The dicarboxylate dianion in the title compound, [Cd(C
5H
5N)
3(C
11H
9NO
6)]·2H
2O, uses its carboxylatomethyl arm to chelate to one Cd atom and the 1-carboxylatoethyl arm to bind to another Cd atom through only one terminal O atom, giving rise to a polymeric chain that runs along [10
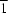
]. The octahedral geometry of the Cd atom is completed by the N atoms of three pyridine N-atom donors.
Supporting information
CCDC reference: 254257
Key indicators
- Single-crystal X-ray study
- T = 295 K
- Mean
![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.006 Å
(C-C) = 0.006 Å
- R factor = 0.037
- wR factor = 0.094
- Data-to-parameter ratio = 17.3
checkCIF/PLATON results
No syntax errors found
 Alert level C
ABSTM02_ALERT_3_C The ratio of expected to reported Tmax/Tmin(RR') is < 0.90
Tmin and Tmax reported: 0.594 0.865
Tmin' and Tmax expected: 0.744 0.862
RR' = 0.795
Please check that your absorption correction is appropriate.
PLAT042_ALERT_1_C Calc. and Rep. MoietyFormula Strings Differ .... ?
PLAT061_ALERT_3_C Tmax/Tmin Range Test RR' too Large ............. 0.79
PLAT241_ALERT_2_C Check High U(eq) as Compared to Neighbors for C14
PLAT242_ALERT_2_C Check Low U(eq) as Compared to Neighbors for N4
PLAT242_ALERT_2_C Check Low U(eq) as Compared to Neighbors for C26
PLAT301_ALERT_3_C Main Residue Disorder ......................... 3.00 Perc.
PLAT720_ALERT_4_C Number of Unusual/Non-Standard Label(s) ........ 4
PLAT731_ALERT_1_C Bond Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Rat
O1W -H1W1 1.555 1.555
PLAT731_ALERT_1_C Bond Calc 0.86(3), Rep 0.850(10) ...... 3.00 su-Rat
O1W -H1W2 1.555 1.555
PLAT731_ALERT_1_C Bond Calc 0.86(3), Rep 0.860(10) ...... 3.00 su-Rat
O2W -H2W1 1.555 1.555
PLAT731_ALERT_1_C Bond Calc 0.86(4), Rep 0.860(10) ...... 4.00 su-Rat
O2W -H2W2 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Rat
O1W -H1# 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.86(3), Rep 0.850(10) ...... 3.00 su-Rat
O1W -H2# 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.86(3), Rep 0.860(10) ...... 3.00 su-Rat
O2W -H3# 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.86(4), Rep 0.860(10) ...... 4.00 su-Rat
O2W -H4# 1.555 1.555
PLAT736_ALERT_1_C H...A Calc 2.03(3), Rep 2.040(10) ...... 3.00 su-Rat
H2# -O4 1.555 1.555
PLAT736_ALERT_1_C H...A Calc 2.00(3), Rep 2.000(10) ...... 3.00 su-Rat
H3# -O1W 1.555 1.555
Alert level C
ABSTM02_ALERT_3_C The ratio of expected to reported Tmax/Tmin(RR') is < 0.90
Tmin and Tmax reported: 0.594 0.865
Tmin' and Tmax expected: 0.744 0.862
RR' = 0.795
Please check that your absorption correction is appropriate.
PLAT042_ALERT_1_C Calc. and Rep. MoietyFormula Strings Differ .... ?
PLAT061_ALERT_3_C Tmax/Tmin Range Test RR' too Large ............. 0.79
PLAT241_ALERT_2_C Check High U(eq) as Compared to Neighbors for C14
PLAT242_ALERT_2_C Check Low U(eq) as Compared to Neighbors for N4
PLAT242_ALERT_2_C Check Low U(eq) as Compared to Neighbors for C26
PLAT301_ALERT_3_C Main Residue Disorder ......................... 3.00 Perc.
PLAT720_ALERT_4_C Number of Unusual/Non-Standard Label(s) ........ 4
PLAT731_ALERT_1_C Bond Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Rat
O1W -H1W1 1.555 1.555
PLAT731_ALERT_1_C Bond Calc 0.86(3), Rep 0.850(10) ...... 3.00 su-Rat
O1W -H1W2 1.555 1.555
PLAT731_ALERT_1_C Bond Calc 0.86(3), Rep 0.860(10) ...... 3.00 su-Rat
O2W -H2W1 1.555 1.555
PLAT731_ALERT_1_C Bond Calc 0.86(4), Rep 0.860(10) ...... 4.00 su-Rat
O2W -H2W2 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Rat
O1W -H1# 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.86(3), Rep 0.850(10) ...... 3.00 su-Rat
O1W -H2# 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.86(3), Rep 0.860(10) ...... 3.00 su-Rat
O2W -H3# 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.86(4), Rep 0.860(10) ...... 4.00 su-Rat
O2W -H4# 1.555 1.555
PLAT736_ALERT_1_C H...A Calc 2.03(3), Rep 2.040(10) ...... 3.00 su-Rat
H2# -O4 1.555 1.555
PLAT736_ALERT_1_C H...A Calc 2.00(3), Rep 2.000(10) ...... 3.00 su-Rat
H3# -O1W 1.555 1.555
0 ALERT level A = In general: serious problem
0 ALERT level B = Potentially serious problem
18 ALERT level C = Check and explain
0 ALERT level G = General alerts; check
11 ALERT type 1 CIF construction/syntax error, inconsistent or missing data
3 ALERT type 2 Indicator that the structure model may be wrong or deficient
3 ALERT type 3 Indicator that the structure quality may be low
1 ALERT type 4 Improvement, methodology, query or suggestion
Data collection: RAPID-AUTO (Rigaku, 1998); cell refinement: RAPID-AUTO; data reduction: CrystalStructure (Rigaku/MSC, 2002); program(s) used to solve structure: SHELXS97 (Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: ORTEPII (Johnson, 1976); software used to prepare material for publication: SHELXL97.
catena-Poly[[tris(pyridine-
κN)cadmium(II)]-µ-{2-[3-(1-carboxylatomethyl)- 4-nitrophenyl]propionato-
κ2O,
O'}
dihydrate]
top
Crystal data top
| [Cd(C5H5N)3(C11H9NO6)]·2H2O | F(000) = 1296 |
| Mr = 636.92 | Dx = 1.500 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 23092 reflections |
| a = 9.748 (2) Å | θ = 3.2–27.5° |
| b = 19.923 (4) Å | µ = 0.83 mm−1 |
| c = 14.598 (3) Å | T = 295 K |
| β = 95.92 (3)° | Prism, yellow |
| V = 2820 (1) Å3 | 0.35 × 0.24 × 0.18 mm |
| Z = 4 | |
Data collection top
Rigaku R-AXIS RAPID
diffractometer | 6413 independent reflections |
| Radiation source: fine-focus sealed tube | 4950 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.032 |
| ω scans | θmax = 27.5°, θmin = 3.2° |
Absorption correction: multi-scan
(ABSCOR; Higashi, 1995) | h = −10→12 |
| Tmin = 0.594, Tmax = 0.865 | k = −25→25 |
| 25939 measured reflections | l = −18→18 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.037 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.094 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0464P)2 + 1.0608P]
where P = (Fo2 + 2Fc2)/3 |
| 6413 reflections | (Δ/σ)max = 0.001 |
| 371 parameters | Δρmax = 0.63 e Å−3 |
| 20 restraints | Δρmin = −0.52 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Cd1 | 0.66726 (2) | 0.33543 (1) | 0.51903 (1) | 0.04599 (8) | |
| O1 | 0.6146 (3) | 0.4342 (1) | 0.5931 (1) | 0.0631 (6) | |
| O2 | 0.5953 (3) | 0.3475 (1) | 0.6834 (2) | 0.0633 (6) | |
| O3 | 0.1557 (2) | 0.2777 (1) | 1.0461 (2) | 0.0634 (6) | |
| O4 | 0.2480 (2) | 0.2667 (1) | 0.9164 (2) | 0.0625 (6) | |
| O5 | 0.8746 (3) | 0.3804 (2) | 0.8894 (3) | 0.119 (1) | |
| O6 | 0.8010 (3) | 0.4578 (2) | 0.7987 (2) | 0.0965 (9) | |
| O1w | 0.4515 (3) | 0.2543 (1) | 0.7905 (2) | 0.0675 (6) | |
| O2w | 0.4122 (3) | 0.1834 (2) | 0.6195 (2) | 0.0936 (9) | |
| N1 | 0.9018 (3) | 0.3281 (1) | 0.5783 (2) | 0.0523 (6) | |
| N2 | 0.7520 (3) | 0.4075 (1) | 0.4054 (2) | 0.0533 (6) | |
| N3 | 0.4546 (3) | 0.3399 (1) | 0.4303 (2) | 0.0535 (6) | |
| N4 | 0.7834 (3) | 0.4165 (2) | 0.8571 (2) | 0.072 (1) | |
| C1 | 0.9373 (4) | 0.3294 (2) | 0.6682 (2) | 0.070 (1) | |
| C2 | 1.0687 (5) | 0.3183 (3) | 0.7071 (3) | 0.092 (1) | |
| C3 | 1.1681 (5) | 0.3051 (3) | 0.6517 (4) | 0.096 (1) | |
| C4 | 1.1342 (4) | 0.3042 (2) | 0.5591 (3) | 0.083 (1) | |
| C5 | 1.0017 (4) | 0.3162 (2) | 0.5242 (3) | 0.065 (1) | |
| C6 | 0.8103 (4) | 0.4651 (2) | 0.4341 (2) | 0.068 (1) | |
| C7 | 0.8791 (4) | 0.5056 (2) | 0.3794 (3) | 0.083 (1) | |
| C8 | 0.8901 (4) | 0.4871 (2) | 0.2910 (3) | 0.085 (1) | |
| C9 | 0.8301 (5) | 0.4292 (2) | 0.2603 (3) | 0.089 (1) | |
| C10 | 0.7618 (4) | 0.3904 (2) | 0.3195 (2) | 0.072 (1) | |
| C11 | 0.3894 (3) | 0.3982 (2) | 0.4170 (2) | 0.059 (1) | |
| C12 | 0.2697 (4) | 0.4059 (2) | 0.3595 (3) | 0.067 (1) | |
| C13 | 0.2143 (4) | 0.3511 (2) | 0.3136 (3) | 0.082 (1) | |
| C14 | 0.2800 (4) | 0.2907 (2) | 0.3266 (4) | 0.097 (2) | |
| C15 | 0.3988 (4) | 0.2872 (2) | 0.3849 (3) | 0.081 (1) | |
| C16 | 0.5840 (3) | 0.4085 (2) | 0.6667 (2) | 0.051 (1) | |
| C17 | 0.5259 (4) | 0.4551 (2) | 0.7357 (2) | 0.060 (1) | |
| C18 | 0.5277 (3) | 0.4271 (1) | 0.8320 (2) | 0.052 (1) | |
| C19 | 0.6452 (3) | 0.4081 (2) | 0.8884 (2) | 0.056 (1) | |
| C20 | 0.6394 (4) | 0.3801 (2) | 0.9740 (2) | 0.074 (1) | |
| C21 | 0.5113 (5) | 0.3707 (2) | 1.0054 (3) | 0.079 (1) | |
| C22 | 0.3916 (4) | 0.3882 (2) | 0.9519 (3) | 0.067 (1) | |
| C23 | 0.4031 (4) | 0.4160 (2) | 0.8667 (2) | 0.058 (1) | |
| C24 | 0.2496 (5) | 0.3751 (2) | 0.9822 (3) | 0.096 (2) | |
| C25 | 0.229 (1) | 0.4085 (5) | 1.0704 (7) | 0.109 (3) | 0.50 |
| C25' | 0.164 (1) | 0.4271 (4) | 1.0034 (9) | 0.109 (3) | 0.50 |
| C26 | 0.2165 (3) | 0.3008 (2) | 0.9817 (2) | 0.0534 (7) | |
| H1w1 | 0.495 (4) | 0.288 (1) | 0.772 (3) | 0.09 (2)* | |
| H1w2 | 0.390 (3) | 0.268 (2) | 0.824 (2) | 0.08 (1)* | |
| H2w1 | 0.428 (4) | 0.204 (2) | 0.671 (2) | 0.10 (2)* | |
| H2w2 | 0.480 (4) | 0.192 (3) | 0.588 (2) | 0.13 (2)* | |
| H1 | 0.8694 | 0.3383 | 0.7068 | 0.084* | |
| H2 | 1.0895 | 0.3199 | 0.7707 | 0.110* | |
| H3 | 1.2582 | 0.2966 | 0.6765 | 0.115* | |
| H4 | 1.2013 | 0.2955 | 0.5198 | 0.099* | |
| H5 | 0.9799 | 0.3161 | 0.4607 | 0.078* | |
| H6 | 0.8038 | 0.4784 | 0.4945 | 0.082* | |
| H7 | 0.9181 | 0.5456 | 0.4023 | 0.099* | |
| H8 | 0.9379 | 0.5137 | 0.2526 | 0.102* | |
| H9 | 0.8346 | 0.4156 | 0.1997 | 0.107* | |
| H10 | 0.7211 | 0.3505 | 0.2976 | 0.086* | |
| H11 | 0.4267 | 0.4358 | 0.4483 | 0.071* | |
| H12 | 0.2273 | 0.4476 | 0.3521 | 0.081* | |
| H13 | 0.1334 | 0.3547 | 0.2743 | 0.098* | |
| H14 | 0.2443 | 0.2526 | 0.2962 | 0.117* | |
| H15 | 0.4427 | 0.2459 | 0.3933 | 0.097* | |
| H17a | 0.5784 | 0.4965 | 0.7386 | 0.072* | |
| H17b | 0.4315 | 0.4660 | 0.7131 | 0.072* | |
| H20 | 0.7197 | 0.3678 | 1.0101 | 0.089* | |
| H21 | 0.5059 | 0.3522 | 1.0634 | 0.095* | |
| H23 | 0.3226 | 0.4279 | 0.8305 | 0.070* | |
| H24 | 0.1833 | 0.3956 | 0.9354 | 0.115* | 0.50 |
| H25a | 0.1351 | 0.4027 | 1.0833 | 0.164* | 0.50 |
| H25b | 0.2489 | 0.4555 | 1.0660 | 0.164* | 0.50 |
| H25c | 0.2896 | 0.3889 | 1.1192 | 0.164* | 0.50 |
| H24' | 0.2889 | 0.3723 | 1.0467 | 0.115* | 0.50 |
| H25d | 0.0902 | 0.4095 | 1.0356 | 0.164* | 0.50 |
| H25e | 0.1252 | 0.4485 | 0.9476 | 0.164* | 0.50 |
| H25f | 0.2154 | 0.4593 | 1.0418 | 0.164* | 0.50 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cd1 | 0.0431 (1) | 0.0434 (1) | 0.0520 (1) | −0.0012 (1) | 0.0077 (1) | 0.0006 (1) |
| O1 | 0.082 (2) | 0.060 (1) | 0.051 (1) | 0.000 (1) | 0.022 (1) | 0.002 (1) |
| O2 | 0.083 (2) | 0.047 (1) | 0.065 (1) | 0.005 (1) | 0.028 (1) | −0.002 (1) |
| O3 | 0.071 (2) | 0.051 (1) | 0.074 (1) | 0.000 (1) | 0.035 (1) | 0.006 (1) |
| O4 | 0.067 (1) | 0.063 (1) | 0.060 (1) | 0.002 (1) | 0.017 (1) | −0.005 (1) |
| O5 | 0.076 (2) | 0.143 (3) | 0.138 (3) | 0.023 (2) | 0.016 (2) | 0.030 (3) |
| O6 | 0.080 (2) | 0.109 (2) | 0.105 (2) | −0.026 (2) | 0.030 (2) | 0.014 (2) |
| O1w | 0.071 (2) | 0.060 (2) | 0.073 (2) | 0.004 (1) | 0.019 (1) | −0.003 (1) |
| O2w | 0.088 (2) | 0.118 (3) | 0.079 (2) | −0.026 (2) | 0.030 (2) | −0.019 (2) |
| N1 | 0.049 (1) | 0.057 (2) | 0.050 (1) | 0.002 (1) | 0.003 (1) | 0.002 (1) |
| N2 | 0.054 (2) | 0.053 (2) | 0.054 (1) | −0.003 (1) | 0.009 (1) | 0.001 (1) |
| N3 | 0.046 (1) | 0.048 (1) | 0.067 (2) | −0.004 (1) | 0.006 (1) | −0.004 (1) |
| N4 | 0.067 (2) | 0.069 (2) | 0.080 (2) | −0.008 (2) | 0.013 (2) | −0.008 (2) |
| C1 | 0.068 (2) | 0.083 (3) | 0.057 (2) | 0.004 (2) | 0.006 (2) | 0.006 (2) |
| C2 | 0.086 (3) | 0.116 (4) | 0.068 (2) | 0.008 (3) | −0.016 (2) | 0.018 (2) |
| C3 | 0.064 (3) | 0.102 (3) | 0.115 (4) | 0.018 (2) | −0.022 (3) | 0.007 (3) |
| C4 | 0.054 (2) | 0.094 (3) | 0.100 (3) | 0.008 (2) | 0.009 (2) | −0.013 (2) |
| C5 | 0.052 (2) | 0.077 (2) | 0.066 (2) | 0.003 (2) | 0.008 (2) | −0.011 (2) |
| C6 | 0.082 (2) | 0.063 (2) | 0.060 (2) | −0.018 (2) | 0.008 (2) | 0.000 (2) |
| C7 | 0.083 (3) | 0.072 (3) | 0.092 (3) | −0.023 (2) | 0.008 (2) | 0.018 (2) |
| C8 | 0.075 (3) | 0.089 (3) | 0.097 (3) | 0.008 (2) | 0.035 (2) | 0.034 (3) |
| C9 | 0.123 (4) | 0.087 (3) | 0.062 (2) | 0.025 (3) | 0.035 (2) | 0.013 (2) |
| C10 | 0.102 (3) | 0.058 (2) | 0.056 (2) | 0.004 (2) | 0.012 (2) | −0.001 (2) |
| C11 | 0.057 (2) | 0.048 (2) | 0.071 (2) | −0.004 (1) | 0.000 (2) | 0.002 (2) |
| C12 | 0.058 (2) | 0.058 (2) | 0.084 (2) | 0.003 (2) | −0.002 (2) | 0.0097 (18) |
| C13 | 0.054 (2) | 0.089 (3) | 0.098 (3) | 0.000 (2) | −0.013 (2) | −0.010 (2) |
| C14 | 0.059 (2) | 0.083 (3) | 0.142 (4) | −0.001 (2) | −0.022 (2) | −0.048 (3) |
| C15 | 0.050 (2) | 0.060 (2) | 0.129 (3) | 0.005 (2) | −0.004 (2) | −0.029 (2) |
| C16 | 0.049 (2) | 0.052 (2) | 0.052 (2) | 0.000 (1) | 0.010 (1) | −0.003 (1) |
| C17 | 0.077 (2) | 0.049 (2) | 0.058 (2) | 0.0072 (16) | 0.023 (2) | 0.003 (1) |
| C18 | 0.066 (2) | 0.036 (1) | 0.056 (2) | −0.003 (1) | 0.021 (1) | −0.004 (1) |
| C19 | 0.062 (2) | 0.049 (2) | 0.059 (2) | −0.009 (2) | 0.019 (1) | −0.006 (1) |
| C20 | 0.091 (3) | 0.073 (2) | 0.057 (2) | −0.003 (2) | 0.002 (2) | 0.004 (2) |
| C21 | 0.114 (3) | 0.065 (2) | 0.064 (2) | −0.003 (2) | 0.040 (2) | 0.013 (2) |
| C22 | 0.083 (3) | 0.041 (2) | 0.083 (2) | −0.002 (2) | 0.041 (2) | 0.002 (2) |
| C23 | 0.065 (2) | 0.042 (2) | 0.071 (2) | 0.000 (1) | 0.021 (2) | −0.003 (1) |
| C24 | 0.115 (3) | 0.046 (2) | 0.143 (4) | −0.005 (2) | 0.085 (3) | −0.003 (2) |
| C25 | 0.105 (6) | 0.064 (4) | 0.169 (7) | −0.014 (4) | 0.062 (5) | −0.018 (5) |
| C25' | 0.105 (6) | 0.064 (4) | 0.169 (7) | −0.014 (4) | 0.062 (5) | −0.018 (5) |
| C26 | 0.056 (2) | 0.045 (2) | 0.061 (2) | 0.002 (1) | 0.017 (1) | 0.003 (1) |
Geometric parameters (Å, º) top
| Cd1—O1 | 2.328 (2) | C18—C23 | 1.381 (4) |
| Cd1—O2 | 2.579 (2) | C18—C19 | 1.392 (5) |
| Cd1—O3i | 2.293 (2) | C19—C20 | 1.375 (5) |
| Cd1—N1 | 2.364 (3) | C20—C21 | 1.387 (6) |
| Cd1—N2 | 2.403 (3) | C21—C22 | 1.380 (6) |
| Cd1—N3 | 2.330 (3) | C22—C23 | 1.376 (5) |
| O1—C16 | 1.254 (3) | C22—C24 | 1.519 (5) |
| O2—C16 | 1.241 (4) | C24—C25 | 1.481 (8) |
| O3—C26 | 1.249 (4) | C24—C26 | 1.515 (5) |
| O4—C26 | 1.236 (4) | C1—H1 | 0.9300 |
| O5—N4 | 1.201 (4) | C2—H2 | 0.9300 |
| O6—N4 | 1.209 (4) | C3—H3 | 0.9300 |
| O1w—H1w1 | 0.85 (1) | C4—H4 | 0.9300 |
| O1w—H1w2 | 0.85 (1) | C5—H5 | 0.9300 |
| O2w—H2w1 | 0.86 (1) | C6—H6 | 0.9300 |
| O2w—H2w2 | 0.86 (1) | C7—H7 | 0.9300 |
| N1—C1 | 1.322 (4) | C8—H8 | 0.9300 |
| N1—C5 | 1.337 (4) | C9—H9 | 0.9300 |
| N2—C10 | 1.313 (4) | C10—H10 | 0.9300 |
| N2—C6 | 1.330 (4) | C11—H11 | 0.9300 |
| N3—C15 | 1.327 (4) | C12—H12 | 0.9300 |
| N3—C11 | 1.329 (4) | C13—H13 | 0.9300 |
| N4—C19 | 1.477 (4) | C14—H14 | 0.9300 |
| C1—C2 | 1.364 (6) | C15—H15 | 0.9300 |
| C2—C3 | 1.351 (7) | C17—H17a | 0.9700 |
| C3—C4 | 1.357 (6) | C17—H17b | 0.9700 |
| C4—C5 | 1.359 (5) | C20—H20 | 0.9300 |
| C6—C7 | 1.360 (5) | C21—H21 | 0.9300 |
| C7—C8 | 1.356 (6) | C23—H23 | 0.9300 |
| C8—C9 | 1.349 (6) | C24—H24 | 0.9800 |
| C9—C10 | 1.381 (5) | C25—H25a | 0.9600 |
| C11—C12 | 1.374 (5) | C25—H25b | 0.9600 |
| C12—C13 | 1.362 (5) | C25—H25c | 0.9600 |
| C13—C14 | 1.366 (6) | C25'—H25d | 0.9600 |
| C14—C15 | 1.367 (5) | C25'—H25e | 0.9600 |
| C16—C17 | 1.521 (4) | C25'—H25F | 0.9600 |
| C17—C18 | 1.511 (4) | | |
| | | |
| O1—Cd1—O2 | 52.75 (7) | C9—C10—H10 | 118.6 |
| O1—Cd1—O3i | 137.18 (7) | N3—C11—C12 | 123.2 (3) |
| O1—Cd1—N1 | 97.66 (9) | N3—C11—H11 | 118.4 |
| O1—Cd1—N2 | 85.65 (8) | C12—C11—H11 | 118.4 |
| O1—Cd1—N3 | 90.01 (9) | C13—C12—C11 | 118.7 (3) |
| O2—Cd1—O3i | 84.75 (7) | C13—C12—H12 | 120.6 |
| O2—Cd1—N1 | 90.61 (9) | C11—C12—H12 | 120.6 |
| O2—Cd1—N2 | 137.06 (8) | C12—C13—C14 | 118.7 (4) |
| O2—Cd1—N3 | 101.64 (9) | C12—C13—H13 | 120.6 |
| O3i—Cd1—N1 | 86.50 (9) | C14—C13—H13 | 120.6 |
| O3i—Cd1—N2 | 137.08 (8) | C15—C14—C13 | 119.1 (4) |
| O3i—Cd1—N3 | 94.37 (9) | C15—C14—H14 | 120.4 |
| N1—Cd1—N2 | 84.35 (9) | C13—C14—H14 | 120.4 |
| N1—Cd1—N3 | 167.75 (9) | N3—C15—C14 | 123.2 (4) |
| N2—Cd1—N3 | 86.74 (9) | N3—C15—H15 | 118.4 |
| C16—O1—Cd1 | 97.9 (2) | C14—C15—H15 | 118.4 |
| C16—O2—Cd1 | 86.4 (2) | O2—C16—O1 | 122.9 (3) |
| C26—O3—Cd1ii | 101.3 (2) | O2—C16—C17 | 120.0 (3) |
| H1w1—O1w—H1w2 | 110 (2) | O1—C16—C17 | 117.1 (3) |
| H2w1—O2w—H2w2 | 107 (2) | C18—C17—C16 | 114.9 (3) |
| C1—N1—C5 | 117.2 (3) | C18—C17—H17a | 108.5 |
| C1—N1—Cd1 | 120.4 (2) | C16—C17—H17a | 108.5 |
| C5—N1—Cd1 | 122.0 (2) | C18—C17—H17b | 108.5 |
| C10—N2—C6 | 116.9 (3) | C16—C17—H17b | 108.5 |
| C10—N2—Cd1 | 124.7 (2) | H17a—C17—H17b | 107.5 |
| C6—N2—Cd1 | 117.7 (2) | C23—C18—C19 | 116.1 (3) |
| C15—N3—C11 | 117.0 (3) | C23—C18—C17 | 118.3 (3) |
| C15—N3—Cd1 | 122.8 (2) | C19—C18—C17 | 125.5 (3) |
| C11—N3—Cd1 | 119.9 (2) | C20—C19—C18 | 122.7 (3) |
| O5—N4—O6 | 122.1 (4) | C20—C19—N4 | 116.8 (3) |
| O5—N4—C19 | 118.2 (3) | C18—C19—N4 | 120.5 (3) |
| O6—N4—C19 | 119.7 (3) | C19—C20—C21 | 118.5 (4) |
| N1—C1—C2 | 123.3 (4) | C19—C20—H20 | 120.7 |
| N1—C1—H1 | 118.4 | C21—C20—H20 | 120.7 |
| C2—C1—H1 | 118.4 | C20—C21—C22 | 121.1 (3) |
| C3—C2—C1 | 118.8 (4) | C20—C21—H21 | 119.5 |
| C3—C2—H2 | 120.6 | C22—C21—H21 | 119.5 |
| C1—C2—H2 | 120.6 | C23—C22—C21 | 118.0 (3) |
| C2—C3—C4 | 118.9 (4) | C23—C22—C24 | 119.7 (4) |
| C2—C3—H3 | 120.6 | C21—C22—C24 | 122.2 (4) |
| C4—C3—H3 | 120.6 | C18—C23—C22 | 123.5 (3) |
| C5—C4—C3 | 119.6 (4) | C18—C23—H23 | 118.2 |
| C5—C4—H4 | 120.2 | C22—C23—H23 | 118.2 |
| C3—C4—H4 | 120.2 | C25—C24—C26 | 113.3 (5) |
| N1—C5—C4 | 122.1 (4) | C25—C24—C22 | 112.7 (5) |
| N1—C5—H5 | 118.9 | C26—C24—C22 | 111.5 (3) |
| C4—C5—H5 | 118.9 | C25—C24—H24 | 106.2 |
| N2—C6—C7 | 123.3 (4) | C26—C24—H24 | 106.2 |
| N2—C6—H6 | 118.4 | C22—C24—H24 | 106.2 |
| C7—C6—H6 | 118.4 | C24—C25—H25a | 109.5 |
| C8—C7—C6 | 119.3 (4) | C24—C25—H25b | 109.5 |
| C8—C7—H7 | 120.3 | H25a—C25—H25b | 109.5 |
| C6—C7—H7 | 120.3 | C24—C25—H25c | 109.5 |
| C9—C8—C7 | 118.3 (4) | H25a—C25—H25c | 109.5 |
| C9—C8—H8 | 120.8 | H25b—C25—H25c | 109.5 |
| C7—C8—H8 | 120.8 | H25d—C25'—H25e | 109.5 |
| C8—C9—C10 | 119.4 (4) | H25d—C25'—H25F | 109.5 |
| C8—C9—H9 | 120.3 | H25e—C25'—H25F | 109.5 |
| C10—C9—H9 | 120.3 | O4—C26—O3 | 123.6 (3) |
| N2—C10—C9 | 122.7 (4) | O4—C26—C24 | 118.1 (3) |
| N2—C10—H10 | 118.6 | O3—C26—C24 | 118.2 (3) |
| | | |
| O3i—Cd1—O1—C16 | 6.1 (3) | N2—C6—C7—C8 | −0.2 (7) |
| N3—Cd1—O1—C16 | 102.5 (2) | C6—C7—C8—C9 | 1.0 (7) |
| N1—Cd1—O1—C16 | −87.1 (2) | C7—C8—C9—C10 | −1.1 (7) |
| N2—Cd1—O1—C16 | −170.8 (2) | C6—N2—C10—C9 | 0.6 (6) |
| O2—Cd1—O1—C16 | −2.16 (18) | Cd1—N2—C10—C9 | −169.9 (3) |
| O3i—Cd1—O2—C16 | −172.2 (2) | C8—C9—C10—N2 | 0.2 (7) |
| O1—Cd1—O2—C16 | 2.16 (18) | C15—N3—C11—C12 | −0.1 (5) |
| N3—Cd1—O2—C16 | −78.8 (2) | Cd1—N3—C11—C12 | 174.4 (3) |
| N1—Cd1—O2—C16 | 101.3 (2) | N3—C11—C12—C13 | 0.1 (6) |
| N2—Cd1—O2—C16 | 19.0 (3) | C11—C12—C13—C14 | 0.0 (7) |
| O3i—Cd1—N1—C1 | −82.8 (3) | C12—C13—C14—C15 | 0.0 (8) |
| O1—Cd1—N1—C1 | 54.4 (3) | C11—N3—C15—C14 | 0.1 (6) |
| N3—Cd1—N1—C1 | −177.3 (4) | Cd1—N3—C15—C14 | −174.3 (4) |
| N2—Cd1—N1—C1 | 139.2 (3) | C13—C14—C15—N3 | 0.0 (8) |
| O2—Cd1—N1—C1 | 1.9 (3) | Cd1—O2—C16—O1 | −3.8 (3) |
| O3i—Cd1—N1—C5 | 90.8 (3) | Cd1—O2—C16—C17 | 173.9 (3) |
| O1—Cd1—N1—C5 | −132.0 (3) | Cd1—O1—C16—O2 | 4.2 (4) |
| N3—Cd1—N1—C5 | −3.7 (6) | Cd1—O1—C16—C17 | −173.5 (2) |
| N2—Cd1—N1—C5 | −47.2 (3) | O2—C16—C17—C18 | 18.3 (5) |
| O2—Cd1—N1—C5 | 175.5 (3) | O1—C16—C17—C18 | −163.8 (3) |
| O3i—Cd1—N2—C10 | 27.4 (3) | C16—C17—C18—C23 | −115.7 (3) |
| O1—Cd1—N2—C10 | −155.7 (3) | C16—C17—C18—C19 | 61.0 (4) |
| N3—Cd1—N2—C10 | −65.5 (3) | C23—C18—C19—C20 | −0.3 (5) |
| N1—Cd1—N2—C10 | 106.1 (3) | C17—C18—C19—C20 | −177.0 (3) |
| O2—Cd1—N2—C10 | −169.1 (3) | C23—C18—C19—N4 | 178.7 (3) |
| O3i—Cd1—N2—C6 | −143.0 (2) | C17—C18—C19—N4 | 1.9 (5) |
| O1—Cd1—N2—C6 | 33.8 (3) | O5—N4—C19—C20 | 24.5 (5) |
| N3—Cd1—N2—C6 | 124.1 (3) | O6—N4—C19—C20 | −157.3 (4) |
| N1—Cd1—N2—C6 | −64.3 (3) | O5—N4—C19—C18 | −154.5 (4) |
| O2—Cd1—N2—C6 | 20.5 (3) | O6—N4—C19—C18 | 23.7 (5) |
| O3i—Cd1—N3—C15 | −25.2 (3) | C18—C19—C20—C21 | −0.2 (5) |
| O1—Cd1—N3—C15 | −162.6 (3) | N4—C19—C20—C21 | −179.2 (3) |
| N1—Cd1—N3—C15 | 68.5 (5) | C19—C20—C21—C22 | 0.7 (6) |
| N2—Cd1—N3—C15 | 111.8 (3) | C20—C21—C22—C23 | −0.7 (6) |
| O2—Cd1—N3—C15 | −110.7 (3) | C20—C21—C22—C24 | 176.7 (3) |
| O3i—Cd1—N3—C11 | 160.6 (3) | C19—C18—C23—C22 | 0.3 (5) |
| O1—Cd1—N3—C11 | 23.2 (3) | C17—C18—C23—C22 | 177.2 (3) |
| N1—Cd1—N3—C11 | −105.7 (4) | C21—C22—C23—C18 | 0.2 (5) |
| N2—Cd1—N3—C11 | −62.4 (3) | C24—C22—C23—C18 | −177.3 (3) |
| O2—Cd1—N3—C11 | 75.1 (3) | C23—C22—C24—C25 | −123.6 (6) |
| C5—N1—C1—C2 | −1.0 (6) | C21—C22—C24—C25 | 59.0 (7) |
| Cd1—N1—C1—C2 | 172.9 (3) | C23—C22—C24—C26 | 107.6 (4) |
| N1—C1—C2—C3 | −0.3 (7) | C21—C22—C24—C26 | −69.7 (5) |
| C1—C2—C3—C4 | 1.0 (8) | Cd1ii—O3—C26—O4 | 10.5 (4) |
| C2—C3—C4—C5 | −0.4 (8) | Cd1ii—O3—C26—C24 | −171.4 (3) |
| C1—N1—C5—C4 | 1.6 (5) | C25—C24—C26—O4 | −170.3 (6) |
| Cd1—N1—C5—C4 | −172.2 (3) | C22—C24—C26—O4 | −41.9 (5) |
| C3—C4—C5—N1 | −0.9 (7) | C25—C24—C26—O3 | 11.5 (7) |
| C10—N2—C6—C7 | −0.7 (6) | C22—C24—C26—O3 | 139.9 (4) |
| Cd1—N2—C6—C7 | 170.5 (3) | | |
| Symmetry codes: (i) x+1/2, −y+1/2, z−1/2; (ii) x−1/2, −y+1/2, z+1/2. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1w—H1w1···O2 | 0.85 (1) | 2.08 (2) | 2.882 (4) | 159 (4) |
| O1w—H1w2···O4 | 0.85 (1) | 2.04 (1) | 2.851 (3) | 160 (3) |
| O2w—H2w1···O1w | 0.86 (1) | 2.00 (1) | 2.860 (4) | 176 (4) |
| O2w—H2w2···O3i | 0.86 (1) | 1.98 (2) | 2.812 (4) | 165 (4) |
| Symmetry code: (i) x+1/2, −y+1/2, z−1/2. |
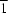 ]. The octahedral geometry of the Cd atom is completed by the N atoms of three pyridine N-atom donors.
]. The octahedral geometry of the Cd atom is completed by the N atoms of three pyridine N-atom donors. 
 journal menu
journal menu











 Alert level C
Alert level C



