500 | 0.63206 (10) | 0.0465 (5) | |
| O1W | 0.2668 (3) | 0.7500 | 0.28182 (14) | 0.0585 (6) | |
| H1W1 | 0.379 (4) | 0.7500 | 0.313 (2) | 0.060 (9)* | |
| H2W1 | 0.208 (6) | 0.7500 | 0.318 (3) | 0.109 (18)* | |
| C1 | 0.3505 (3) | 0.2500 | 0.34722 (14) | 0.0315 (5) | |
| C2 | 0.2036 (3) | 0.2500 | 0.39233 (15) | 0.0341 (6) | |
| H2A | 0.100 (3) | 0.2500 | 0.3631 (18) | 0.040 (7)* | |
| C3 | 0.2154 (3) | 0.2500 | 0.48886 (14) | 0.0312 (5) | |
| C4 | 0.3758 (3) | 0.2500 | 0.54426 (14) | 0.0317 (5) | |
| C5 | 0.5219 (3) | 0.2500 | 0.49069 (16) | 0.0401 (6) | |
| H5A | 0.623 (4) | 0.2500 | 0.522 (2) | 0.049 (8)* | |
| C6 | 0.5104 (3) | 0.2500 | 0.39662 (15) | 0.0356 (6) | |
| H6A | 0.607 (3) | 0.2500 | 0.3653 (18) | 0.045 (7)* | |
| C7 | 0.5830 (3) | 0.7500 | 0.14533 (17) | 0.0393 (6) | |
| H7A | 0.516 (2) | 0.628 (3) | 0.1585 (13) | 0.045 (5)* | |
| C8 | 0.7247 (2) | 0.5697 (3) | 0.03182 (12) | 0.0390 (4) | |
| H8A | 0.655 (2) | 0.442 (3) | 0.0453 (13) | 0.047 (5)* | |
| C9 | 0.8499 (2) | 0.5642 (3) | 0.18767 (11) | 0.0353 (4) | |
| H9B | 0.957 (2) | 0.563 (3) | 0.2306 (13) | 0.045* | |
| C10 | 0.9820 (3) | 0.7500 | 0.07362 (16) | 0.0368 (6) | |
| H11A | 1.009 (3) | 0.7500 | 0.0117 (19) | 0.044* | |
| H8B | 0.754 (2) | 0.579 (3) | −0.0316 (15) | 0.048 (5)* | |
| H9A | 0.784 (3) | 0.453 (3) | 0.2013 (14) | 0.053 (6)* | |
| H11B | 1.088 (4) | 0.7500 | 0.1155 (19) | 0.045 (7)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N1 | 0.0491 (13) | 0.0472 (13) | 0.0248 (10) | 0.000 | 0.0058 (9) | 0.000 |
| N2 | 0.0238 (10) | 0.0765 (16) | 0.0296 (10) | 0.000 | 0.0044 (8) | 0.000 |
| N3 | 0.0361 (11) | 0.0453 (12) | 0.0165 (9) | 0.000 | 0.0049 (8) | 0.000 |
| N4 | 0.0412 (8) | 0.0327 (8) | 0.0269 (7) | 0.0056 (6) | 0.0007 (6) | −0.0029 (5) |
| N5 | 0.0344 (11) | 0.0420 (12) | 0.0276 (10) | 0.000 | −0.0053 (8) | 0.000 |
| O1 | 0.0588 (12) | 0.0720 (14) | 0.0295 (9) | 0.000 | 0.0190 (8) | 0.000 |
| O2 | 0.0564 (13) | 0.1013 (18) | 0.0269 (9) | 0.000 | −0.0061 (9) | 0.000 |
| O3 | 0.0228 (9) | 0.156 (2) | 0.0465 (12) | 0.000 | −0.0018 (8) | 0.000 |
| O4 | 0.0330 (11) | 0.182 (3) | 0.0295 (10) | 0.000 | 0.0110 (8) | 0.000 |
| O5 | 0.0292 (9) | 0.0894 (15) | 0.0210 (8) | 0.000 | 0.0022 (6) | 0.000 |
| O1W | 0.0396 (12) | 0.0976 (18) | 0.0368 (11) | 0.000 | −0.0054 (9) | 0.000 |
| C1 | 0.0385 (13) | 0.0358 (13) | 0.0209 (10) | 0.000 | 0.0059 (9) | 0.000 |
| C2 | 0.0276 (12) | 0.0477 (15) | 0.0266 (11) | 0.000 | −0.0009 (9) | 0.000 |
| C3 | 0.0248 (11) | 0.0451 (14) | 0.0244 (11) | 0.000 | 0.0063 (8) | 0.000 |
| C4 | 0.0277 (11) | 0.0450 (14) | 0.0228 (10) | 0.000 | 0.0043 (8) | 0.000 |
| C5 | 0.0236 (11) | 0.0676 (18) | 0.0292 (12) | 0.000 | 0.0023 (9) | 0.000 |
| C6 | 0.0301 (12) | 0.0501 (15) | 0.0281 (11) | 0.000 | 0.0110 (9) | 0.000 |
| C7 | 0.0291 (12) | 0.0535 (17) | 0.0357 (13) | 0.000 | 0.0045 (10) | 0.000 |
| C8 | 0.0489 (11) | 0.0368 (10) | 0.0301 (9) | −0.0013 (8) | −0.0043 (7) | −0.0063 (7) |
| C9 | 0.0438 (10) | 0.0329 (9) | 0.0289 (9) | 0.0001 (7) | 0.0010 (7) | 0.0064 (7) |
| C10 | 0.0341 (13) | 0.0516 (16) | 0.0252 (11) | 0.000 | 0.0064 (10) | 0.000 |
Geometric parameters (Å, º) top
| N1—O2 | 1.234 (3) | O1W—H2W1 | 0.73 (5) |
| N1—O1 | 1.243 (3) | C1—C2 | 1.374 (3) |
| N1—C1 | 1.441 (3) | C1—C6 | 1.391 (3) |
| N2—O4 | 1.213 (3) | C2—C3 | 1.388 (3) |
| N2—O3 | 1.219 (3) | C2—H2A | 0.88 (3) |
| N2—C3 | 1.440 (3) | C3—C4 | 1.434 (3) |
| N3—C9i | 1.510 (2) | C4—C5 | 1.438 (3) |
| N3—C9 | 1.510 (2) | C5—C6 | 1.353 (3) |
| N3—C7 | 1.519 (3) | C5—H5A | 0.87 (3) |
| N3—H1N3 | 0.85 (3) | C6—H6A | 0.91 (3) |
| N4—C9 | 1.438 (2) | C7—H7A | 0.989 (19) |
| N4—C10 | 1.4666 (19) | C8—H8A | 1.029 (19) |
| N4—C8 | 1.468 (2) | C8—H8B | 0.97 (2) |
| N5—C7 | 1.446 (3) | C9—H9B | 1.00 (2) |
| N5—C8i | 1.473 (2) | C9—H9A | 0.93 (2) |
| N5—C8 | 1.473 (2) | C10—N4i | 1.4666 (19) |
| O5—C4 | 1.264 (3) | C10—H11A | 0.94 (3) |
| O1W—H1W1 | 0.96 (3) | C10—H11B | 0.99 (3) |
| | | |
| O2—N1—O1 | 122.4 (2) | O5—C4—C5 | 119.4 (2) |
| O2—N1—C1 | 119.8 (2) | C3—C4—C5 | 113.87 (19) |
| O1—N1—C1 | 117.9 (2) | C6—C5—C4 | 123.5 (2) |
| O4—N2—O3 | 120.4 (2) | C6—C5—H5A | 119.5 (18) |
| O4—N2—C3 | 120.4 (2) | C4—C5—H5A | 117.1 (18) |
| O3—N2—C3 | 119.20 (19) | C5—C6—C1 | 119.6 (2) |
| C9i—N3—C9 | 108.61 (18) | C5—C6—H6A | 120.6 (17) |
| C9i—N3—C7 | 108.71 (11) | C1—C6—H6A | 119.8 (17) |
| C9—N3—C7 | 108.71 (11) | N5—C7—N3 | 109.74 (18) |
| C9i—N3—H1N3 | 110.0 (10) | N5—C7—H7A | 110.1 (11) |
| C9—N3—H1N3 | 110.0 (10) | N3—C7—H7A | 109.0 (11) |
| C7—N3—H1N3 | 111 (2) | N4—C8—N5 | 111.82 (13) |
| C9—N4—C10 | 108.88 (14) | N4—C8—H8A | 108.6 (11) |
| C9—N4—C8 | 109.75 (14) | N5—C8—H8A | 108.6 (10) |
| C10—N4—C8 | 108.35 (15) | N4—C8—H8B | 106.9 (12) |
| C7—N5—C8i | 109.10 (12) | N5—C8—H8B | 106.9 (11) |
| C7—N5—C8 | 109.10 (12) | H8A—C8—H8B | 114.0 (15) |
| C8i—N5—C8 | 107.76 (19) | N4—C9—N3 | 109.65 (13) |
| H1W1—O1W—H2W1 | 106 (4) | N4—C9—H9B | 112.0 (10) |
| C2—C1—C6 | 121.11 (19) | N3—C9—H9B | 108.3 (11) |
| C2—C1—N1 | 118.4 (2) | N4—C9—H9A | 111.7 (12) |
| C6—C1—N1 | 120.51 (19) | N3—C9—H9A | 106.4 (12) |
| C1—C2—C3 | 119.3 (2) | H9B—C9—H9A | 108.7 (16) |
| C1—C2—H2A | 123.4 (16) | N4—C10—N4i | 111.77 (18) |
| C3—C2—H2A | 117.4 (16) | N4—C10—H11A | 109.4 (8) |
| C2—C3—C4 | 122.66 (19) | N4i—C10—H11A | 109.4 (8) |
| C2—C3—N2 | 115.9 (2) | N4—C10—H11B | 108.3 (7) |
| C4—C3—N2 | 121.47 (18) | N4i—C10—H11B | 108.3 (7) |
| O5—C4—C3 | 126.68 (19) | H11A—C10—H11B | 110 (2) |
| | | |
| O2—N1—C1—C2 | 0.0 | C4—C5—C6—C1 | 0.0 |
| O1—N1—C1—C2 | 180.0 | C2—C1—C6—C5 | 0.0 |
| O2—N1—C1—C6 | 180.0 | N1—C1—C6—C5 | 180.0 |
| O1—N1—C1—C6 | 0.0 | C8i—N5—C7—N3 | −58.75 (12) |
| C6—C1—C2—C3 | 0.0 | C8—N5—C7—N3 | 58.75 (12) |
| N1—C1—C2—C3 | 180.0 | C9i—N3—C7—N5 | 59.03 (10) |
| C1—C2—C3—C4 | 0.0 | C9—N3—C7—N5 | −59.03 (10) |
| C1—C2—C3—N2 | 180.0 | C9—N4—C8—N5 | 59.88 (18) |
| O4—N2—C3—C2 | 180.0 | C10—N4—C8—N5 | −58.89 (18) |
| O3—N2—C3—C2 | 0.0 | C7—N5—C8—N4 | −59.55 (19) |
| O4—N2—C3—C4 | 0.0 | C8i—N5—C8—N4 | 58.8 (2) |
| O3—N2—C3—C4 | 180.0 | C10—N4—C9—N3 | 59.37 (19) |
| C2—C3—C4—O5 | 180.0 | C8—N4—C9—N3 | −59.07 (18) |
| N2—C3—C4—O5 | 0.0 | C9i—N3—C9—N4 | −59.2 (2) |
| C2—C3—C4—C5 | 0.0 | C7—N3—C9—N4 | 58.92 (18) |
| N2—C3—C4—C5 | 180.0 | C9—N4—C10—N4i | −60.7 (2) |
| O5—C4—C5—C6 | 180.0 | C8—N4—C10—N4i | 58.6 (2) |
| C3—C4—C5—C6 | 0.0 | | |
| Symmetry code: (i) x, −y+3/2, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H13N···O5ii | 0.85 (3) | 1.85 (3) | 2.657 (2) | 159 (3) |
| O1W—H11W···O5ii | 0.96 (3) | 1.86 (3) | 2.816 (3) | 177 (3) |
| O1W—H21W···O4iii | 0.73 (5) | 2.35 (5) | 3.037 (3) | 159 (5) |
| C5—H5A···O3iv | 0.88 (3) | 2.48 (3) | 3.164 (3) | 135 (2) |
| Symmetry codes: (ii) −x+1, y+1/2, −z+1; (iii) −x, y+1/2, −z+1; (iv) x+1, y, z. |
(Iat143K) Hexamethylenetetramine-2,4-dinitrophenol Hydrate
top
Crystal data top
| C6H13N4+·C6H3N2O5−·H2O | Z = 2 |
| Mr = 342.32 | F(000) = 360 |
| Triclinic, P1 | Dx = 1.574 Mg m−3 |
| a = 6.3877 (4) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 7.9017 (5) Å | Cell parameters from 3560 reflections |
| c = 14.4042 (9) Å | θ = 1.4–29.6° |
| α = 84.412 (1)° | µ = 0.13 mm−1 |
| β = 86.367 (1)° | T = 143 K |
| γ = 89.304 (1)° | Block, yellow |
| V = 722.11 (8) Å3 | 0.42 × 0.24 × 0.20 mm |
Data collection top
Siemens SMART CCD area-detector
diffractometer | 3227 independent reflections |
| Radiation source: fine-focus sealed tube | 2201 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.073 |
| Detector resolution: 8.33 pixels mm-1 | θmax = 28.0°, θmin = 2.6° |
| ω scans | h = −8→6 |
Absorption correction: empirical (using intensity measurements)
(SADABS; Sheldrick, 1996) | k = −10→10 |
| Tmin = 0.948, Tmax = 0.975 | l = −18→18 |
| 4841 measured reflections | |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.087 | All H-atom parameters refined |
| wR(F2) = 0.206 | w = 1/[σ2(Fo2) + (0.077P)2]
where P = (Fo2 + 2Fc2)/3 |
| S = 0.96 | (Δ/σ)max < 0.001 |
| 3227 reflections | Δρmax = 0.54 e Å−3 |
| 281 parameters | Δρmin = −0.54 e Å−3 |
| 0 restraints | Extinction correction: SHELXTL, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.099 (15) |
Crystal data top
| C6H13N4+·C6H3N2O5−·H2O | γ = 89.304 (1)° |
| Mr = 342.32 | V = 722.11 (8) Å3 |
| Triclinic, P1 | Z = 2 |
| a = 6.3877 (4) Å | Mo Kα radiation |
| b = 7.9017 (5) Å | µ = 0.13 mm−1 |
| c = 14.4042 (9) Å | T = 143 K |
| α = 84.412 (1)° | 0.42 × 0.24 × 0.20 mm |
| β = 86.367 (1)° | |
Data collection top
Siemens SMART CCD area-detector
diffractometer | 3227 independent reflections |
Absorption correction: empirical (using intensity measurements)
(SADABS; Sheldrick, 1996) | 2201 reflections with I > 2σ(I) |
| Tmin = 0.948, Tmax = 0.975 | Rint = 0.073 |
| 4841 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.087 | 0 restraints |
| wR(F2) = 0.206 | All H-atom parameters refined |
| S = 0.96 | Δρmax = 0.54 e Å−3 |
| 3227 reflections | Δρmin = −0.54 e Å−3 |
| 281 parameters | |
Special details top
Experimental. The data collection covered over a hemisphere of reciprocal space by a
combination of three sets of exposures; each set had a different ϕ angle (0,
88 and 180°) for the crystal and each exposure of 10 s covered 0.3° in ω.
The crystal-to-detector distance was 4 cm and the detector swing angle was
-35°. Crystal decay was monitored by repeating fifty initial frames at the
end of data collection and analysing the intensity of duplicate reflections,
and was found to be negligible. |
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. # loop_ # _publ_manuscript_incl_extra_item # # '_geom_extra_tableA_col_1' #
'_geom_extra_tableA_col_2' # '_geom_extra_table_head_A' #
'_geom_table_footnote_A' # # _geom_extra_table_head_A #; # Average C—N bond
distances (Å) along with the puckering parameters (Å, °) # of the
six-membered C—N—C—N—C—N rings of the HMT #; # # loop_ #
_geom_extra_tableA_col_1 # _geom_extra_tableA_col_2 # 'Scheme 2 here' ? # ? ?
# 'T = 300K' 'T = 143K' # ? ? # 'Average C—N = 1.474' 'Average C—N =
1.480 (3)' # 'Q2 = 0.014 (2)' 'Q2 = 0.009 (3)' # 'Q3 = 0.592 (2)' 'Q3 =
0.596 (3)' # 'QT = 0.593 (2)' 'QT = 0.596 (3)' # 'θ = 0.4 (2)' 'θ = 0.0 (3)'
# ? ? # 'Average C—N = 1.474 (3)' 'Average C—N = 1.480 (3)' # 'Q2 =
0.014 (2)' 'Q2 = 0.017 (3)' # 'Q3 = 0.592 (2)' 'Q3 = 0.600 (3)' # 'QT =
0.593 (2)' 'QT = 0.600 (3)' # 'θ = 0.4 (2)' 'θ = 1.8 (3)' # ? ? # 'Average
C—N = 1.471 (3)' 'Average C—N = 1.479 (3)' # 'Q2 = 0.008 (2)' 'Q2 =
0.007 (2)' # 'Q3 = 0.598 (2)' 'Q3 = 0.597 (2)' # 'QT = 0.598 (2)' 'QT =
0.597 (3)' # 'θ = 1.6 (2)' 'θ = 2.1 (3)' # ? ? # 'Average C—N = 1.469 (3)'
'Average C—N = 1.473 (3)' # 'Q2 = 0.004 (2)' 'Q2 = 0.009 (3)' # 'Q3 =
0.584 (2)' 'Q3 = 0.578 (3)' # 'QT = 0.585 (2)' 'QT = 0.578 (3)' # 'θ =
1.1 (2)' 'θ = 0.0 (3)' # ? ? # # _geom_table_footnote_A #; # Note: All the
rings adopt a chair conformation. #; # |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N1 | 0.2752 (3) | 0.6767 (3) | −0.25439 (15) | 0.0154 (5) | |
| N2 | 0.2440 (3) | 0.9482 (3) | 0.03112 (14) | 0.0147 (5) | |
| N3 | 0.2238 (3) | 0.7463 (3) | 0.29237 (15) | 0.0133 (5) | |
| H1N3 | 0.219 (5) | 0.733 (4) | 0.239 (2) | 0.025* | |
| N4 | 0.4515 (3) | 0.8628 (3) | 0.39867 (15) | 0.0140 (5) | |
| N4A | 0.0720 (3) | 0.9031 (3) | 0.41653 (15) | 0.0145 (5) | |
| N5 | 0.2253 (3) | 0.6220 (3) | 0.45392 (15) | 0.0145 (5) | |
| O1 | 0.2775 (3) | 0.5438 (2) | −0.29495 (13) | 0.0222 (5) | |
| O2 | 0.2812 (3) | 0.8190 (3) | −0.29746 (13) | 0.0233 (5) | |
| O3 | 0.2575 (4) | 1.0828 (2) | −0.01981 (14) | 0.0272 (5) | |
| O4 | 0.2269 (4) | 0.9473 (3) | 0.11750 (14) | 0.0288 (6) | |
| O5 | 0.2311 (3) | 0.6037 (2) | 0.13357 (12) | 0.0178 (5) | |
| O1W | −0.2302 (3) | 0.7318 (3) | −0.22082 (14) | 0.0242 (5) | |
| H1W1 | −0.234 (5) | 0.622 (5) | −0.189 (2) | 0.028* | |
| H2W1 | −0.240 (5) | 0.796 (5) | −0.175 (2) | 0.029* | |
| C1 | 0.2629 (4) | 0.6592 (3) | −0.15335 (16) | 0.0127 (5) | |
| C2 | 0.2576 (4) | 0.8054 (3) | −0.10769 (18) | 0.0140 (6) | |
| H2 | 0.256 (5) | 0.919 (5) | −0.140 (2) | 0.035 (10)* | |
| C3 | 0.2473 (4) | 0.7898 (3) | −0.01062 (16) | 0.0115 (5) | |
| C4 | 0.2418 (4) | 0.6288 (3) | 0.04504 (17) | 0.0123 (5) | |
| C5 | 0.2503 (4) | 0.4839 (3) | −0.00912 (18) | 0.0156 (6) | |
| H5A | 0.247 (5) | 0.373 (4) | 0.020 (2) | 0.022 (8)* | |
| C6 | 0.2602 (4) | 0.4979 (3) | −0.10400 (18) | 0.0136 (5) | |
| H6A | 0.263 (4) | 0.404 (4) | −0.139 (2) | 0.021 (8)* | |
| C7 | 0.2059 (4) | 0.5831 (3) | 0.35838 (18) | 0.0156 (6) | |
| H7B | 0.069 (5) | 0.532 (4) | 0.348 (2) | 0.020 (8)* | |
| H7A | 0.322 (5) | 0.505 (4) | 0.334 (2) | 0.024* | |
| C8 | 0.4297 (4) | 0.7033 (3) | 0.45981 (19) | 0.0162 (6) | |
| H8B | 0.554 (5) | 0.625 (4) | 0.451 (2) | 0.022* | |
| H8A | 0.438 (5) | 0.727 (4) | 0.519 (2) | 0.028 (9)* | |
| C8A | 0.0584 (4) | 0.7438 (3) | 0.47820 (18) | 0.0152 (6) | |
| H8AB | −0.084 (5) | 0.689 (4) | 0.469 (2) | 0.022* | |
| H8AA | 0.072 (5) | 0.767 (4) | 0.547 (2) | 0.027 (8)* | |
| C9 | 0.4339 (4) | 0.8273 (3) | 0.30282 (18) | 0.0148 (6) | |
| H9B | 0.448 (5) | 0.931 (4) | 0.259 (2) | 0.021 (8)* | |
| H9A | 0.545 (5) | 0.749 (4) | 0.283 (2) | 0.028* | |
| C9A | 0.0505 (4) | 0.8664 (3) | 0.32122 (18) | 0.0150 (5) | |
| H9AB | 0.069 (5) | 0.961 (4) | 0.278 (2) | 0.024* | |
| H9AA | −0.084 (5) | 0.801 (4) | 0.314 (2) | 0.019 (8)* | |
| C10 | 0.2787 (4) | 0.9795 (3) | 0.42406 (18) | 0.0149 (6) | |
| H10B | 0.290 (5) | 1.087 (4) | 0.385 (2) | 0.029* | |
| H10A | 0.279 (4) | 1.002 (4) | 0.486 (2) | 0.019 (8)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N1 | 0.0139 (11) | 0.0187 (11) | 0.0140 (11) | 0.0035 (9) | −0.0013 (8) | −0.0036 (9) |
| N2 | 0.0242 (12) | 0.0093 (10) | 0.0112 (10) | 0.0044 (9) | −0.0043 (8) | −0.0023 (8) |
| N3 | 0.0184 (12) | 0.0132 (11) | 0.0091 (10) | 0.0033 (8) | −0.0036 (8) | −0.0040 (9) |
| N4 | 0.0137 (11) | 0.0131 (11) | 0.0158 (11) | 0.0003 (8) | −0.0034 (8) | −0.0029 (9) |
| N4A | 0.0141 (11) | 0.0139 (11) | 0.0162 (11) | 0.0024 (8) | −0.0015 (8) | −0.0045 (9) |
| N5 | 0.0174 (12) | 0.0117 (10) | 0.0145 (11) | 0.0015 (8) | −0.0016 (8) | −0.0020 (9) |
| O1 | 0.0297 (12) | 0.0217 (11) | 0.0172 (10) | 0.0072 (8) | −0.0035 (8) | −0.0115 (8) |
| O2 | 0.0368 (13) | 0.0184 (10) | 0.0144 (10) | 0.0032 (8) | −0.0017 (8) | 0.0000 (8) |
| O3 | 0.0528 (14) | 0.0087 (9) | 0.0203 (11) | 0.0015 (9) | −0.0044 (9) | −0.0005 (8) |
| O4 | 0.0617 (16) | 0.0135 (10) | 0.0122 (9) | 0.0042 (9) | −0.0043 (9) | −0.0053 (8) |
| O5 | 0.0334 (12) | 0.0110 (9) | 0.0092 (9) | 0.0013 (8) | −0.0029 (7) | −0.0020 (7) |
| O1W | 0.0399 (13) | 0.0158 (10) | 0.0165 (10) | 0.0018 (9) | −0.0022 (8) | −0.0001 (9) |
| C1 | 0.0142 (13) | 0.0163 (13) | 0.0083 (11) | 0.0035 (10) | −0.0020 (9) | −0.0043 (10) |
| C2 | 0.0152 (13) | 0.0122 (12) | 0.0148 (12) | 0.0024 (10) | −0.0013 (9) | −0.0021 (10) |
| C3 | 0.0158 (13) | 0.0074 (11) | 0.0121 (12) | 0.0024 (9) | −0.0043 (9) | −0.0033 (10) |
| C4 | 0.0137 (12) | 0.0099 (12) | 0.0144 (12) | 0.0022 (9) | −0.0030 (9) | −0.0050 (10) |
| C5 | 0.0235 (14) | 0.0083 (12) | 0.0155 (13) | 0.0022 (10) | −0.0022 (9) | −0.0026 (10) |
| C6 | 0.0156 (13) | 0.0120 (12) | 0.0142 (12) | 0.0026 (9) | −0.0017 (9) | −0.0055 (10) |
| C7 | 0.0207 (14) | 0.0090 (12) | 0.0184 (13) | 0.0021 (10) | −0.0052 (10) | −0.0050 (10) |
| C8 | 0.0170 (14) | 0.0162 (13) | 0.0155 (12) | 0.0003 (10) | −0.0051 (10) | 0.0007 (11) |
| C8A | 0.0165 (13) | 0.0146 (13) | 0.0145 (12) | 0.0020 (10) | −0.0005 (9) | −0.0025 (10) |
| C9 | 0.0135 (13) | 0.0156 (13) | 0.0150 (12) | 0.0008 (10) | 0.0010 (9) | −0.0018 (11) |
| C9A | 0.0128 (13) | 0.0166 (13) | 0.0165 (12) | 0.0045 (10) | −0.0035 (9) | −0.0059 (11) |
| C10 | 0.0180 (14) | 0.0136 (13) | 0.0141 (12) | 0.0017 (10) | −0.0029 (9) | −0.0057 (11) |
Geometric parameters (Å, º) top
| N1—O2 | 1.230 (3) | C1—C2 | 1.383 (3) |
| N1—O1 | 1.250 (3) | C1—C6 | 1.398 (4) |
| N1—C1 | 1.446 (3) | C2—C3 | 1.389 (3) |
| N2—O3 | 1.232 (3) | C2—H2 | 0.97 (4) |
| N2—O4 | 1.241 (3) | C3—C4 | 1.436 (3) |
| N2—C3 | 1.440 (3) | C4—C5 | 1.446 (3) |
| N3—C9A | 1.514 (3) | C5—C6 | 1.358 (3) |
| N3—C9 | 1.516 (3) | C5—H5A | 0.93 (3) |
| N3—C7 | 1.526 (3) | C6—H6A | 0.94 (3) |
| N3—H1N3 | 0.79 (3) | C7—H7B | 1.00 (3) |
| N4—C9 | 1.447 (3) | C7—H7A | 1.03 (3) |
| N4—C8 | 1.469 (3) | C8—H8B | 1.01 (3) |
| N4—C10 | 1.480 (3) | C8—H8A | 0.90 (3) |
| N4A—C9A | 1.446 (3) | C8A—H8AB | 1.03 (3) |
| N4A—C8A | 1.468 (3) | C8A—H8AA | 1.03 (3) |
| N4A—C10 | 1.473 (3) | C9—H9B | 0.99 (3) |
| N5—C7 | 1.452 (3) | C9—H9A | 0.98 (3) |
| N5—C8 | 1.473 (3) | C9A—H9AB | 0.93 (3) |
| N5—C8A | 1.476 (3) | C9A—H9AA | 1.03 (3) |
| O5—C4 | 1.269 (3) | C10—H10B | 0.97 (3) |
| O1W—H1W1 | 0.94 (3) | C10—H10A | 0.92 (3) |
| O1W—H2W1 | 0.87 (4) | | |
| | | |
| O2—N1—O1 | 122.3 (2) | C5—C6—H6A | 123.3 (19) |
| O2—N1—C1 | 120.0 (2) | C1—C6—H6A | 117.3 (19) |
| O1—N1—C1 | 117.8 (2) | N5—C7—N3 | 109.75 (19) |
| O3—N2—O4 | 121.1 (2) | N5—C7—H7B | 113.7 (17) |
| O3—N2—C3 | 119.2 (2) | N3—C7—H7B | 106.0 (18) |
| O4—N2—C3 | 119.7 (2) | N5—C7—H7A | 114.4 (18) |
| C9A—N3—C9 | 108.9 (2) | N3—C7—H7A | 104.7 (17) |
| C9A—N3—C7 | 108.2 (2) | H7B—C7—H7A | 108 (3) |
| C9—N3—C7 | 108.6 (2) | N4—C8—N5 | 112.3 (2) |
| C9A—N3—H1N3 | 110 (3) | N4—C8—H8B | 111.7 (17) |
| C9—N3—H1N3 | 107 (2) | N5—C8—H8B | 114.1 (18) |
| C7—N3—H1N3 | 115 (2) | N4—C8—H8A | 108 (2) |
| C9—N4—C8 | 109.0 (2) | N5—C8—H8A | 107 (2) |
| C9—N4—C10 | 108.4 (2) | H8B—C8—H8A | 102 (3) |
| C8—N4—C10 | 108.8 (2) | N4A—C8A—N5 | 112.07 (19) |
| C9A—N4A—C8A | 109.2 (2) | N4A—C8A—H8AB | 107.4 (18) |
| C9A—N4A—C10 | 109.48 (19) | N5—C8A—H8AB | 107.8 (17) |
| C8A—N4A—C10 | 108.5 (2) | N4A—C8A—H8AA | 110.5 (18) |
| C7—N5—C8 | 108.7 (2) | N5—C8A—H8AA | 107.6 (19) |
| C7—N5—C8A | 108.8 (2) | H8AB—C8A—H8AA | 112 (2) |
| C8—N5—C8A | 108.3 (2) | N4—C9—N3 | 110.08 (19) |
| H1W1—O1W—H2W1 | 102 (3) | N4—C9—H9B | 112.0 (18) |
| C2—C1—C6 | 121.4 (2) | N3—C9—H9B | 108.9 (18) |
| C2—C1—N1 | 118.3 (2) | N4—C9—H9A | 111 (2) |
| C6—C1—N1 | 120.3 (2) | N3—C9—H9A | 108 (2) |
| C1—C2—C3 | 118.7 (2) | H9B—C9—H9A | 107 (2) |
| C1—C2—H2 | 123 (2) | N4A—C9A—N3 | 109.7 (2) |
| C3—C2—H2 | 118 (2) | N4A—C9A—H9AB | 113 (2) |
| C2—C3—C4 | 123.2 (2) | N3—C9A—H9AB | 103.5 (19) |
| C2—C3—N2 | 115.0 (2) | N4A—C9A—H9AA | 112.2 (16) |
| C4—C3—N2 | 121.8 (2) | N3—C9A—H9AA | 103.7 (17) |
| O5—C4—C3 | 127.1 (2) | H9AB—C9A—H9AA | 114 (3) |
| O5—C4—C5 | 119.0 (2) | N4A—C10—N4 | 111.5 (2) |
| C3—C4—C5 | 113.9 (2) | N4A—C10—H10B | 110 (2) |
| C6—C5—C4 | 123.3 (2) | N4—C10—H10B | 110.8 (19) |
| C6—C5—H5A | 115 (2) | N4A—C10—H10A | 104.1 (18) |
| C4—C5—H5A | 122 (2) | N4—C10—H10A | 111.9 (19) |
| C5—C6—C1 | 119.4 (2) | H10B—C10—H10A | 108 (3) |
| | | |
| O2—N1—C1—C2 | 0.2 (3) | C9A—N3—C7—N5 | −59.4 (3) |
| O1—N1—C1—C2 | −179.2 (2) | C9—N3—C7—N5 | 58.7 (3) |
| O2—N1—C1—C6 | −178.7 (2) | C9—N4—C8—N5 | −60.5 (3) |
| O1—N1—C1—C6 | 1.9 (3) | C10—N4—C8—N5 | 57.6 (3) |
| C6—C1—C2—C3 | −0.6 (4) | C7—N5—C8—N4 | 60.6 (3) |
| N1—C1—C2—C3 | −179.5 (2) | C8A—N5—C8—N4 | −57.5 (3) |
| C1—C2—C3—C4 | −0.1 (4) | C9A—N4A—C8A—N5 | 60.4 (3) |
| C1—C2—C3—N2 | 179.5 (2) | C10—N4A—C8A—N5 | −58.9 (3) |
| O3—N2—C3—C2 | −2.5 (4) | C7—N5—C8A—N4A | −59.9 (3) |
| O4—N2—C3—C2 | 177.3 (2) | C8—N5—C8A—N4A | 58.1 (3) |
| O3—N2—C3—C4 | 177.1 (2) | C8—N4—C9—N3 | 58.9 (3) |
| O4—N2—C3—C4 | −3.1 (4) | C10—N4—C9—N3 | −59.5 (3) |
| C2—C3—C4—O5 | −179.5 (2) | C9A—N3—C9—N4 | 59.0 (3) |
| N2—C3—C4—O5 | 0.9 (4) | C7—N3—C9—N4 | −58.5 (3) |
| C2—C3—C4—C5 | 0.7 (4) | C8A—N4A—C9A—N3 | −59.8 (3) |
| N2—C3—C4—C5 | −178.9 (2) | C10—N4A—C9A—N3 | 58.8 (3) |
| O5—C4—C5—C6 | 179.4 (2) | C9—N3—C9A—N4A | −58.2 (3) |
| C3—C4—C5—C6 | −0.7 (4) | C7—N3—C9A—N4A | 59.6 (3) |
| C4—C5—C6—C1 | 0.1 (4) | C9A—N4A—C10—N4 | −60.7 (3) |
| C2—C1—C6—C5 | 0.6 (4) | C8A—N4A—C10—N4 | 58.4 (3) |
| N1—C1—C6—C5 | 179.5 (2) | C9—N4—C10—N4A | 60.7 (3) |
| C8—N5—C7—N3 | −59.0 (3) | C8—N4—C10—N4A | −57.8 (3) |
| C8A—N5—C7—N3 | 58.8 (3) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H13N···O5 | 0.79 (3) | 1.91 (3) | 2.644 (3) | 155 (3) |
| O1W—H11W···O5i | 0.94 (4) | 1.88 (4) | 2.821 (3) | 176 (3) |
| O1W—H21W···O4ii | 0.87 (3) | 2.27 (4) | 3.064 (3) | 152 (3) |
| C5—H5A···O3iii | 0.93 (3) | 2.42 (3) | 3.188 (3) | 140 (2) |
| C7—H7B···O1i | 0.99 (3) | 2.49 (3) | 3.462 (3) | 168 (2) |
| C9—H9A···O1iv | 0.98 (3) | 2.56 (3) | 3.452 (3) | 151 (2) |
| Symmetry codes: (i) −x, −y+1, −z; (ii) −x, −y+2, −z; (iii) x, y−1, z; (iv) −x+1, −y+1, −z. |
Experimental details
| | (Iat300K) | (Iat143K) |
| Crystal data |
| Chemical formula | C6H13N4+·C6H3N2O5−·H2O | C6H13N4+·C6H3N2O5−·H2O |
| Mr | 342.32 | 342.32 |
| Crystal system, space group | Monoclinic, P21/m | Triclinic, P1 |
| Temperature (K) | 300 | 143 |
| a, b, c (Å) | 7.8610 (5), 6.5980 (4), 14.4339 (9) | 6.3877 (4), 7.9017 (5), 14.4042 (9) |
| α, β, γ (°) | 90, 94.915 (1), 90 | 84.412 (1), 86.367 (1), 89.304 (1) |
| V (Å3) | 745.89 (8) | 722.11 (8) |
| Z | 2 | 2 |
| Radiation type | Mo Kα | Mo Kα |
| µ (mm−1) | 0.12 | 0.13 |
| Crystal size (mm) | 0.42 × 0.24 × 0.20 | 0.42 × 0.24 × 0.20 |
| |
| Data collection |
| Diffractometer | Siemens SMART CCD area-detector
diffractometer | Siemens SMART CCD area-detector
diffractometer |
| Absorption correction | Empirical (using intensity measurements)
(SADABS; Sheldrick, 1996) | Empirical (using intensity measurements)
(SADABS; Sheldrick, 1996) |
| Tmin, Tmax | 0.950, 0.976 | 0.948, 0.975 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 5088, 1900, 1288 | 4841, 3227, 2201 |
| Rint | 0.069 | 0.073 |
| (sin θ/λ)max (Å−1) | 0.661 | 0.661 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.059, 0.144, 0.95 | 0.087, 0.206, 0.96 |
| No. of reflections | 1900 | 3227 |
| No. of parameters | 178 | 281 |
| H-atom treatment | All H-atom parameters refined | All H-atom parameters refined |
| Δρmax, Δρmin (e Å−3) | 0.33, −0.31 | 0.54, −0.54 |
Selected geometric parameters (Å, º) for (Iat300K) top| N1—O2 | 1.234 (3) | N3—C7 | 1.519 (3) |
| N1—O1 | 1.243 (3) | N4—C9 | 1.438 (2) |
| N1—C1 | 1.441 (3) | N4—C10 | 1.4666 (19) |
| N2—O4 | 1.213 (3) | N4—C8 | 1.468 (2) |
| N2—O3 | 1.219 (3) | N5—C7 | 1.446 (3) |
| N2—C3 | 1.440 (3) | N5—C8 | 1.473 (2) |
| N3—C9 | 1.510 (2) | O5—C4 | 1.264 (3) |
| | | |
| O2—N1—O1 | 122.4 (2) | C10—N4—C8 | 108.35 (15) |
| O4—N2—O3 | 120.4 (2) | C7—N5—C8 | 109.10 (12) |
| C9i—N3—C9 | 108.61 (18) | C8i—N5—C8 | 107.76 (19) |
| C9i—N3—C7 | 108.71 (11) | N5—C7—N3 | 109.74 (18) |
| C9—N3—C7 | 108.71 (11) | N4—C8—N5 | 111.82 (13) |
| C9—N4—C10 | 108.88 (14) | N4—C9—N3 | 109.65 (13) |
| C9—N4—C8 | 109.75 (14) | N4—C10—N4i | 111.77 (18) |
| Symmetry code: (i) x, −y+3/2, z. |
Hydrogen-bond geometry (Å, º) for (Iat300K) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H13N···O5ii | 0.85 (3) | 1.85 (3) | 2.657 (2) | 159 (3) |
| O1W—H11W···O5ii | 0.96 (3) | 1.86 (3) | 2.816 (3) | 177 (3) |
| O1W—H21W···O4iii | 0.73 (5) | 2.35 (5) | 3.037 (3) | 159 (5) |
| C5—H5A···O3iv | 0.88 (3) | 2.48 (3) | 3.164 (3) | 135 (2) |
| Symmetry codes: (ii) −x+1, y+1/2, −z+1; (iii) −x, y+1/2, −z+1; (iv) x+1, y, z. |
Selected geometric parameters (Å, º) for (Iat143K) top| N1—O2 | 1.230 (3) | N4—C8 | 1.469 (3) |
| N1—O1 | 1.250 (3) | N4—C10 | 1.480 (3) |
| N1—C1 | 1.446 (3) | N4A—C9A | 1.446 (3) |
| N2—O3 | 1.232 (3) | N4A—C8A | 1.468 (3) |
| N2—O4 | 1.241 (3) | N4A—C10 | 1.473 (3) |
| N2—C3 | 1.440 (3) | N5—C7 | 1.452 (3) |
| N3—C9A | 1.514 (3) | N5—C8 | 1.473 (3) |
| N3—C9 | 1.516 (3) | N5—C8A | 1.476 (3) |
| N3—C7 | 1.526 (3) | O5—C4 | 1.269 (3) |
| N4—C9 | 1.447 (3) | | |
| | | |
| O2—N1—O1 | 122.3 (2) | C8A—N4A—C10 | 108.5 (2) |
| O3—N2—O4 | 121.1 (2) | C7—N5—C8 | 108.7 (2) |
| C9A—N3—C9 | 108.9 (2) | C7—N5—C8A | 108.8 (2) |
| C9A—N3—C7 | 108.2 (2) | C8—N5—C8A | 108.3 (2) |
| C9—N3—C7 | 108.6 (2) | N5—C7—N3 | 109.75 (19) |
| C9—N4—C8 | 109.0 (2) | N4—C8—N5 | 112.3 (2) |
| C9—N4—C10 | 108.4 (2) | N4A—C8A—N5 | 112.07 (19) |
| C8—N4—C10 | 108.8 (2) | N4—C9—N3 | 110.08 (19) |
| C9A—N4A—C8A | 109.2 (2) | N4A—C9A—N3 | 109.7 (2) |
| C9A—N4A—C10 | 109.48 (19) | N4A—C10—N4 | 111.5 (2) |
Hydrogen-bond geometry (Å, º) for (Iat143K) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H13N···O5 | 0.79 (3) | 1.91 (3) | 2.644 (3) | 155 (3) |
| O1W—H11W···O5i | 0.94 (4) | 1.88 (4) | 2.821 (3) | 176 (3) |
| O1W—H21W···O4ii | 0.87 (3) | 2.27 (4) | 3.064 (3) | 152 (3) |
| C5—H5A···O3iii | 0.93 (3) | 2.42 (3) | 3.188 (3) | 140 (2) |
| C7—H7B···O1i | 0.99 (3) | 2.49 (3) | 3.462 (3) | 168 (2) |
| C9—H9A···O1iv | 0.98 (3) | 2.56 (3) | 3.452 (3) | 151 (2) |
| Symmetry codes: (i) −x, −y+1, −z; (ii) −x, −y+2, −z; (iii) x, y−1, z; (iv) −x+1, −y+1, −z. |
 O hydrogen bonds which, together with C-H
O hydrogen bonds which, together with C-H O hydrogen bonds, bridge the adducts into molecular ribbons. Extra hydrogen bonds and weak interactions exist for the low-temperature polymorph and these interconnect the molecular ribbons into a three-dimensional packing structure. Also in these two temperature-dependent polymorphs, dinitrophenol acts as a hydrogen-bond acceptor and HMT acts as a hydrogen-bond donor.
O hydrogen bonds, bridge the adducts into molecular ribbons. Extra hydrogen bonds and weak interactions exist for the low-temperature polymorph and these interconnect the molecular ribbons into a three-dimensional packing structure. Also in these two temperature-dependent polymorphs, dinitrophenol acts as a hydrogen-bond acceptor and HMT acts as a hydrogen-bond donor.
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2001/12/00/sk1495/sk1495fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2001/12/00/sk1495/sk1495fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2001/12/00/sk1495/sk1495fig3thm.gif)




The N—H···O-type of hydrogen bond is a versatile synthon in crystal engineering (Fan et al., 1994; Desiraju, 1995). Phenols, in general, are strong acids and tend to form N—H···O hydrogen bonds with aromatic or tertiary amines. Therefore, a number of studies have been conducted into phenol–hexamethylenetetramine adducts (with various ratios) to design and construct a variety of hydrogen-bonding systems. In general, in the adducts of hexamethylenetetramine (HMT) with simple phenols, HMT acts as either a mono-, bis- or tris-acceptor of hydrogen bonds (Jordan & Mak, 1970; Mak et al., 1977, 1978; Mahmoud & Wallwork, 1979; Coupar, Glidewell & Ferguson, 1997; Coupar, Ferguson et al., 1997).
We have now isolated the crystalline form of the hydrate, (I), of the 1:1 stoichiometric adduct of 2,4-dinitrophenol with HMT, and one water molecule is incorporated in the adduct during the crystallization.
The adduct undergoes a phase transition when the temperature is lowered. The room-temperature monoclinic P21/m phase transforms into a low-temperature triclinic P1 phase upon lowering the temperature below 273 K. The phase transition is reversible. We report here the crystal structures of these two temperature-dependent polymorphs, the monoclinic polymorph at 300 K and the triclinic polymorph at 143 K.
In these two temperature-dependent polymorphs, HMT, in an unusual manner, acts as a hydrogen-bond donor of the N—H···O hydrogen bonds observed in these structures.
At 300 K, the asymmetric unit contains one half of HMT, and the monoclinic unit-cell volume contains two adducts. One half of the HMT is related to the other by a centre of inversion passing through atoms N5, N3 and C10. At 143 K, the asymmetric unit contains the complete adduc and the triclinic unit-cell volume also contains two adducts. In both these two space groups, the geometry of dinitrophenol and HMT are as expected. The dinitrophenol transfers an H atom from the hydroxy group to the HMT and becomes negatively charged, with the transfered H atom being localized at the N3 atom of HMT, making it postively charged. A similar effect was also observed in the adduct of HMT with azelaic acid (Hostettler et al., 1999).
In the present adduct, the transfer of the H atom from the hydroxy group was followed by an increase in the delocalization of the π-electron resulting in slight distortions in the N—O, N—C and O—C bond distances of the functional groups within the dinitrophenol moiety, whereas due to the localization of the positive charge at the N3 atom, the C—N3 bond distances are as expected for C—N single bond in an ammonium ion (Allen et al., 1987). All the other bond lengths and angles in the adduct (I) show normal values and are listed in Table 1 for the monoclinic and Table 3 for the triclinic polymorphs.
In both these polymorphs, the dinitrophenol is essentially planar, with the O3 atom deviating by a maximum of 0.0 Å at 300 K and 0.080 (3) Å at 143 K. All the six-membered C—N—C—N—C—N rings of HMT adopt a chair conformation with almost the same puckering parameters (Cremer & Pople, 1975); see _geom_special_details of the archived CIF. These facts rule out that the temperature phase transformation is due to conformational changes since there are not much differences in the structures of these two temperature-dependent polymorphs. However, there are changes in the hydrogen-bond motifs of these two polymorphs.
In both these polymorphs, there are conventional hydrogen bonds (Table 2 and Table 4) and C9—H9B···O4 and N3—H13N···O4 weak interactions [C9···O4i and C9—H9B···O4i = 3.136 (2) Å and 117 (1)° at 300 K, and 3.114 (3) Å and 117 (2)° at 143 K; N3···O4ii and N3—H13N···O4ii = 2.873 (2) Å and 122 (3)° at 300 K, and 2.843 (3) Å and 125 (3)° at 143 K; symmetry codes: (i) 1 - x, 1/2 + y, 1 - z; (ii) 1 - x, 1/2 + y, 1 - z]. Figs. 2 and 3 show the packing diagrams with hydrogen bonds and weak interactions. The crystal structure of the adduct in both the polymorphs is built from molecular sheets of HMT and molecular sheets dinitrophenol alternatingly stacked along the c axis. HMT unusually takes a role as a donor of these intermolecular N—H···O hydrogen bonds, i.e. the N3 atom acts as donor to the O4 and O5 atoms in the dinitrophenol. The N···O distances within the N—H···O hydrogen bonds in these two polymorphs of adduct (I) are comparable with those observed for the O—H···N hydrogen bonds in other HMT—phenol adducts (Mak et al., 1978; Mahmoud & Wallock, 1979; Coupar, Glidewell & Ferguson, 1997; Coupar, Ferguson et al., 1997).
The water molecule in the adduct plays an important role forming O—H···O hydrogen bonds, which together with an intermolecular C—H···O hydrogen bond, i.e. C5—H5A···O3, bridge the dinitrophenols and links the adducts into molecular ribbons parallel to the b axis. These molecular ribbons pack on top of one another.
At low temperature, two extra intramolecular weak interactions [C9A—H9AB···O4 123 (2)° and C6—H6A···O1 101 (2)°] and two extra intermolecular C—H···O hydrogen bonds (Table 4) were observed; the extra weak interactions and hydrogen bonds link the four C atoms of the HMT and the two O atoms of the dinitrophenol, i.e. they link the C9 and C9A atoms and the O4 atom, and they link the C7 and C9 atoms and the O1 atom. These extra intermolecular C—H···O hydrogen bonds interconnects the molecular ribbons into a three-dimentional molecular packing, and stabilize the triclinic polymorphic structure.
Since the only difference in these two polymorphs are the packing structures which are governed by the different hydrogen-bonding and weak interaction patterns, we can attribute this reversible temperature phase transition, which we named as Fun-Anwar-Suchada-Transition (FAST), as due to the presence or absence of these extra hydrogen bonding and weak interactions.
In conclusion, the FAST observed in the 1:1 HMT–2,4-dinitrophenol hydrate adduct is a new type of reversible temperature phase transistion governed by hydrogen bonding and weak interactions. We have also observed similar FAST phenomena in other complexes which are presently under investigation in our research laboratory.