Buy article online - an online subscription or single-article purchase is required to access this article.
A new azaanthracene alkaloid, namely 9,10-dimethoxy-4-methyl-1,2-dihydro-1-azaanthracen-2-one (kalasinamide), C
16H
15NO
3, has been isolated from the stems of
Polyalthia suberosa collected in the northeastern part of Thailand. Each of the aromatic rings in the molecule is planar within
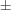
0.021 (2) Å. Molecules are linked to form centrosymmetric dimers by N-H

O hydrogen bonds [N

O 2.941 (4) Å].
Supporting information
CCDC reference: 147701
The orange title compound was crystallized from EtOH–CH2Cl2.
Based on a statistical analysis of intensity distribution, the space group was determined to be P21/a and has been transformed to the conventional space group of P21/c. Friedel opposites were collected and merged. The H atoms were allowed in the refinement at geometrically idealized positions.
Data collection: CAD-4 Software (Enraf-Nonius, 1989); cell refinement: CAD-4 Software; data reduction: maXus (Mackay et al., 1999); program(s) used to solve structure: SHELXS97 (Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997).
Crystal data top
| C16H15NO3 | Dx = 1.380 Mg m−3 |
| Mr = 269.30 | Mo Kα radiation, λ = 0.70930 Å |
| Monoclinic, P21/c | Cell parameters from 9 reflections |
| a = 10.005 (5) Å | θ = 8.8–11.7° |
| b = 16.449 (5) Å | µ = 0.10 mm−1 |
| c = 8.186 (5) Å | T = 298 K |
| β = 105.8 (2)° | Needle, orange |
| V = 1297.0 (10) Å3 | 0.25 × 0.04 × 0.03 mm |
| Z = 4 | |
Data collection top
Enraf-Nonius CAD-4
diffractometer | θmax = 24.9° |
| Radiation source: rotating anode generator | h = −11→11 |
| ω–2θ scans | k = −19→19 |
| 9602 measured reflections | l = −9→9 |
| 2272 independent reflections | 3 standard reflections every 2608 min |
| 1785 reflections with I > 2σ(I) | intensity decay: 0.8% |
| Rint = 0.066 | |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.053 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.163 | H-atom parameters constrained |
| S = 1.07 | Calculated w = 1/[σ2(Fo2) + (0.0824P)2 + 0.7373P]
where P = (Fo2 + 2Fc2)/3 |
| 2272 reflections | (Δ/σ)max = 0.001 |
| 184 parameters | Δρmax = 0.24 e Å−3 |
| 0 restraints | Δρmin = −0.29 e Å−3 |
Crystal data top
| C16H15NO3 | V = 1297.0 (10) Å3 |
| Mr = 269.30 | Z = 4 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 10.005 (5) Å | µ = 0.10 mm−1 |
| b = 16.449 (5) Å | T = 298 K |
| c = 8.186 (5) Å | 0.25 × 0.04 × 0.03 mm |
| β = 105.8 (2)° | |
Data collection top
Enraf-Nonius CAD-4
diffractometer | Rint = 0.066 |
| 9602 measured reflections | 3 standard reflections every 2608 min |
| 2272 independent reflections | intensity decay: 0.8% |
| 1785 reflections with I > 2σ(I) | |
Refinement top
| R[F2 > 2σ(F2)] = 0.053 | 0 restraints |
| wR(F2) = 0.163 | H-atom parameters constrained |
| S = 1.07 | Δρmax = 0.24 e Å−3 |
| 2272 reflections | Δρmin = −0.29 e Å−3 |
| 184 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.19412 (15) | 0.53442 (9) | 0.14924 (18) | 0.0430 (4) | |
| O2 | 0.66117 (17) | 0.52286 (12) | 0.4852 (2) | 0.0579 (5) | |
| O3 | 0.48398 (17) | 0.70615 (9) | −0.22291 (19) | 0.0487 (4) | |
| N1 | 0.47139 (18) | 0.55055 (11) | 0.2710 (2) | 0.0407 (5) | |
| H1 | 0.4206 | 0.5260 | 0.3250 | 0.049* | |
| C2 | 0.6097 (2) | 0.55317 (14) | 0.3438 (3) | 0.0450 (5) | |
| C3 | 0.6901 (2) | 0.59236 (15) | 0.2461 (3) | 0.0496 (6) | |
| H3 | 0.7860 | 0.5936 | 0.2908 | 0.060* | |
| C4 | 0.6361 (2) | 0.62745 (14) | 0.0946 (3) | 0.0445 (5) | |
| C5 | 0.1947 (3) | 0.69683 (14) | −0.3442 (3) | 0.0494 (6) | |
| H5 | 0.2425 | 0.7285 | −0.4038 | 0.059* | |
| C6 | 0.0547 (3) | 0.68918 (16) | −0.4028 (3) | 0.0576 (7) | |
| H6 | 0.0077 | 0.7157 | −0.5020 | 0.069* | |
| C7 | −0.0195 (3) | 0.64184 (18) | −0.3157 (3) | 0.0589 (7) | |
| H7 | −0.1149 | 0.6357 | −0.3596 | 0.071* | |
| C8 | 0.0469 (2) | 0.60464 (15) | −0.1668 (3) | 0.0489 (6) | |
| H8 | −0.0036 | 0.5742 | −0.1086 | 0.059* | |
| C9 | 0.2642 (2) | 0.57739 (12) | 0.0551 (3) | 0.0374 (5) | |
| C10 | 0.4140 (2) | 0.66153 (12) | −0.1314 (3) | 0.0393 (5) | |
| C11 | 0.7350 (3) | 0.6661 (2) | 0.0077 (4) | 0.0697 (8) | |
| H11A | 0.8264 | 0.6677 | 0.0847 | 0.104* | |
| H11B | 0.7047 | 0.7204 | −0.0263 | 0.104* | |
| H11C | 0.7368 | 0.6347 | −0.0905 | 0.104* | |
| C12 | 0.1369 (3) | 0.58426 (17) | 0.2573 (3) | 0.0548 (6) | |
| H12A | 0.2072 | 0.6205 | 0.3209 | 0.082* | |
| H12B | 0.1046 | 0.5503 | 0.3340 | 0.082* | |
| H12C | 0.0607 | 0.6153 | 0.1893 | 0.082* | |
| C13 | 0.5062 (4) | 0.66468 (17) | −0.3667 (4) | 0.0671 (8) | |
| H13A | 0.5741 | 0.6227 | −0.3289 | 0.101* | |
| H13B | 0.5388 | 0.7026 | −0.4362 | 0.101* | |
| H13C | 0.4205 | 0.6410 | −0.4314 | 0.101* | |
| C41 | 0.4869 (2) | 0.62533 (12) | 0.0213 (3) | 0.0377 (5) | |
| C81 | 0.1926 (2) | 0.61220 (13) | −0.1007 (3) | 0.0396 (5) | |
| C91 | 0.4058 (2) | 0.58486 (12) | 0.1147 (2) | 0.0360 (5) | |
| C101 | 0.2686 (2) | 0.65706 (12) | −0.1931 (3) | 0.0404 (5) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0419 (8) | 0.0495 (8) | 0.0414 (8) | −0.0065 (6) | 0.0178 (7) | 0.0028 (6) |
| O2 | 0.0444 (9) | 0.0837 (13) | 0.0440 (9) | 0.0057 (8) | 0.0092 (7) | 0.0177 (9) |
| O3 | 0.0615 (10) | 0.0490 (9) | 0.0432 (9) | −0.0067 (7) | 0.0274 (8) | 0.0038 (7) |
| N1 | 0.0371 (10) | 0.0512 (10) | 0.0361 (9) | 0.0001 (8) | 0.0136 (8) | 0.0078 (8) |
| C2 | 0.0392 (12) | 0.0562 (13) | 0.0405 (12) | 0.0033 (10) | 0.0121 (10) | 0.0025 (10) |
| C3 | 0.0339 (11) | 0.0658 (15) | 0.0498 (13) | −0.0012 (10) | 0.0125 (10) | 0.0021 (11) |
| C4 | 0.0405 (12) | 0.0513 (12) | 0.0465 (13) | −0.0001 (10) | 0.0202 (10) | 0.0024 (10) |
| C5 | 0.0629 (15) | 0.0504 (13) | 0.0355 (11) | 0.0038 (11) | 0.0146 (11) | 0.0018 (10) |
| C6 | 0.0631 (16) | 0.0673 (16) | 0.0375 (12) | 0.0133 (13) | 0.0052 (11) | 0.0063 (11) |
| C7 | 0.0458 (13) | 0.0809 (17) | 0.0437 (13) | 0.0043 (12) | 0.0013 (11) | −0.0008 (12) |
| C8 | 0.0435 (12) | 0.0611 (14) | 0.0417 (12) | −0.0025 (11) | 0.0105 (10) | −0.0038 (10) |
| C9 | 0.0390 (11) | 0.0413 (10) | 0.0345 (10) | −0.0031 (9) | 0.0145 (9) | 0.0001 (8) |
| C10 | 0.0484 (12) | 0.0397 (11) | 0.0347 (11) | −0.0018 (9) | 0.0198 (9) | −0.0005 (8) |
| C11 | 0.0439 (14) | 0.096 (2) | 0.0766 (19) | −0.0019 (14) | 0.0284 (13) | 0.0244 (16) |
| C12 | 0.0500 (14) | 0.0719 (16) | 0.0491 (14) | 0.0083 (12) | 0.0249 (11) | 0.0060 (12) |
| C13 | 0.095 (2) | 0.0667 (16) | 0.0551 (16) | −0.0054 (15) | 0.0461 (15) | 0.0016 (13) |
| C41 | 0.0406 (11) | 0.0396 (11) | 0.0370 (11) | −0.0009 (9) | 0.0173 (9) | −0.0032 (8) |
| C81 | 0.0417 (11) | 0.0445 (11) | 0.0331 (11) | −0.0014 (9) | 0.0110 (9) | −0.0052 (9) |
| C91 | 0.0405 (11) | 0.0387 (10) | 0.0301 (10) | 0.0012 (9) | 0.0120 (8) | −0.0015 (8) |
| C101 | 0.0501 (13) | 0.0405 (11) | 0.0319 (11) | 0.0007 (9) | 0.0130 (9) | −0.0021 (8) |
Geometric parameters (Å, º) top
| O1—C9 | 1.371 (3) | C7—H7 | 0.9300 |
| O1—C12 | 1.435 (3) | C8—C81 | 1.416 (4) |
| O2—C2 | 1.237 (3) | C8—H8 | 0.9300 |
| O3—C10 | 1.371 (3) | C9—C91 | 1.374 (3) |
| O3—C13 | 1.429 (3) | C9—C81 | 1.404 (4) |
| N1—C2 | 1.351 (4) | C10—C41 | 1.398 (4) |
| N1—C91 | 1.389 (3) | C10—C101 | 1.406 (4) |
| N1—H1 | 0.8600 | C11—H11A | 0.9600 |
| C2—C3 | 1.433 (4) | C11—H11B | 0.9600 |
| C3—C4 | 1.342 (4) | C11—H11C | 0.9600 |
| C3—H3 | 0.9300 | C12—H12A | 0.9600 |
| C4—C41 | 1.451 (4) | C12—H12B | 0.9600 |
| C4—C11 | 1.508 (4) | C12—H12C | 0.9600 |
| C5—C6 | 1.357 (4) | C13—H13A | 0.9600 |
| C5—C101 | 1.417 (4) | C13—H13B | 0.9600 |
| C5—H5 | 0.9300 | C13—H13C | 0.9600 |
| C6—C7 | 1.398 (4) | C41—C91 | 1.423 (3) |
| C6—H6 | 0.9300 | C81—C101 | 1.417 (3) |
| C7—C8 | 1.365 (4) | | |
| | | |
| C9—O1—C12 | 113.78 (18) | C41—C10—C101 | 122.4 (2) |
| C10—O3—C13 | 114.35 (19) | C4—C11—H11A | 109.5 |
| C2—N1—C91 | 124.3 (2) | C4—C11—H11B | 109.5 |
| C2—N1—H1 | 117.8 | H11A—C11—H11B | 109.5 |
| C91—N1—H1 | 117.8 | C4—C11—H11C | 109.5 |
| O2—C2—N1 | 121.1 (3) | H11A—C11—H11C | 109.5 |
| O2—C2—C3 | 123.4 (2) | H11B—C11—H11C | 109.5 |
| N1—C2—C3 | 115.6 (2) | O1—C12—H12A | 109.5 |
| C4—C3—C2 | 124.3 (2) | O1—C12—H12B | 109.5 |
| C4—C3—H3 | 117.8 | H12A—C12—H12B | 109.5 |
| C2—C3—H3 | 117.8 | O1—C12—H12C | 109.5 |
| C3—C4—C41 | 118.8 (3) | H12A—C12—H12C | 109.5 |
| C3—C4—C11 | 117.8 (2) | H12B—C12—H12C | 109.5 |
| C41—C4—C11 | 123.3 (2) | O3—C13—H13A | 109.5 |
| C6—C5—C101 | 120.6 (3) | O3—C13—H13B | 109.5 |
| C6—C5—H5 | 119.7 | H13A—C13—H13B | 109.5 |
| C101—C5—H5 | 119.7 | O3—C13—H13C | 109.5 |
| C5—C6—C7 | 120.8 (2) | H13A—C13—H13C | 109.5 |
| C5—C6—H6 | 119.6 | H13B—C13—H13C | 109.5 |
| C7—C6—H6 | 119.6 | C10—C41—C91 | 116.4 (2) |
| C8—C7—C6 | 120.6 (2) | C10—C41—C4 | 126.0 (2) |
| C8—C7—H7 | 119.7 | C91—C41—C4 | 117.5 (2) |
| C6—C7—H7 | 119.7 | C9—C81—C8 | 122.0 (3) |
| C7—C8—C81 | 120.2 (3) | C9—C81—C101 | 118.8 (2) |
| C7—C8—H8 | 119.9 | C8—C81—C101 | 119.2 (2) |
| C81—C8—H8 | 119.9 | C9—C91—N1 | 118.3 (2) |
| O1—C9—C91 | 118.9 (2) | C9—C91—C41 | 122.3 (2) |
| O1—C9—C81 | 120.6 (2) | N1—C91—C41 | 119.3 (2) |
| C91—C9—C81 | 120.6 (2) | C10—C101—C5 | 122.2 (3) |
| O3—C10—C41 | 119.8 (2) | C10—C101—C81 | 119.2 (2) |
| O3—C10—C101 | 117.7 (2) | C5—C101—C81 | 118.6 (2) |
| | | |
| C91—N1—C2—O2 | 178.4 (2) | C91—C9—C81—C101 | −1.7 (3) |
| C91—N1—C2—C3 | −2.0 (3) | C7—C8—C81—C9 | 177.6 (2) |
| O2—C2—C3—C4 | −178.2 (2) | C7—C8—C81—C101 | −1.6 (3) |
| N1—C2—C3—C4 | 2.2 (4) | O1—C9—C91—N1 | −2.0 (3) |
| C2—C3—C4—C41 | −0.9 (4) | C81—C9—C91—N1 | 178.85 (18) |
| C2—C3—C4—C11 | −179.9 (2) | O1—C9—C91—C41 | 177.16 (17) |
| C101—C5—C6—C7 | −0.1 (4) | C81—C9—C91—C41 | −2.0 (3) |
| C5—C6—C7—C8 | 2.2 (4) | C2—N1—C91—C9 | 179.7 (2) |
| C6—C7—C8—C81 | −1.4 (4) | C2—N1—C91—C41 | 0.5 (3) |
| C12—O1—C9—C91 | 95.8 (2) | C10—C41—C91—C9 | 3.5 (3) |
| C12—O1—C9—C81 | −85.0 (2) | C4—C41—C91—C9 | −178.24 (19) |
| C13—O3—C10—C41 | 102.3 (3) | C10—C41—C91—N1 | −177.30 (17) |
| C13—O3—C10—C101 | −80.4 (3) | C4—C41—C91—N1 | 0.9 (3) |
| O3—C10—C41—C91 | 175.73 (17) | O3—C10—C101—C5 | −0.1 (3) |
| C101—C10—C41—C91 | −1.5 (3) | C41—C10—C101—C5 | 177.21 (19) |
| O3—C10—C41—C4 | −2.3 (3) | O3—C10—C101—C81 | −179.29 (17) |
| C101—C10—C41—C4 | −179.5 (2) | C41—C10—C101—C81 | −2.0 (3) |
| C3—C4—C41—C10 | 177.3 (2) | C6—C5—C101—C10 | 178.0 (2) |
| C11—C4—C41—C10 | −3.7 (4) | C6—C5—C101—C81 | −2.8 (3) |
| C3—C4—C41—C91 | −0.7 (3) | C9—C81—C101—C10 | 3.6 (3) |
| C11—C4—C41—C91 | 178.2 (2) | C8—C81—C101—C10 | −177.1 (2) |
| O1—C9—C81—C8 | 0.0 (3) | C9—C81—C101—C5 | −175.63 (19) |
| C91—C9—C81—C8 | 179.1 (2) | C8—C81—C101—C5 | 3.6 (3) |
| O1—C9—C81—C101 | 179.19 (18) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O2i | 0.86 | 2.10 | 2.941 (4) | 164 |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Experimental details
| Crystal data |
| Chemical formula | C16H15NO3 |
| Mr | 269.30 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 298 |
| a, b, c (Å) | 10.005 (5), 16.449 (5), 8.186 (5) |
| β (°) | 105.8 (2) |
| V (Å3) | 1297.0 (10) |
| Z | 4 |
| Radiation type | Mo Kα |
| µ (mm−1) | 0.10 |
| Crystal size (mm) | 0.25 × 0.04 × 0.03 |
| |
| Data collection |
| Diffractometer | Enraf-Nonius CAD-4
diffractometer |
| Absorption correction | – |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 9602, 2272, 1785 |
| Rint | 0.066 |
| (sin θ/λ)max (Å−1) | 0.594 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.053, 0.163, 1.07 |
| No. of reflections | 2272 |
| No. of parameters | 184 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.24, −0.29 |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O2i | 0.86 | 2.10 | 2.941 (4) | 164.3 |
| Symmetry code: (i) −x+1, −y+1, −z+1. |

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.
 O hydrogen bonds [N
O hydrogen bonds [N O 2.941 (4) Å].
O 2.941 (4) Å].
 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
 journal menu
journal menu









 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry



Polyalthia suberosa (Roxb.), a shrubby tree native to southeast Asia and South China, has been under wide investigation since 1985. Various parts of the tree, leaves, stems and barks have been investigated. The compounds identified are coumarin, sterols (Dan et al., 1985; Goyal et al., 1986), alkaloids (Ferdous et al., 1992; Sahai et al., 1996), triterpenes (Goyal et al., 1986; Li et al., 1993) and nitrogen heterocyclic compounds (Sahai et al., 1996, Tuchinda et al., 2000). Studies of suberosa has shown an anti-HIV principle (Li et al.,1993).
The title compound, (I), extracted from the stem of the tree from Kalasin Province in the northeastern part of Thailand was found to be a new azaanthracene alkaloid (Tuchinda et al., 2000). We therefore report the structure of the title compound as this might lead to a new anti-HIV active compound.
Bond lengths and angles within the anthracene rings system of (I) are in good agreement with literature values (Brown et al., 1964). However, the C2═O2 bond length of 1.237 (6) Å is slightly longer than that normally found in ketones. Also, the N1—C2 and N1—C91 mean distance of 1.373 (6) Å is slightly shorter than C—N bond length in heteroaromatic systems (Allen et al., 1987). Appreciable electron delocalizations therefore occur here. Noticable electron delocalizations also occur at O1 and O3; the C9—O1 and O1—C12 bond lengths are 1.373 (6) and 1.435 (1) Å, and C10—O3 and O3—C13 are 1.369 (5) and 1.430 (2) Å, respectively. Mean planes through each of the three fused six-membered rings show that they are planar within ±0.02 Å. The dihedral angles A/B 3.00, B/C 2.2 and A/C 5.15° [A = C5—C6—C7—C8—C81—C101, B = C81—C9—C91—C41—C10—C101 and C = C91—N1—C2—C3—C4—C41] indicate that the planes are not quite coplanar. Mean-plane calculation through the 14-membered fused A/B/C ring shows a total puckering amplitude of 0.166 (3) Å. The torsion angles C13—O3—C10—C41 and C12—O1—C9—C91 of 102.5 (3) and 94.8 (3)°, respectively, are as expected. The methyl group at C4 has a steric effect on the methoxy group at C10. Molecules are linked by N—H···O hydrogen bonds about inversion centres to form centrosymmetric dimers with an N···O distance of 2.941 (4) Å.