
The title compound, C
6H
12N
4
C
9H
16O
4, undergoes several thermotropic phase transitions. The crystalline structure is layered, with sheets of azelaic acid linked to sheets of hexamethylenetetramine by hydrogen bonds. In the room-temperature phase, the azelaic acid molecules are disordered. By lowering the temperature, this disorder partially disappears. The ordering is clearly observed in reciprocal space where on the rods of diffuse scattering, present in the room-temperature phase, a series of superstructure reflections emerges. This phase transition leads to twin-lattice quasi-symmetry (TLQS) twinning. The structure of this twinned phase is explored in this paper. There are two orientational domains linked by a mirror plane which relates disordered orientations of the acid molecules above the phase transition. A single domain has space group
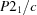
. The structure has been solved and refined on the complete set of data to
R1 = 0.0469. The chains remain partially disordered, showing two acid groups with unequal population: the major form corresponding to a carboxylic acid and the minor to a carboxylate. The ordering of the structure, when going through the phase transition, is interpreted in terms of stabilization by C—H

O hydrogen bonding. A least-squares estimator of the twinning volume ratio is developed that gives an expression for the twinning ratio in terms of the intensities of nonoverlapping reflections. The twinning ratio obtained in the structure refinement compares very well with that obtained from this estimator.
Supporting information
 | Crystallographic Information File (CIF)
Contains datablocks HMTA-PHASE-3, I |
 | Structure factor file (CIF format)
Contains datablock I |
CCDC references: 1157220; 1157221
Data collection: EXPOSE (STOE & CIE GmbH (1997) IPDS 2.87 Software Manual); cell refinement: INDEX, CELL (STOE & CIE GmbH (1997) IPDS 2.87 Software Manual); data reduction: PROFILE, EMS, INTEGRATE (STOE & CIE GmbH (1997) IPDS 2.87 Software
Manual), XPREP Version 5.04, Siemens Analytical X-ray Insts. (1996)
and home written programs; program(s) used to solve structure: SHELXS97 (Sheldrick, 1990); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: XP Version 5.04, Siemens Analytical X-ray Insts. (1996).
Crystal data top
| C6H12N4·C9H16O4 | Z = 4 |
| Mr = 328.41 | F(000) = 712 |
| Monoclinic, P21/c | Dx = 1.244 Mg m−3 |
| a = 5.8907 (7) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 26.071 (2) Å | µ = 0.09 mm−1 |
| c = 11.8868 (11) Å | T = 258 K |
| β = 106.118 (9)° | Platelet, colourless |
| V = 1753.8 (3) Å3 | 0.3 × 0.2 × 0.02 mm |
Data collection top
Stoe IPDS imaging plate system
diffractometer | 1443 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = see _diffrn_special_details |
| Graphite monochromator | θmax = 20.8°, θmin = 2.4° |
| oscillation scans | h = −5→5 |
| 6256 measured reflections | k = −25→25 |
| 1443 independent reflections | l = −11→11 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.047 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.136 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.15 | Calculated w = 1/[σ2(Fo2) + (0.0908P)2 + 0.0702P]
where P = (Fo2 + 2Fc2)/3 |
| 1443 reflections | (Δ/σ)max = 0.001 |
| 246 parameters | Δρmax = 0.17 e Å−3 |
| 8 restraints | Δρmin = −0.12 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The structure is
non-merohedral twin. All reflections, i.e. both common and single
domain reflections are used in the refinement. The refinement was performed
using the HKLF 5 option in SHELXL. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| O1A | −0.025 (7) | 0.875 (2) | 0.098 (4) | 0.108 (12) | 0.29 (4) |
| O1B | −0.056 (2) | 0.8552 (4) | 0.0475 (16) | 0.076 (3) | 0.71 (4) |
| O2A | 0.291 (7) | 0.8681 (16) | 0.048 (4) | 0.090 (11) | 0.29 (4) |
| O2B | 0.310 (2) | 0.8795 (6) | 0.0680 (14) | 0.075 (4) | 0.71 (4) |
| H2O | 0.3406 | 0.8557 | 0.1214 | 0.112* | 0.71 (4) |
| C11 | 0.0823 (9) | 0.88405 (15) | 0.0257 (4) | 0.0509 (12) | |
| C12 | 0.0111 (8) | 0.92394 (16) | −0.0675 (4) | 0.0568 (13) | |
| H12A | 0.0357 | 0.9101 | −0.1389 | 0.068* | |
| H12B | 0.1164 | 0.9530 | −0.0446 | 0.068* | |
| C13 | −0.2383 (8) | 0.94317 (15) | −0.0946 (4) | 0.0575 (13) | |
| H13A | −0.2578 | 0.9624 | −0.0281 | 0.069* | |
| H13B | −0.3452 | 0.9141 | −0.1068 | 0.069* | |
| C14 | −0.3034 (9) | 0.97712 (18) | −0.2026 (5) | 0.0692 (15) | |
| H14A | −0.1861 | 1.0041 | −0.1919 | 0.083* | |
| H14B | −0.2920 | 0.9567 | −0.2690 | 0.083* | |
| C15 | −0.5437 (8) | 1.00182 (18) | −0.2334 (4) | 0.0607 (12) | |
| H15A | −0.5499 | 1.0265 | −0.1732 | 0.073* | |
| H15B | −0.6623 | 0.9758 | −0.2354 | 0.073* | |
| C16 | −0.6007 (9) | 1.02870 (18) | −0.3506 (4) | 0.0666 (15) | |
| H16A | −0.4717 | 1.0519 | −0.3502 | 0.080* | |
| H16B | −0.6058 | 1.0031 | −0.4105 | 0.080* | |
| C17 | −0.8278 (9) | 1.05880 (16) | −0.3857 (4) | 0.0596 (14) | |
| H17A | −0.8190 | 1.0869 | −0.3312 | 0.072* | |
| H17B | −0.9577 | 1.0368 | −0.3814 | 0.072* | |
| C18 | −0.8753 (8) | 1.07994 (17) | −0.5081 (4) | 0.0569 (13) | |
| H18A | −0.7358 | 1.0984 | −0.5134 | 0.068* | |
| H18B | −0.8967 | 1.0512 | −0.5620 | 0.068* | |
| C19 | −1.0829 (9) | 1.11470 (15) | −0.5487 (5) | 0.0546 (12) | |
| O3A | −1.237 (8) | 1.1182 (16) | −0.499 (5) | 0.072 (9) | 0.29 (4) |
| O3B | −1.178 (4) | 1.1367 (7) | −0.486 (2) | 0.088 (5) | 0.71 (4) |
| O4A | −1.119 (6) | 1.1335 (18) | −0.652 (4) | 0.078 (11) | 0.29 (4) |
| O4B | −1.130 (2) | 1.1250 (7) | −0.6618 (14) | 0.067 (4) | 0.71 (4) |
| H4O | −1.2724 | 1.1373 | −0.6868 | 0.101* | 0.71 (4) |
| N1 | 0.4557 (6) | 0.79563 (12) | 0.2069 (3) | 0.0460 (9) | |
| H1 | 0.3891 | 0.8230 | 0.1622 | 0.055* | 0.29 (4) |
| N2 | 0.4336 (6) | 0.70364 (11) | 0.2341 (3) | 0.0449 (9) | |
| H2 | 0.3519 | 0.6746 | 0.2058 | 0.054* | 0.29 (4) |
| N3 | 0.8159 (6) | 0.74534 (13) | 0.2758 (3) | 0.0495 (10) | |
| N4 | 0.5608 (7) | 0.75981 (13) | 0.4034 (3) | 0.0509 (10) | |
| C1 | 0.3276 (8) | 0.74864 (15) | 0.1633 (4) | 0.0486 (10) | |
| H1A | 0.3282 | 0.7432 | 0.0826 | 0.058* | |
| H1B | 0.1645 | 0.7522 | 0.1648 | 0.058* | |
| C2 | 0.7049 (7) | 0.78895 (16) | 0.2057 (4) | 0.0507 (12) | |
| H2A | 0.7106 | 0.7841 | 0.1256 | 0.061* | |
| H2B | 0.7930 | 0.8198 | 0.2356 | 0.061* | |
| C3 | 0.4510 (9) | 0.80276 (16) | 0.3295 (4) | 0.0566 (13) | |
| H3A | 0.5338 | 0.8342 | 0.3600 | 0.068* | |
| H3B | 0.2885 | 0.8062 | 0.3319 | 0.068* | |
| C4 | 0.6794 (8) | 0.69955 (16) | 0.2305 (4) | 0.0548 (13) | |
| H4A | 0.7522 | 0.6701 | 0.2764 | 0.066* | |
| H4B | 0.6825 | 0.6939 | 0.1503 | 0.066* | |
| C5 | 0.8048 (8) | 0.75397 (17) | 0.3963 (5) | 0.0589 (12) | |
| H5A | 0.8938 | 0.7846 | 0.4271 | 0.071* | |
| H5B | 0.8783 | 0.7253 | 0.4448 | 0.071* | |
| C6 | 0.4308 (7) | 0.71313 (15) | 0.3560 (4) | 0.0483 (12) | |
| H6A | 0.2685 | 0.7163 | 0.3590 | 0.058* | |
| H6B | 0.5010 | 0.6841 | 0.4042 | 0.058* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1A | 0.098 (17) | 0.16 (3) | 0.08 (2) | 0.007 (19) | 0.044 (17) | 0.057 (19) |
| O1B | 0.065 (4) | 0.069 (5) | 0.084 (9) | −0.014 (3) | 0.007 (5) | 0.036 (4) |
| O2A | 0.116 (17) | 0.09 (2) | 0.100 (17) | 0.062 (18) | 0.086 (17) | 0.028 (14) |
| O2B | 0.066 (5) | 0.054 (5) | 0.092 (8) | −0.008 (4) | 0.003 (5) | 0.033 (5) |
| C11 | 0.058 (3) | 0.041 (3) | 0.053 (4) | −0.004 (2) | 0.015 (3) | 0.002 (2) |
| C12 | 0.071 (3) | 0.046 (2) | 0.052 (3) | −0.002 (2) | 0.014 (3) | 0.010 (2) |
| C13 | 0.071 (3) | 0.043 (2) | 0.060 (3) | 0.004 (2) | 0.020 (3) | 0.006 (2) |
| C14 | 0.085 (4) | 0.064 (3) | 0.061 (4) | 0.015 (3) | 0.023 (3) | 0.018 (2) |
| C15 | 0.074 (3) | 0.054 (2) | 0.055 (3) | 0.010 (2) | 0.019 (3) | 0.009 (2) |
| C16 | 0.080 (4) | 0.061 (3) | 0.061 (4) | 0.012 (3) | 0.023 (3) | 0.014 (2) |
| C17 | 0.075 (3) | 0.045 (2) | 0.060 (4) | 0.010 (2) | 0.022 (3) | 0.006 (2) |
| C18 | 0.070 (3) | 0.049 (2) | 0.052 (3) | 0.011 (2) | 0.017 (3) | 0.002 (2) |
| C19 | 0.071 (4) | 0.038 (2) | 0.059 (4) | 0.004 (2) | 0.025 (3) | −0.003 (2) |
| O3A | 0.081 (15) | 0.07 (2) | 0.071 (18) | 0.031 (14) | 0.037 (13) | 0.023 (17) |
| O3B | 0.129 (13) | 0.079 (9) | 0.066 (5) | 0.048 (8) | 0.044 (8) | 0.004 (8) |
| O4A | 0.101 (18) | 0.09 (2) | 0.077 (14) | −0.003 (12) | 0.074 (15) | 0.014 (13) |
| O4B | 0.082 (7) | 0.062 (5) | 0.050 (5) | 0.039 (5) | 0.006 (5) | 0.008 (3) |
| N1 | 0.051 (2) | 0.0436 (19) | 0.045 (3) | 0.0034 (17) | 0.0157 (18) | 0.0069 (16) |
| N2 | 0.047 (2) | 0.0436 (19) | 0.044 (3) | −0.0037 (16) | 0.0125 (18) | −0.0031 (16) |
| N3 | 0.038 (2) | 0.063 (2) | 0.049 (3) | 0.0004 (17) | 0.0128 (18) | 0.0026 (18) |
| N4 | 0.057 (2) | 0.058 (2) | 0.040 (3) | −0.0041 (19) | 0.017 (2) | −0.0025 (18) |
| C1 | 0.045 (3) | 0.058 (3) | 0.040 (3) | 0.001 (2) | 0.007 (2) | 0.000 (2) |
| C2 | 0.049 (3) | 0.058 (3) | 0.047 (3) | −0.005 (2) | 0.017 (2) | 0.005 (2) |
| C3 | 0.070 (3) | 0.049 (2) | 0.056 (4) | 0.001 (2) | 0.026 (3) | −0.008 (2) |
| C4 | 0.060 (3) | 0.054 (3) | 0.055 (3) | 0.015 (2) | 0.023 (3) | 0.002 (2) |
| C5 | 0.050 (3) | 0.071 (3) | 0.048 (4) | −0.008 (2) | 0.002 (2) | 0.003 (2) |
| C6 | 0.046 (3) | 0.056 (3) | 0.046 (3) | −0.002 (2) | 0.018 (2) | 0.007 (2) |
Geometric parameters (Å, º) top
| N1—C1 | 1.457 (5) | C15—C16 | 1.512 (6) |
| N1—C3 | 1.477 (6) | C15—H15A | 0.9700 |
| N1—C2 | 1.482 (5) | C15—H15B | 0.9700 |
| N1—H1 | 0.9100 | C16—C17 | 1.507 (6) |
| N2—C4 | 1.464 (5) | C16—H16A | 0.9700 |
| N2—C6 | 1.475 (6) | C16—H16B | 0.9700 |
| N2—C1 | 1.477 (5) | C17—C18 | 1.507 (6) |
| N2—H2 | 0.9100 | C17—H17A | 0.9700 |
| N3—C2 | 1.454 (6) | C17—H17B | 0.9700 |
| N3—C4 | 1.457 (6) | C18—C19 | 1.490 (6) |
| N3—C5 | 1.469 (7) | C18—H18A | 0.9700 |
| N4—C3 | 1.459 (6) | C18—H18B | 0.9700 |
| N4—C6 | 1.464 (5) | C19—O3B | 1.20 (2) |
| N4—C5 | 1.471 (6) | C19—O3A | 1.22 (5) |
| O1A—C11 | 1.22 (3) | C19—O4A | 1.29 (4) |
| O1B—C11 | 1.190 (11) | C19—O4B | 1.322 (15) |
| O2A—C11 | 1.25 (3) | O4B—H4O | 0.8700 |
| O2B—C11 | 1.298 (14) | C1—H1A | 0.9700 |
| O2B—H2O | 0.8700 | C1—H1B | 0.9700 |
| C11—C12 | 1.492 (6) | C2—H2A | 0.9700 |
| C12—C13 | 1.500 (6) | C2—H2B | 0.9700 |
| C12—H12A | 0.9700 | C3—H3A | 0.9700 |
| C12—H12B | 0.9700 | C3—H3B | 0.9700 |
| C13—C14 | 1.518 (7) | C4—H4A | 0.9700 |
| C13—H13A | 0.9700 | C4—H4B | 0.9700 |
| C13—H13B | 0.9700 | C5—H5A | 0.9700 |
| C14—C15 | 1.505 (6) | C5—H5B | 0.9700 |
| C14—H14A | 0.9700 | C6—H6A | 0.9700 |
| C14—H14B | 0.9700 | C6—H6B | 0.9700 |
| | | |
| C11—O2B—H2O | 109.5 | O3B—C19—C18 | 124.9 (12) |
| O1B—C11—O1A | 37 (3) | O3A—C19—C18 | 123 (2) |
| O1B—C11—O2A | 115.7 (19) | O4A—C19—C18 | 116.5 (14) |
| O1A—C11—O2A | 117 (2) | O4B—C19—C18 | 112.0 (8) |
| O1B—C11—O2B | 123.6 (9) | C19—O4B—H4O | 109.5 |
| O1A—C11—O2B | 113 (2) | C1—N1—C3 | 107.9 (3) |
| O2A—C11—O2B | 17 (2) | C1—N1—C2 | 108.3 (3) |
| O1B—C11—C12 | 122.2 (7) | C3—N1—C2 | 108.3 (4) |
| O1A—C11—C12 | 124.2 (17) | C1—N1—H1 | 110.7 |
| O2A—C11—C12 | 116.7 (17) | C3—N1—H1 | 110.7 |
| O2B—C11—C12 | 113.5 (8) | C2—N1—H1 | 110.7 |
| C11—C12—C13 | 116.7 (4) | C4—N2—C6 | 108.7 (3) |
| C11—C12—H12A | 108.1 | C4—N2—C1 | 107.9 (3) |
| C13—C12—H12A | 108.1 | C6—N2—C1 | 108.0 (3) |
| C11—C12—H12B | 108.1 | C4—N2—H2 | 110.7 |
| C13—C12—H12B | 108.1 | C6—N2—H2 | 110.7 |
| H12A—C12—H12B | 107.3 | C1—N2—H2 | 110.7 |
| C12—C13—C14 | 112.3 (4) | C2—N3—C4 | 108.3 (4) |
| C12—C13—H13A | 109.1 | C2—N3—C5 | 107.8 (3) |
| C14—C13—H13A | 109.1 | C4—N3—C5 | 108.4 (3) |
| C12—C13—H13B | 109.1 | C3—N4—C6 | 108.0 (4) |
| C14—C13—H13B | 109.1 | C3—N4—C5 | 108.8 (3) |
| H13A—C13—H13B | 107.9 | C6—N4—C5 | 107.9 (3) |
| C15—C14—C13 | 117.1 (4) | N1—C1—N2 | 111.7 (3) |
| C15—C14—H14A | 108.0 | N1—C1—H1A | 109.3 |
| C13—C14—H14A | 108.0 | N2—C1—H1A | 109.3 |
| C15—C14—H14B | 108.0 | N1—C1—H1B | 109.3 |
| C13—C14—H14B | 108.0 | N2—C1—H1B | 109.3 |
| H14A—C14—H14B | 107.3 | H1A—C1—H1B | 107.9 |
| C14—C15—C16 | 112.1 (4) | N3—C2—N1 | 112.0 (3) |
| C14—C15—H15A | 109.2 | N3—C2—H2A | 109.2 |
| C16—C15—H15A | 109.2 | N1—C2—H2A | 109.2 |
| C14—C15—H15B | 109.2 | N3—C2—H2B | 109.2 |
| C16—C15—H15B | 109.2 | N1—C2—H2B | 109.2 |
| H15A—C15—H15B | 107.9 | H2A—C2—H2B | 107.9 |
| C17—C16—C15 | 116.7 (4) | N4—C3—N1 | 111.8 (3) |
| C17—C16—H16A | 108.1 | N4—C3—H3A | 109.3 |
| C15—C16—H16A | 108.1 | N1—C3—H3A | 109.3 |
| C17—C16—H16B | 108.1 | N4—C3—H3B | 109.3 |
| C15—C16—H16B | 108.1 | N1—C3—H3B | 109.3 |
| H16A—C16—H16B | 107.3 | H3A—C3—H3B | 107.9 |
| C16—C17—C18 | 111.7 (4) | N3—C4—N2 | 112.2 (3) |
| C16—C17—H17A | 109.3 | N3—C4—H4A | 109.2 |
| C18—C17—H17A | 109.3 | N2—C4—H4A | 109.2 |
| C16—C17—H17B | 109.3 | N3—C4—H4B | 109.2 |
| C18—C17—H17B | 109.3 | N2—C4—H4B | 109.2 |
| H17A—C17—H17B | 107.9 | H4A—C4—H4B | 107.9 |
| C19—C18—C17 | 116.8 (4) | N3—C5—N4 | 112.4 (3) |
| C19—C18—H18A | 108.1 | N3—C5—H5A | 109.1 |
| C17—C18—H18A | 108.1 | N4—C5—H5A | 109.1 |
| C19—C18—H18B | 108.1 | N3—C5—H5B | 109.1 |
| C17—C18—H18B | 108.1 | N4—C5—H5B | 109.1 |
| H18A—C18—H18B | 107.3 | H5A—C5—H5B | 107.9 |
| O3B—C19—O3A | 28.1 (18) | N4—C6—N2 | 111.7 (3) |
| O3B—C19—O4A | 116 (2) | N4—C6—H6A | 109.3 |
| O3A—C19—O4A | 119 (3) | N2—C6—H6A | 109.3 |
| O3B—C19—O4B | 122.4 (13) | N4—C6—H6B | 109.3 |
| O3A—C19—O4B | 121 (3) | N2—C6—H6B | 109.3 |
| O4A—C19—O4B | 11 (3) | H6A—C6—H6B | 107.9 |
| | | |
| C13—C12—C11—O1A | −18 (4) | C14—C15—C16—C17 | −174.7 (4) |
| C13—C12—C11—O2A | −180 (2) | C15—C16—C17—C18 | −175.1 (4) |
| C13—C12—C11—O1B | 27.2 (12) | C16—C17—C18—C19 | −174.2 (4) |
| C13—C12—C11—O2B | −161.7 (9) | C17—C18—C19—O3A | −15 (3) |
| C11—C12—C13—C14 | −171.1 (4) | C17—C18—C19—O4A | 179 (2) |
| C12—C13—C14—C15 | −175.7 (4) | C17—C18—C19—O3B | 18.9 (16) |
| C13—C14—C15—C16 | −172.3 (4) | C17—C18—C19—O4B | −170.7 (10) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2B—H2O···N1 | 0.87 | 1.89 | 2.730 (16) | 163 |
| O4B—H4O···N2i | 0.87 | 1.98 | 2.699 (14) | 140 |
| C2—H2A···O1Bii | 0.97 | 2.63 | 3.157 (14) | 115 |
| C3—H3B···O3Biii | 0.97 | 2.58 | 3.19 (2) | 121 |
| C5—H5A···O3Biv | 0.97 | 2.62 | 3.58 (2) | 170 |
| C5—H5B···O1Bv | 0.97 | 2.40 | 3.345 (15) | 163 |
| C12—H12A···O4Bvi | 0.97 | 2.74 | 3.706 (18) | 176 |
| N1—H1···O2A | 0.91 | 1.76 | 2.65 (4) | 165 |
| N1—H1···O1A | 0.91 | 2.71 | 3.46 (3) | 140 |
| N2—H2···O3Avii | 0.91 | 2.79 | 3.51 (5) | 138 |
| N2—H2···O4Avii | 0.91 | 1.72 | 2.59 (4) | 160 |
| C2—H2B···O1Aii | 0.97 | 2.62 | 3.21 (4) | 120 |
| C6—H6B···O2Aviii | 0.97 | 2.74 | 3.38 (4) | 124 |
| C2—H2A···N4ix | 0.97 | 2.79 | 3.682 (6) | 154 |
| C1—H1B···N3x | 0.97 | 2.74 | 3.622 (6) | 152 |
| C6—H6A···N3x | 0.97 | 2.68 | 3.580 (6) | 154 |
| Symmetry codes: (i) −x−1, y+1/2, −z−1/2; (ii) x+1, y, z; (iii) −x−1, −y+2, −z; (iv) −x, −y+2, −z; (v) x+1, −y+3/2, z+1/2; (vi) −x−1, −y+2, −z−1; (vii) −x−1, y−1/2, −z−1/2; (viii) x, −y+3/2, z+1/2; (ix) x, −y+3/2, z−1/2; (x) x−1, y, z. |
Bond distances and angles in the HMT molecule top| N1-C1 | 1.457 (5) | C1-N1-C2 | 108.3 (3) |
| N1-C2 | 1.482 (5) | C1-N1-C3 | 107.9 (3) |
| N1-C3 | 1.477 (6) | C3-N1-C2 | 108.3 (4) |
| N2-C1 | 1.477 (5) | C4-N2-C6 | 108.7 (3) |
| N2-C4 | 1.464 (5) | C4-N2-C1 | 107.9 (3) |
| N2-C6 | 1.475 (6) | C6-N2-C1 | 108.0 (3) |
| N3-C2 | 1.454 (6) | C2-N3-C4 | 108.3 (4) |
| N3-C4 | 1.457 (6) | C2-N3-C5 | 107.8 (3) |
| N3-C5 | 1.469 (7) | C4-N3-C5 | 108.4 (3) |
| N4-C3 | 1.459 (6) | C3-N4-C5 | 108.8 (3) |
| N4-C5 | 1.471 (6) | C3-N4-C6 | 108.0 (4) |
| N4-C6 | 1.464 (5) | C6-N4-C5 | 107.9 (3) |
| | N1-C1-N2 | 111.7 (3) |
| | N3-C2-N1 | 112.0 (3) |
| | N4-C3-N1 | 111.8 (3) |
| | N3-C4-N2 | 112.2 (3) |
| | N3-C5-N4 | 112.4 (3) |
| | N4-C6-N2 | 111.7 (3) |
Bond distances and angles in the acid molecule top| O1A-C11 | 1.22 (3) | O1A-C11-O2A | 117 (2) |
| O2A-C11 | 1.25 (3) | O1A-C11-C12 | 124.2 (17) |
| O1B-C11 | 1.190 (11) | O2A-C11-C12 | 116.7 (17) |
| O2B-C11 | 1.298 (14) | O1B-C11-O2B | 123.6 (9) |
| C11-C12 | 1.492 (6) | O1B-C11-C12 | 122.2 (7) |
| C12-C13 | 1.500 (6) | O2B-C11-C12 | 113.5 (8) |
| C13-C14 | 1.518 (7) | C11-C12-C13 | 116.7 (4) |
| C14-C15 | 1.505 (6) | C12-C13-C14 | 112.3 (4) |
| C15-C16 | 1.512 (6) | C15-C14-C13 | 117.1 (4) |
| C16-C17 | 1.507 (6) | C14-C15-C16 | 112.1 (4) |
| C17-C18 | 1.507 (6) | C17-C16-C15 | 116.7 (4) |
| C18-C19 | 1.490 (6) | C16-C17-C18 | 111.7 (4) |
| C19-O3A | 1.22 (5) | C19-C18-C17 | 116.8 (4) |
| C19-O4A | 1.29 (4) | O3A-C19-O4A | 119 (3) |
| C19-O3B | 1.20 (2) | O3A-C19-C18 | 123 (2) |
| C19-O4B | 1.322 (15) | O4A-C19-C18 | 116.5 (14) |
| | O3B-C19-O4B | 122.4 (13) |
| | O3B-C19-C18 | 124.9 (12) |
| | O4B-C19-C18 | 112.0 (8) |
 O hydrogen bonding. A least-squares estimator of the twinning volume ratio is developed that gives an expression for the twinning ratio in terms of the intensities of nonoverlapping reflections. The twinning ratio obtained in the structure refinement compares very well with that obtained from this estimator.
O hydrogen bonding. A least-squares estimator of the twinning volume ratio is developed that gives an expression for the twinning ratio in terms of the intensities of nonoverlapping reflections. The twinning ratio obtained in the structure refinement compares very well with that obtained from this estimator.
 journal menu
journal menu











