The title compounds, C
27H
26Cl
2NO
5PS, (I), and C
27H
27BrNO
5PS, (II), respectively, crystallize in the centrosymmetric space group
P2
1/
n with one molecule in the asymmetric unit in each case. The dihedral angle between the benzene and pyrrole rings is 2.1 (1)° in (I) and 0.9 (2)° in (II). The phenylsulfonyl group is orthogonal to the halophenyl moiety, with a dihedral angle of 82.0 (1)° in (I) and 78.7 (2)° in (II). In both compounds, the molecular structures and packing are stabilized by C—H

O and C—H

halogen interactions. The intermolecular hydrogen bonds in (I) form cyclic dimers with graph-set descriptors
R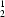
(10) and
R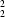
(8) about a 2
1 axis, and those in (II) form a
C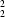
(20) chain.
Supporting information
CCDC references: 193415; 193416
A mixture of 2-bromomethyl-1-phenylsulfonyl-3-(β-arylvinyl)indole (5 mmol) and
triethylphosphite (1.5 g, 9 mmol) was heated under nitrogen at 543 K for 3 h.
The sticky oil was then poured over ice (200 g) and acidified with
concentrated HCl (1 ml). The solid which precipitated was filtered and dried
over calcium chloride. The crude product was recrystallized from ethyl acetate
to give the phosphonate esters, (I) and (II), as colourless crystalline
solids. How were the two compounds separated?
All H atoms were geometrically fixed and allowed to ride on their parent atoms,
with C—H = 0.93–0.97 Å, and with Uiso(H) = 1.5eq(C) for methyl
H atoms and 1.2Ueq(C) for other H atoms.
For both compounds, data collection: CAD-4 EXPRESS (Enraf-Nonius, 1994); cell refinement: CAD-4 EXPRESS; data reduction: XCAD4 (Harms & Wocadlo, 1995); program(s) used to solve structure: SHELXS97 (Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: ZORTEP (Zsolnai, 1997) and PLATON (Spek, 2000); software used to prepare material for publication: SHELX97 and PARST (Nardelli, 1995).
(I) Diethyl
{3-[
β-(2,4-dichlorophenyl)vinyl]-
N-phenylsulfonyl-indol-2-ylmethyl}phosphonate
top
Crystal data top
| C27H26Cl2NO5PS | F(000) = 1200 |
| Mr = 578.42 | Dx = 1.411 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71069 Å |
| a = 16.608 (2) Å | Cell parameters from 25 reflections |
| b = 8.357 (1) Å | θ = 2.1–25.0° |
| c = 19.647 (5) Å | µ = 0.41 mm−1 |
| β = 93.30 (1)° | T = 293 K |
| V = 2722.4 (9) Å3 | Tablet, colourless |
| Z = 4 | 0.36 × 0.36 × 0.21 mm |
Data collection top
Enraf-Nonius CAD-4
diffractometer | 2932 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.030 |
| Graphite monochromator | θmax = 25.0°, θmin = 2.1° |
| non–profiled ω/2θ scans | h = −19→19 |
Absorption correction: ψ scan
(North et al., 1968) | k = −9→0 |
| Tmin = 0.865, Tmax = 0.917 | l = 0→23 |
| 4904 measured reflections | 3 standard reflections every 100 reflections |
| 4759 independent reflections | intensity decay: none |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.059 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.181 | H-atom parameters constrained |
| S = 1.01 | w = 1/[σ2(Fo2) + (0.1P)2]
where P = (Fo2 + 2Fc2)/3 |
| 4759 reflections | (Δ/σ)max < 0.001 |
| 336 parameters | Δρmax = 0.39 e Å−3 |
| 0 restraints | Δρmin = −0.32 e Å−3 |
Crystal data top
| C27H26Cl2NO5PS | V = 2722.4 (9) Å3 |
| Mr = 578.42 | Z = 4 |
| Monoclinic, P21/n | Mo Kα radiation |
| a = 16.608 (2) Å | µ = 0.41 mm−1 |
| b = 8.357 (1) Å | T = 293 K |
| c = 19.647 (5) Å | 0.36 × 0.36 × 0.21 mm |
| β = 93.30 (1)° | |
Data collection top
Enraf-Nonius CAD-4
diffractometer | 2932 reflections with I > 2σ(I) |
Absorption correction: ψ scan
(North et al., 1968) | Rint = 0.030 |
| Tmin = 0.865, Tmax = 0.917 | 3 standard reflections every 100 reflections |
| 4904 measured reflections | intensity decay: none |
| 4759 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.059 | 0 restraints |
| wR(F2) = 0.181 | H-atom parameters constrained |
| S = 1.01 | Δρmax = 0.39 e Å−3 |
| 4759 reflections | Δρmin = −0.32 e Å−3 |
| 336 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| P1 | 0.31476 (6) | −0.40244 (15) | 0.20785 (5) | 0.0529 (3) | |
| S1 | 0.56810 (6) | −0.49475 (14) | 0.26025 (5) | 0.0531 (3) | |
| Cl1 | 0.03728 (8) | −1.20568 (18) | −0.03072 (7) | 0.0844 (5) | |
| Cl2 | 0.31530 (9) | −0.9612 (2) | −0.10733 (6) | 0.0956 (5) | |
| N1 | 0.51749 (18) | −0.5255 (4) | 0.18548 (15) | 0.0450 (8) | |
| O1 | 0.26812 (19) | −0.3479 (4) | 0.26437 (15) | 0.0714 (9) | |
| O2 | 0.64009 (18) | −0.4126 (4) | 0.24564 (16) | 0.0708 (9) | |
| O3 | 0.51363 (18) | −0.4218 (4) | 0.30449 (15) | 0.0671 (9) | |
| O4 | 0.3698 (2) | −0.2740 (4) | 0.17576 (16) | 0.0719 (9) | |
| O5 | 0.26459 (18) | −0.4594 (5) | 0.14249 (17) | 0.0866 (11) | |
| C1 | 0.4410 (2) | −0.6030 (4) | 0.17979 (18) | 0.0419 (9) | |
| C2 | 0.4326 (2) | −0.6851 (5) | 0.11981 (18) | 0.0419 (9) | |
| C3 | 0.5067 (2) | −0.6632 (5) | 0.08469 (18) | 0.0432 (9) | |
| C4 | 0.5578 (2) | −0.5671 (5) | 0.12538 (19) | 0.0475 (10) | |
| C5 | 0.6326 (2) | −0.5164 (6) | 0.1048 (2) | 0.0642 (13) | |
| H5 | 0.6655 | −0.4485 | 0.1316 | 0.077* | |
| C6 | 0.6557 (3) | −0.5718 (7) | 0.0426 (3) | 0.0774 (15) | |
| H6 | 0.7062 | −0.5439 | 0.0283 | 0.093* | |
| C7 | 0.6061 (3) | −0.6668 (7) | 0.0015 (2) | 0.0742 (14) | |
| H7 | 0.6229 | −0.6995 | −0.0407 | 0.089* | |
| C8 | 0.5322 (3) | −0.7139 (6) | 0.0219 (2) | 0.0611 (12) | |
| H8 | 0.4992 | −0.7792 | −0.0061 | 0.073* | |
| C9 | 0.5921 (2) | −0.6862 (5) | 0.2918 (2) | 0.0502 (10) | |
| C10 | 0.5567 (3) | −0.7392 (7) | 0.3484 (3) | 0.0860 (17) | |
| H10 | 0.5200 | −0.6750 | 0.3697 | 0.103* | |
| C11 | 0.5760 (4) | −0.8894 (8) | 0.3737 (3) | 0.114 (2) | |
| H11 | 0.5531 | −0.9258 | 0.4130 | 0.137* | |
| C12 | 0.6287 (4) | −0.9853 (7) | 0.3412 (3) | 0.0975 (19) | |
| H12 | 0.6401 | −1.0878 | 0.3575 | 0.117* | |
| C13 | 0.6642 (3) | −0.9304 (7) | 0.2854 (3) | 0.0803 (15) | |
| H13 | 0.7006 | −0.9948 | 0.2638 | 0.096* | |
| C14 | 0.6466 (3) | −0.7804 (6) | 0.2607 (2) | 0.0611 (12) | |
| H14 | 0.6717 | −0.7425 | 0.2227 | 0.073* | |
| C15 | 0.3776 (2) | −0.5725 (5) | 0.22920 (19) | 0.0484 (10) | |
| H15A | 0.4036 | −0.5561 | 0.2741 | 0.058* | |
| H15B | 0.3437 | −0.6668 | 0.2314 | 0.058* | |
| C16 | 0.4035 (4) | −0.1393 (7) | 0.2100 (3) | 0.0906 (17) | |
| H16A | 0.4166 | −0.1662 | 0.2574 | 0.109* | |
| H16B | 0.3644 | −0.0530 | 0.2087 | 0.109* | |
| C17 | 0.4773 (4) | −0.0859 (7) | 0.1778 (4) | 0.117 (2) | |
| H17A | 0.5167 | −0.1701 | 0.1806 | 0.175* | |
| H17B | 0.4988 | 0.0070 | 0.2012 | 0.175* | |
| H17C | 0.4644 | −0.0601 | 0.1309 | 0.175* | |
| C18 | 0.1903 (4) | −0.4472 (13) | 0.1260 (4) | 0.183 (5) | |
| H18A | 0.1804 | −0.3347 | 0.1167 | 0.220* | |
| H18B | 0.1623 | −0.4726 | 0.1666 | 0.220* | |
| C19 | 0.1507 (4) | −0.5314 (8) | 0.0726 (3) | 0.113 (2) | |
| H19A | 0.1897 | −0.5859 | 0.0468 | 0.170* | |
| H19B | 0.1205 | −0.4580 | 0.0435 | 0.170* | |
| H19C | 0.1147 | −0.6081 | 0.0908 | 0.170* | |
| C20 | 0.3606 (2) | −0.7712 (5) | 0.0984 (2) | 0.0480 (10) | |
| H20 | 0.3218 | −0.7783 | 0.1306 | 0.058* | |
| C21 | 0.3420 (2) | −0.8415 (5) | 0.0395 (2) | 0.0553 (11) | |
| H21 | 0.3790 | −0.8328 | 0.0059 | 0.066* | |
| C22 | 0.2681 (2) | −0.9320 (5) | 0.0224 (2) | 0.0526 (11) | |
| C23 | 0.2112 (3) | −0.9613 (6) | 0.0703 (2) | 0.0683 (13) | |
| H23 | 0.2209 | −0.9241 | 0.1147 | 0.082* | |
| C24 | 0.1414 (3) | −1.0431 (6) | 0.0539 (2) | 0.0718 (14) | |
| H24 | 0.1043 | −1.0594 | 0.0869 | 0.086* | |
| C25 | 0.1258 (3) | −1.1009 (5) | −0.0101 (2) | 0.0598 (11) | |
| C26 | 0.1792 (3) | −1.0764 (5) | −0.0599 (2) | 0.0592 (12) | |
| H26 | 0.1686 | −1.1155 | −0.1038 | 0.071* | |
| C27 | 0.2494 (2) | −0.9919 (5) | −0.0432 (2) | 0.0541 (11) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| P1 | 0.0492 (6) | 0.0631 (8) | 0.0476 (6) | 0.0046 (6) | 0.0149 (5) | −0.0059 (5) |
| S1 | 0.0511 (6) | 0.0574 (7) | 0.0501 (6) | −0.0019 (5) | −0.0033 (5) | −0.0076 (5) |
| Cl1 | 0.0758 (8) | 0.0931 (10) | 0.0822 (9) | −0.0294 (7) | −0.0151 (7) | 0.0024 (7) |
| Cl2 | 0.0972 (10) | 0.1380 (13) | 0.0536 (7) | −0.0354 (10) | 0.0225 (7) | −0.0246 (8) |
| N1 | 0.0407 (17) | 0.055 (2) | 0.0393 (18) | −0.0023 (16) | 0.0038 (14) | −0.0019 (15) |
| O1 | 0.077 (2) | 0.077 (2) | 0.0630 (19) | 0.0183 (18) | 0.0294 (16) | −0.0064 (17) |
| O2 | 0.0604 (19) | 0.074 (2) | 0.077 (2) | −0.0223 (17) | −0.0056 (16) | −0.0020 (18) |
| O3 | 0.073 (2) | 0.071 (2) | 0.0568 (18) | 0.0150 (17) | −0.0024 (15) | −0.0225 (16) |
| O4 | 0.087 (2) | 0.062 (2) | 0.069 (2) | −0.0063 (18) | 0.0265 (17) | −0.0052 (17) |
| O5 | 0.0495 (18) | 0.137 (3) | 0.072 (2) | 0.016 (2) | −0.0050 (16) | −0.031 (2) |
| C1 | 0.040 (2) | 0.046 (2) | 0.040 (2) | 0.0026 (18) | 0.0046 (16) | 0.0041 (18) |
| C2 | 0.042 (2) | 0.048 (2) | 0.036 (2) | 0.0035 (18) | 0.0031 (16) | 0.0038 (18) |
| C3 | 0.045 (2) | 0.047 (2) | 0.038 (2) | 0.0074 (19) | 0.0075 (17) | 0.0037 (18) |
| C4 | 0.046 (2) | 0.055 (3) | 0.042 (2) | 0.003 (2) | 0.0066 (18) | 0.0086 (19) |
| C5 | 0.043 (2) | 0.085 (3) | 0.065 (3) | −0.006 (2) | 0.011 (2) | 0.014 (3) |
| C6 | 0.053 (3) | 0.115 (4) | 0.067 (3) | 0.000 (3) | 0.025 (3) | 0.022 (3) |
| C7 | 0.066 (3) | 0.105 (4) | 0.054 (3) | 0.012 (3) | 0.019 (2) | −0.002 (3) |
| C8 | 0.057 (3) | 0.083 (3) | 0.044 (2) | 0.006 (2) | 0.011 (2) | −0.003 (2) |
| C9 | 0.041 (2) | 0.065 (3) | 0.044 (2) | 0.001 (2) | −0.0017 (18) | 0.001 (2) |
| C10 | 0.075 (3) | 0.106 (4) | 0.080 (4) | 0.031 (3) | 0.028 (3) | 0.024 (3) |
| C11 | 0.114 (5) | 0.126 (6) | 0.107 (5) | 0.035 (5) | 0.043 (4) | 0.058 (4) |
| C12 | 0.104 (4) | 0.088 (4) | 0.101 (5) | 0.033 (4) | 0.015 (4) | 0.037 (4) |
| C13 | 0.070 (3) | 0.086 (4) | 0.085 (4) | 0.026 (3) | 0.004 (3) | 0.002 (3) |
| C14 | 0.054 (3) | 0.075 (3) | 0.055 (3) | 0.011 (2) | 0.007 (2) | 0.004 (2) |
| C15 | 0.050 (2) | 0.058 (3) | 0.038 (2) | −0.005 (2) | 0.0091 (17) | −0.0047 (19) |
| C16 | 0.105 (4) | 0.063 (3) | 0.105 (4) | 0.000 (3) | 0.016 (4) | −0.013 (3) |
| C17 | 0.089 (4) | 0.074 (4) | 0.184 (7) | −0.013 (3) | −0.016 (5) | 0.035 (4) |
| C18 | 0.074 (4) | 0.319 (13) | 0.150 (7) | 0.056 (6) | −0.032 (5) | −0.121 (8) |
| C19 | 0.085 (4) | 0.132 (6) | 0.117 (5) | −0.002 (4) | −0.035 (4) | 0.009 (4) |
| C20 | 0.046 (2) | 0.052 (2) | 0.047 (2) | 0.005 (2) | 0.0079 (18) | −0.001 (2) |
| C21 | 0.053 (2) | 0.072 (3) | 0.042 (2) | −0.007 (2) | 0.0063 (19) | −0.005 (2) |
| C22 | 0.060 (3) | 0.055 (3) | 0.043 (2) | −0.004 (2) | 0.002 (2) | −0.002 (2) |
| C23 | 0.070 (3) | 0.086 (4) | 0.049 (3) | −0.021 (3) | 0.005 (2) | −0.010 (2) |
| C24 | 0.072 (3) | 0.095 (4) | 0.049 (3) | −0.025 (3) | 0.008 (2) | −0.003 (3) |
| C25 | 0.053 (3) | 0.062 (3) | 0.063 (3) | −0.007 (2) | −0.005 (2) | 0.008 (2) |
| C26 | 0.071 (3) | 0.055 (3) | 0.050 (2) | −0.003 (2) | −0.011 (2) | −0.005 (2) |
| C27 | 0.058 (3) | 0.058 (3) | 0.046 (2) | 0.003 (2) | 0.003 (2) | −0.002 (2) |
Geometric parameters (Å, º) top
| P1—O1 | 1.463 (3) | C11—H11 | 0.9300 |
| P1—O4 | 1.566 (3) | C12—C13 | 1.355 (7) |
| P1—O5 | 1.564 (3) | C12—H12 | 0.9300 |
| P1—C15 | 1.798 (4) | C13—C14 | 1.370 (7) |
| S1—O2 | 1.422 (3) | C13—H13 | 0.9300 |
| S1—O3 | 1.427 (3) | C14—H14 | 0.9300 |
| S1—N1 | 1.670 (3) | C15—H15A | 0.9700 |
| S1—C9 | 1.754 (4) | C15—H15B | 0.9700 |
| Cl1—C25 | 1.739 (4) | C16—C17 | 1.480 (8) |
| Cl2—C27 | 1.735 (4) | C16—H16A | 0.9700 |
| N1—C1 | 1.424 (5) | C16—H16B | 0.9700 |
| N1—C4 | 1.433 (5) | C17—H17A | 0.9600 |
| O4—C16 | 1.410 (6) | C17—H17B | 0.9600 |
| O5—C18 | 1.261 (6) | C17—H17C | 0.9600 |
| C1—C2 | 1.364 (5) | C18—C19 | 1.395 (9) |
| C1—C15 | 1.495 (5) | C18—H18A | 0.9700 |
| C2—C20 | 1.438 (5) | C18—H18B | 0.9700 |
| C2—C3 | 1.457 (5) | C19—H19A | 0.9600 |
| C3—C4 | 1.387 (5) | C19—H19B | 0.9600 |
| C3—C8 | 1.394 (5) | C19—H19C | 0.9600 |
| C4—C5 | 1.396 (5) | C20—C21 | 1.320 (5) |
| C5—C6 | 1.380 (6) | C20—H20 | 0.9300 |
| C5—H5 | 0.9300 | C21—C22 | 1.464 (6) |
| C6—C7 | 1.373 (7) | C21—H21 | 0.9300 |
| C6—H6 | 0.9300 | C22—C23 | 1.395 (6) |
| C7—C8 | 1.371 (6) | C22—C27 | 1.400 (5) |
| C7—H7 | 0.9300 | C23—C24 | 1.368 (6) |
| C8—H8 | 0.9300 | C23—H23 | 0.9300 |
| C9—C10 | 1.361 (6) | C24—C25 | 1.358 (6) |
| C9—C14 | 1.371 (6) | C24—H24 | 0.9300 |
| C10—C11 | 1.382 (8) | C25—C26 | 1.373 (6) |
| C10—H10 | 0.9300 | C26—C27 | 1.385 (6) |
| C11—C12 | 1.371 (8) | C26—H26 | 0.9300 |
| | | |
| O1—P1—O4 | 116.1 (2) | C9—C14—H14 | 120.0 |
| O1—P1—O5 | 116.0 (2) | C13—C14—H14 | 120.0 |
| O4—P1—O5 | 100.0 (2) | C1—C15—P1 | 113.8 (3) |
| O1—P1—C15 | 113.2 (2) | C1—C15—H15A | 108.8 |
| O4—P1—C15 | 106.9 (2) | P1—C15—H15A | 108.8 |
| O5—P1—C15 | 103.0 (2) | C1—C15—H15B | 108.8 |
| O2—S1—O3 | 119.1 (2) | P1—C15—H15B | 108.8 |
| O2—S1—N1 | 106.5 (2) | H15A—C15—H15B | 107.7 |
| O3—S1—N1 | 107.3 (2) | O4—C16—C17 | 110.7 (5) |
| O2—S1—C9 | 109.6 (2) | O4—C16—H16A | 109.5 |
| O3—S1—C9 | 108.3 (2) | C17—C16—H16A | 109.5 |
| N1—S1—C9 | 105.26 (18) | O4—C16—H16B | 109.5 |
| C1—N1—C4 | 106.3 (3) | C17—C16—H16B | 109.5 |
| C1—N1—S1 | 122.7 (2) | H16A—C16—H16B | 108.1 |
| C4—N1—S1 | 121.8 (2) | C16—C17—H17A | 109.5 |
| C16—O4—P1 | 125.4 (3) | C16—C17—H17B | 109.5 |
| C18—O5—P1 | 130.8 (4) | H17A—C17—H17B | 109.5 |
| C2—C1—N1 | 110.1 (3) | C16—C17—H17C | 109.5 |
| C2—C1—C15 | 127.3 (3) | H17A—C17—H17C | 109.5 |
| N1—C1—C15 | 122.0 (3) | H17B—C17—H17C | 109.5 |
| C1—C2—C20 | 123.1 (3) | O5—C18—C19 | 124.3 (7) |
| C1—C2—C3 | 107.4 (3) | O5—C18—H18A | 106.2 |
| C20—C2—C3 | 129.5 (3) | C19—C18—H18A | 106.2 |
| C4—C3—C8 | 118.7 (4) | O5—C18—H18B | 106.2 |
| C4—C3—C2 | 107.7 (3) | C19—C18—H18B | 106.2 |
| C8—C3—C2 | 133.6 (4) | H18A—C18—H18B | 106.4 |
| C3—C4—C5 | 122.2 (4) | C18—C19—H19A | 109.5 |
| C3—C4—N1 | 108.5 (3) | C18—C19—H19B | 109.5 |
| C5—C4—N1 | 129.1 (4) | H19A—C19—H19B | 109.5 |
| C4—C5—C6 | 116.9 (5) | C18—C19—H19C | 109.5 |
| C4—C5—H5 | 121.6 | H19A—C19—H19C | 109.5 |
| C6—C5—H5 | 121.6 | H19B—C19—H19C | 109.5 |
| C7—C6—C5 | 121.9 (4) | C21—C20—C2 | 128.9 (4) |
| C7—C6—H6 | 119.1 | C21—C20—H20 | 115.5 |
| C5—C6—H6 | 119.1 | C2—C20—H20 | 115.5 |
| C8—C7—C6 | 120.7 (4) | C20—C21—C22 | 125.6 (4) |
| C8—C7—H7 | 119.7 | C20—C21—H21 | 117.2 |
| C6—C7—H7 | 119.7 | C22—C21—H21 | 117.2 |
| C7—C8—C3 | 119.6 (4) | C23—C22—C27 | 115.8 (4) |
| C7—C8—H8 | 120.2 | C23—C22—C21 | 121.8 (4) |
| C3—C8—H8 | 120.2 | C27—C22—C21 | 122.4 (4) |
| C10—C9—C14 | 120.4 (4) | C24—C23—C22 | 121.8 (4) |
| C10—C9—S1 | 119.0 (4) | C24—C23—H23 | 119.1 |
| C14—C9—S1 | 120.6 (3) | C22—C23—H23 | 119.1 |
| C9—C10—C11 | 119.2 (5) | C25—C24—C23 | 120.7 (4) |
| C9—C10—H10 | 120.4 | C25—C24—H24 | 119.7 |
| C11—C10—H10 | 120.4 | C23—C24—H24 | 119.7 |
| C12—C11—C10 | 120.3 (5) | C24—C25—C26 | 120.7 (4) |
| C12—C11—H11 | 119.8 | C24—C25—Cl1 | 120.5 (4) |
| C10—C11—H11 | 119.8 | C26—C25—Cl1 | 118.8 (4) |
| C13—C12—C11 | 119.9 (5) | C27—C26—C25 | 118.4 (4) |
| C13—C12—H12 | 120.1 | C27—C26—H26 | 120.8 |
| C11—C12—H12 | 120.1 | C25—C26—H26 | 120.8 |
| C12—C13—C14 | 120.2 (5) | C26—C27—C22 | 122.7 (4) |
| C12—C13—H13 | 119.9 | C26—C27—Cl2 | 117.4 (3) |
| C14—C13—H13 | 119.9 | C22—C27—Cl2 | 119.9 (3) |
| C9—C14—C13 | 120.0 (4) | | |
| | | |
| O2—S1—N1—C1 | −179.0 (3) | O2—S1—C9—C10 | 133.5 (4) |
| O3—S1—N1—C1 | −50.4 (3) | O3—S1—C9—C10 | 2.0 (4) |
| C9—S1—N1—C1 | 64.7 (3) | N1—S1—C9—C10 | −112.4 (4) |
| O2—S1—N1—C4 | 38.7 (4) | O2—S1—C9—C14 | −45.5 (4) |
| O3—S1—N1—C4 | 167.3 (3) | O3—S1—C9—C14 | −176.9 (3) |
| C9—S1—N1—C4 | −77.6 (3) | N1—S1—C9—C14 | 68.6 (4) |
| O1—P1—O4—C16 | 27.0 (5) | C14—C9—C10—C11 | −0.5 (8) |
| O5—P1—O4—C16 | 152.5 (4) | S1—C9—C10—C11 | −179.4 (5) |
| C15—P1—O4—C16 | −100.5 (4) | C9—C10—C11—C12 | −1.5 (10) |
| O1—P1—O5—C18 | 10.8 (9) | C10—C11—C12—C13 | 2.3 (11) |
| O4—P1—O5—C18 | −114.8 (8) | C11—C12—C13—C14 | −1.0 (10) |
| C15—P1—O5—C18 | 135.1 (8) | C10—C9—C14—C13 | 1.8 (7) |
| C4—N1—C1—C2 | −1.2 (4) | S1—C9—C14—C13 | −179.3 (4) |
| S1—N1—C1—C2 | −148.5 (3) | C12—C13—C14—C9 | −1.0 (8) |
| C4—N1—C1—C15 | −172.7 (3) | C2—C1—C15—P1 | −83.8 (5) |
| S1—N1—C1—C15 | 40.1 (5) | N1—C1—C15—P1 | 86.1 (4) |
| N1—C1—C2—C20 | −177.8 (3) | O1—P1—C15—C1 | −164.6 (3) |
| C15—C1—C2—C20 | −6.9 (6) | O4—P1—C15—C1 | −35.5 (3) |
| N1—C1—C2—C3 | 0.8 (4) | O5—P1—C15—C1 | 69.4 (3) |
| C15—C1—C2—C3 | 171.7 (4) | P1—O4—C16—C17 | 155.1 (4) |
| C1—C2—C3—C4 | −0.1 (4) | P1—O5—C18—C19 | −166.3 (7) |
| C20—C2—C3—C4 | 178.4 (4) | C1—C2—C20—C21 | 172.8 (4) |
| C1—C2—C3—C8 | −178.1 (4) | C3—C2—C20—C21 | −5.6 (7) |
| C20—C2—C3—C8 | 0.5 (7) | C2—C20—C21—C22 | 177.8 (4) |
| C8—C3—C4—C5 | 1.6 (6) | C20—C21—C22—C23 | −4.3 (7) |
| C2—C3—C4—C5 | −176.7 (4) | C20—C21—C22—C27 | 174.5 (4) |
| C8—C3—C4—N1 | 177.7 (3) | C27—C22—C23—C24 | −0.1 (7) |
| C2—C3—C4—N1 | −0.6 (4) | C21—C22—C23—C24 | 178.7 (5) |
| C1—N1—C4—C3 | 1.1 (4) | C22—C23—C24—C25 | 0.9 (8) |
| S1—N1—C4—C3 | 148.7 (3) | C23—C24—C25—C26 | −0.9 (8) |
| C1—N1—C4—C5 | 176.8 (4) | C23—C24—C25—Cl1 | 179.7 (4) |
| S1—N1—C4—C5 | −35.6 (6) | C24—C25—C26—C27 | 0.2 (7) |
| C3—C4—C5—C6 | −2.6 (7) | Cl1—C25—C26—C27 | 179.6 (3) |
| N1—C4—C5—C6 | −177.8 (4) | C25—C26—C27—C22 | 0.5 (7) |
| C4—C5—C6—C7 | 2.7 (7) | C25—C26—C27—Cl2 | −179.4 (3) |
| C5—C6—C7—C8 | −1.8 (8) | C23—C22—C27—C26 | −0.6 (7) |
| C6—C7—C8—C3 | 0.7 (8) | C21—C22—C27—C26 | −179.4 (4) |
| C4—C3—C8—C7 | −0.6 (6) | C23—C22—C27—Cl2 | 179.4 (3) |
| C2—C3—C8—C7 | 177.1 (4) | C21—C22—C27—Cl2 | 0.6 (6) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C5—H5···O2 | 0.93 | 2.32 | 2.896 (6) | 120 |
| C10—H10···O3 | 0.93 | 2.47 | 2.867 (7) | 106 |
| C15—H15A···O3 | 0.97 | 2.20 | 2.915 (5) | 130 |
| C15—H15B···O1i | 0.97 | 2.40 | 3.347 (5) | 165 |
| C23—H23···O1i | 0.93 | 2.46 | 3.382 (5) | 173 |
| C19—H19B···Cl1ii | 0.96 | 2.87 | 3.826 (7) | 172 |
| Symmetry codes: (i) −x+1/2, y−1/2, −z+1/2; (ii) x, y+1, z. |
(II) Diethyl
{3-[
β-(4-bromophenyl)vinyl]-
N-phenylsulfonyl-indol-2-ylmethyl}phosphonate
top
Crystal data top
| C27H27BrNO5PS | F(000) = 1208 |
| Mr = 588.44 | Dx = 1.466 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 16.837 (1) Å | Cell parameters from 25 reflections |
| b = 8.323 (1) Å | θ = 1.4–24.9° |
| c = 19.611 (1) Å | µ = 1.72 mm−1 |
| β = 104.11 (1)° | T = 293 K |
| V = 2665.3 (3) Å3 | Prism, colourless |
| Z = 4 | 0.3 × 0.2 × 0.2 mm |
Data collection top
Enraf-Nonius CAD-4
diffractometer | 2474 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.060 |
| Graphite monochromator | θmax = 25.0°, θmin = 1.4° |
| non–profiled ω/2θ scans | h = −20→19 |
Absorption correction: ψ scan
(North et al., 1968) | k = 0→9 |
| Tmin = 0.461, Tmax = 0.709 | l = 0→23 |
| 4827 measured reflections | 3 standard reflections every 100 reflections |
| 4683 independent reflections | intensity decay: none |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.050 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.144 | H-atom parameters constrained |
| S = 1.01 | |
| 4683 reflections | (Δ/σ)max = 0.012 |
| 327 parameters | Δρmax = 0.36 e Å−3 |
| 0 restraints | Δρmin = −0.28 e Å−3 |
Crystal data top
| C27H27BrNO5PS | V = 2665.3 (3) Å3 |
| Mr = 588.44 | Z = 4 |
| Monoclinic, P21/n | Mo Kα radiation |
| a = 16.837 (1) Å | µ = 1.72 mm−1 |
| b = 8.323 (1) Å | T = 293 K |
| c = 19.611 (1) Å | 0.3 × 0.2 × 0.2 mm |
| β = 104.11 (1)° | |
Data collection top
Enraf-Nonius CAD-4
diffractometer | 2474 reflections with I > 2σ(I) |
Absorption correction: ψ scan
(North et al., 1968) | Rint = 0.060 |
| Tmin = 0.461, Tmax = 0.709 | 3 standard reflections every 100 reflections |
| 4827 measured reflections | intensity decay: none |
| 4683 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.050 | 0 restraints |
| wR(F2) = 0.144 | H-atom parameters constrained |
| S = 1.01 | Δρmax = 0.36 e Å−3 |
| 4683 reflections | Δρmin = −0.28 e Å−3 |
| 327 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Br1 | 0.21126 (4) | −0.48138 (8) | −0.10170 (3) | 0.0981 (3) | |
| S1 | −0.00350 (8) | 0.20548 (15) | 0.36559 (6) | 0.0597 (4) | |
| P1 | 0.15918 (7) | 0.33526 (16) | 0.23152 (7) | 0.0612 (4) | |
| N1 | −0.0179 (2) | 0.1773 (4) | 0.27833 (18) | 0.0520 (9) | |
| O1 | 0.23730 (19) | 0.4025 (5) | 0.26952 (18) | 0.0833 (11) | |
| O2 | −0.0741 (2) | 0.2863 (4) | 0.37432 (18) | 0.0803 (10) | |
| O3 | 0.0749 (2) | 0.2779 (4) | 0.39078 (17) | 0.0745 (10) | |
| O4 | 0.0835 (2) | 0.4509 (4) | 0.21792 (19) | 0.0798 (10) | |
| O5 | 0.1538 (2) | 0.2815 (5) | 0.15409 (17) | 0.0837 (11) | |
| C1 | 0.0437 (2) | 0.1104 (5) | 0.2479 (2) | 0.0479 (11) | |
| C2 | 0.0086 (2) | 0.0200 (5) | 0.1911 (2) | 0.0482 (10) | |
| C3 | −0.0794 (3) | 0.0221 (5) | 0.1847 (2) | 0.0515 (11) | |
| C4 | −0.0941 (3) | 0.1178 (5) | 0.2382 (2) | 0.0537 (11) | |
| C5 | −0.1726 (3) | 0.1447 (6) | 0.2468 (3) | 0.0695 (14) | |
| H5 | −0.1815 | 0.2100 | 0.2827 | 0.083* | |
| C6 | −0.2362 (3) | 0.0716 (7) | 0.2005 (3) | 0.0805 (16) | |
| H6 | −0.2894 | 0.0880 | 0.2049 | 0.097* | |
| C7 | −0.2231 (3) | −0.0262 (7) | 0.1473 (3) | 0.0825 (17) | |
| H7 | −0.2674 | −0.0767 | 0.1173 | 0.099* | |
| C8 | −0.1455 (3) | −0.0501 (6) | 0.1381 (3) | 0.0663 (13) | |
| H8 | −0.1373 | −0.1133 | 0.1013 | 0.080* | |
| C10 | 0.0714 (4) | −0.0660 (8) | 0.4220 (3) | 0.0883 (18) | |
| H10 | 0.1203 | −0.0131 | 0.4229 | 0.106* | |
| C9 | −0.0015 (3) | 0.0095 (5) | 0.3988 (2) | 0.0538 (11) | |
| C11 | 0.0715 (5) | −0.2243 (10) | 0.4445 (4) | 0.121 (3) | |
| H11 | 0.1209 | −0.2782 | 0.4605 | 0.145* | |
| C12 | 0.0005 (6) | −0.3002 (8) | 0.4432 (3) | 0.110 (2) | |
| H12 | 0.0009 | −0.4069 | 0.4573 | 0.132* | |
| C13 | −0.0708 (5) | −0.2214 (9) | 0.4216 (3) | 0.107 (2) | |
| H13 | −0.1197 | −0.2735 | 0.4216 | 0.128* | |
| C14 | −0.0722 (4) | −0.0653 (7) | 0.3997 (3) | 0.0878 (18) | |
| H14 | −0.1218 | −0.0111 | 0.3854 | 0.105* | |
| C15 | 0.1320 (2) | 0.1575 (5) | 0.2721 (2) | 0.0535 (11) | |
| H15A | 0.1654 | 0.0692 | 0.2629 | 0.064* | |
| H15B | 0.1449 | 0.1739 | 0.3226 | 0.064* | |
| C16 | 0.0746 (4) | 0.5820 (7) | 0.2633 (3) | 0.0928 (18) | |
| H16A | 0.1043 | 0.6751 | 0.2530 | 0.111* | |
| H16B | 0.0968 | 0.5521 | 0.3120 | 0.111* | |
| C17 | −0.0129 (4) | 0.6209 (7) | 0.2517 (3) | 0.0998 (19) | |
| H17A | −0.0420 | 0.5279 | 0.2613 | 0.150* | |
| H17B | −0.0341 | 0.6533 | 0.2037 | 0.150* | |
| H17C | −0.0196 | 0.7068 | 0.2824 | 0.150* | |
| C18 | 0.2087 (6) | 0.2973 (16) | 0.1193 (4) | 0.228 (7) | |
| H18A | 0.2607 | 0.2692 | 0.1509 | 0.274* | |
| H18B | 0.2114 | 0.4108 | 0.1088 | 0.274* | |
| C19 | 0.2040 (4) | 0.2139 (10) | 0.0563 (3) | 0.127 (3) | |
| H19A | 0.2028 | 0.2893 | 0.0190 | 0.190* | |
| H19B | 0.1551 | 0.1499 | 0.0453 | 0.190* | |
| H19C | 0.2510 | 0.1454 | 0.0615 | 0.190* | |
| C20 | 0.0543 (3) | −0.0591 (5) | 0.1474 (2) | 0.0505 (11) | |
| H20 | 0.1109 | −0.0587 | 0.1643 | 0.061* | |
| C21 | 0.0265 (3) | −0.1312 (6) | 0.0872 (3) | 0.0671 (14) | |
| H21 | −0.0301 | −0.1300 | 0.0698 | 0.080* | |
| C22 | 0.0728 (3) | −0.2135 (6) | 0.0437 (2) | 0.0573 (12) | |
| C23 | 0.1564 (3) | −0.2252 (7) | 0.0615 (3) | 0.0752 (15) | |
| H23 | 0.1857 | −0.1815 | 0.1039 | 0.090* | |
| C24 | 0.1986 (3) | −0.3007 (7) | 0.0180 (3) | 0.0810 (16) | |
| H24 | 0.2556 | −0.3038 | 0.0301 | 0.097* | |
| C25 | 0.1557 (3) | −0.3703 (6) | −0.0427 (3) | 0.0636 (13) | |
| C26 | 0.0738 (3) | −0.3576 (7) | −0.0627 (3) | 0.0808 (16) | |
| H26 | 0.0449 | −0.4022 | −0.1049 | 0.097* | |
| C27 | 0.0332 (3) | −0.2777 (8) | −0.0199 (3) | 0.0883 (18) | |
| H27 | −0.0234 | −0.2668 | −0.0347 | 0.106* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Br1 | 0.1018 (5) | 0.1097 (6) | 0.0948 (5) | 0.0133 (4) | 0.0473 (4) | −0.0188 (4) |
| S1 | 0.0738 (8) | 0.0523 (7) | 0.0596 (7) | 0.0006 (7) | 0.0290 (6) | −0.0040 (6) |
| P1 | 0.0499 (7) | 0.0706 (9) | 0.0593 (8) | −0.0124 (7) | 0.0058 (6) | 0.0020 (7) |
| N1 | 0.053 (2) | 0.054 (2) | 0.053 (2) | 0.0015 (18) | 0.0208 (18) | 0.0032 (18) |
| O1 | 0.059 (2) | 0.095 (3) | 0.084 (3) | −0.0299 (19) | −0.0033 (18) | 0.002 (2) |
| O2 | 0.094 (3) | 0.076 (2) | 0.082 (2) | 0.024 (2) | 0.042 (2) | −0.0048 (19) |
| O3 | 0.090 (2) | 0.067 (2) | 0.069 (2) | −0.0276 (19) | 0.0245 (19) | −0.0158 (18) |
| O4 | 0.073 (2) | 0.068 (2) | 0.090 (3) | −0.0017 (18) | 0.0018 (19) | −0.010 (2) |
| O5 | 0.065 (2) | 0.129 (3) | 0.059 (2) | −0.031 (2) | 0.0188 (17) | −0.007 (2) |
| C1 | 0.050 (3) | 0.045 (3) | 0.053 (3) | 0.005 (2) | 0.020 (2) | 0.008 (2) |
| C2 | 0.047 (2) | 0.046 (3) | 0.054 (3) | 0.001 (2) | 0.016 (2) | 0.003 (2) |
| C3 | 0.054 (3) | 0.044 (3) | 0.058 (3) | 0.001 (2) | 0.016 (2) | 0.011 (2) |
| C4 | 0.049 (3) | 0.050 (3) | 0.065 (3) | 0.003 (2) | 0.018 (2) | 0.013 (2) |
| C5 | 0.059 (3) | 0.077 (4) | 0.077 (3) | 0.011 (3) | 0.026 (3) | 0.015 (3) |
| C6 | 0.047 (3) | 0.101 (5) | 0.098 (4) | 0.009 (3) | 0.027 (3) | 0.019 (4) |
| C7 | 0.048 (3) | 0.101 (5) | 0.094 (4) | −0.019 (3) | 0.009 (3) | 0.014 (4) |
| C8 | 0.053 (3) | 0.074 (3) | 0.070 (3) | −0.009 (3) | 0.012 (3) | 0.000 (3) |
| C10 | 0.075 (4) | 0.094 (5) | 0.097 (4) | 0.010 (3) | 0.024 (3) | 0.034 (4) |
| C9 | 0.059 (3) | 0.056 (3) | 0.049 (3) | −0.006 (3) | 0.019 (2) | −0.006 (2) |
| C11 | 0.132 (6) | 0.107 (6) | 0.141 (7) | 0.051 (5) | 0.068 (5) | 0.058 (5) |
| C12 | 0.199 (9) | 0.063 (4) | 0.079 (4) | 0.007 (5) | 0.053 (5) | 0.016 (3) |
| C13 | 0.132 (6) | 0.095 (5) | 0.091 (5) | −0.042 (5) | 0.021 (4) | 0.024 (4) |
| C14 | 0.077 (4) | 0.089 (5) | 0.097 (4) | −0.017 (3) | 0.020 (3) | 0.026 (4) |
| C15 | 0.050 (3) | 0.064 (3) | 0.044 (2) | 0.005 (2) | 0.007 (2) | 0.000 (2) |
| C16 | 0.115 (5) | 0.066 (4) | 0.092 (4) | −0.008 (4) | 0.015 (4) | −0.008 (3) |
| C17 | 0.129 (5) | 0.078 (4) | 0.100 (5) | 0.030 (4) | 0.041 (4) | 0.012 (4) |
| C18 | 0.185 (8) | 0.407 (17) | 0.124 (7) | −0.185 (11) | 0.097 (7) | −0.110 (9) |
| C19 | 0.107 (5) | 0.200 (8) | 0.086 (5) | −0.006 (5) | 0.046 (4) | 0.004 (5) |
| C20 | 0.045 (2) | 0.053 (3) | 0.051 (3) | 0.002 (2) | 0.007 (2) | 0.001 (2) |
| C21 | 0.046 (3) | 0.086 (4) | 0.067 (3) | 0.005 (3) | 0.009 (2) | −0.012 (3) |
| C22 | 0.049 (3) | 0.066 (3) | 0.055 (3) | 0.004 (2) | 0.009 (2) | −0.007 (2) |
| C23 | 0.068 (3) | 0.093 (4) | 0.059 (3) | 0.015 (3) | 0.004 (3) | −0.018 (3) |
| C24 | 0.055 (3) | 0.107 (5) | 0.078 (4) | 0.013 (3) | 0.010 (3) | −0.017 (4) |
| C25 | 0.072 (3) | 0.065 (3) | 0.059 (3) | 0.012 (3) | 0.025 (3) | 0.000 (3) |
| C26 | 0.068 (4) | 0.101 (4) | 0.069 (3) | 0.004 (3) | 0.008 (3) | −0.027 (3) |
| C27 | 0.048 (3) | 0.129 (5) | 0.083 (4) | 0.008 (3) | 0.006 (3) | −0.041 (4) |
Geometric parameters (Å, º) top
| Br1—C25 | 1.897 (5) | C12—C13 | 1.343 (9) |
| S1—O2 | 1.412 (3) | C12—H12 | 0.9300 |
| S1—O3 | 1.425 (3) | C13—C14 | 1.367 (8) |
| S1—N1 | 1.685 (4) | C13—H13 | 0.9300 |
| S1—C9 | 1.753 (5) | C14—H14 | 0.9300 |
| P1—O1 | 1.456 (3) | C15—H15A | 0.9700 |
| P1—O5 | 1.564 (4) | C15—H15B | 0.9700 |
| P1—O4 | 1.567 (4) | C16—C17 | 1.470 (8) |
| P1—C15 | 1.792 (4) | C16—H16A | 0.9700 |
| N1—C4 | 1.420 (5) | C16—H16B | 0.9700 |
| N1—C1 | 1.430 (5) | C17—H17A | 0.9600 |
| O4—C16 | 1.439 (6) | C17—H17B | 0.9600 |
| O5—C18 | 1.281 (7) | C17—H17C | 0.9600 |
| C1—C2 | 1.355 (6) | C18—C19 | 1.403 (9) |
| C1—C15 | 1.497 (5) | C18—H18A | 0.9700 |
| C2—C20 | 1.442 (6) | C18—H18B | 0.9700 |
| C2—C3 | 1.457 (6) | C19—H19A | 0.9600 |
| C3—C4 | 1.386 (6) | C19—H19B | 0.9600 |
| C3—C8 | 1.393 (6) | C19—H19C | 0.9600 |
| C4—C5 | 1.391 (6) | C20—C21 | 1.306 (6) |
| C5—C6 | 1.367 (7) | C20—H20 | 0.9300 |
| C5—H5 | 0.9300 | C21—C22 | 1.460 (6) |
| C6—C7 | 1.383 (8) | C21—H21 | 0.9300 |
| C6—H6 | 0.9300 | C22—C23 | 1.369 (6) |
| C7—C8 | 1.376 (7) | C22—C27 | 1.371 (6) |
| C7—H7 | 0.9300 | C23—C24 | 1.387 (7) |
| C8—H8 | 0.9300 | C23—H23 | 0.9300 |
| C10—C9 | 1.356 (7) | C24—C25 | 1.362 (7) |
| C10—C11 | 1.389 (8) | C24—H24 | 0.9300 |
| C10—H10 | 0.9300 | C25—C26 | 1.342 (6) |
| C9—C14 | 1.348 (7) | C26—C27 | 1.376 (7) |
| C11—C12 | 1.347 (9) | C26—H26 | 0.9300 |
| C11—H11 | 0.9300 | C27—H27 | 0.9300 |
| | | |
| O2—S1—O3 | 119.5 (2) | C9—C14—H14 | 120.1 |
| O2—S1—N1 | 105.8 (2) | C13—C14—H14 | 120.1 |
| O3—S1—N1 | 107.62 (18) | C1—C15—P1 | 114.6 (3) |
| O2—S1—C9 | 109.8 (2) | C1—C15—H15A | 108.6 |
| O3—S1—C9 | 109.4 (2) | P1—C15—H15A | 108.6 |
| N1—S1—C9 | 103.46 (19) | C1—C15—H15B | 108.6 |
| O1—P1—O5 | 116.0 (2) | P1—C15—H15B | 108.6 |
| O1—P1—O4 | 116.5 (2) | H15A—C15—H15B | 107.6 |
| O5—P1—O4 | 99.1 (2) | O4—C16—C17 | 108.8 (5) |
| O1—P1—C15 | 112.8 (2) | O4—C16—H16A | 109.9 |
| O5—P1—C15 | 103.6 (2) | C17—C16—H16A | 109.9 |
| O4—P1—C15 | 107.3 (2) | O4—C16—H16B | 109.9 |
| C4—N1—C1 | 106.4 (3) | C17—C16—H16B | 109.9 |
| C4—N1—S1 | 119.8 (3) | H16A—C16—H16B | 108.3 |
| C1—N1—S1 | 122.6 (3) | C16—C17—H17A | 109.5 |
| C16—O4—P1 | 124.3 (3) | C16—C17—H17B | 109.5 |
| C18—O5—P1 | 127.5 (4) | H17A—C17—H17B | 109.5 |
| C2—C1—N1 | 110.1 (4) | C16—C17—H17C | 109.5 |
| C2—C1—C15 | 127.3 (4) | H17A—C17—H17C | 109.5 |
| N1—C1—C15 | 122.0 (4) | H17B—C17—H17C | 109.5 |
| C1—C2—C20 | 123.6 (4) | O5—C18—C19 | 121.7 (7) |
| C1—C2—C3 | 107.1 (4) | O5—C18—H18A | 106.9 |
| C20—C2—C3 | 129.3 (4) | C19—C18—H18A | 106.9 |
| C4—C3—C8 | 118.9 (4) | O5—C18—H18B | 106.9 |
| C4—C3—C2 | 107.9 (4) | C19—C18—H18B | 106.9 |
| C8—C3—C2 | 133.1 (4) | H18A—C18—H18B | 106.7 |
| C3—C4—C5 | 122.3 (4) | C18—C19—H19A | 109.5 |
| C3—C4—N1 | 108.4 (4) | C18—C19—H19B | 109.5 |
| C5—C4—N1 | 129.3 (5) | H19A—C19—H19B | 109.5 |
| C6—C5—C4 | 117.5 (5) | C18—C19—H19C | 109.5 |
| C6—C5—H5 | 121.3 | H19A—C19—H19C | 109.5 |
| C4—C5—H5 | 121.3 | H19B—C19—H19C | 109.5 |
| C5—C6—C7 | 121.3 (5) | C21—C20—C2 | 128.4 (4) |
| C5—C6—H6 | 119.3 | C21—C20—H20 | 115.8 |
| C7—C6—H6 | 119.3 | C2—C20—H20 | 115.8 |
| C8—C7—C6 | 121.1 (5) | C20—C21—C22 | 128.3 (4) |
| C8—C7—H7 | 119.5 | C20—C21—H21 | 115.9 |
| C6—C7—H7 | 119.5 | C22—C21—H21 | 115.9 |
| C7—C8—C3 | 118.9 (5) | C23—C22—C27 | 116.6 (4) |
| C7—C8—H8 | 120.6 | C23—C22—C21 | 123.2 (4) |
| C3—C8—H8 | 120.6 | C27—C22—C21 | 120.2 (4) |
| C9—C10—C11 | 118.5 (6) | C22—C23—C24 | 121.6 (5) |
| C9—C10—H10 | 120.8 | C22—C23—H23 | 119.2 |
| C11—C10—H10 | 120.8 | C24—C23—H23 | 119.2 |
| C14—C9—C10 | 120.8 (5) | C25—C24—C23 | 119.2 (5) |
| C14—C9—S1 | 119.9 (4) | C25—C24—H24 | 120.4 |
| C10—C9—S1 | 119.3 (4) | C23—C24—H24 | 120.4 |
| C12—C11—C10 | 120.5 (7) | C26—C25—C24 | 120.7 (4) |
| C12—C11—H11 | 119.8 | C26—C25—Br1 | 119.0 (4) |
| C10—C11—H11 | 119.8 | C24—C25—Br1 | 120.3 (4) |
| C11—C12—C13 | 119.9 (7) | C25—C26—C27 | 119.2 (5) |
| C11—C12—H12 | 120.1 | C25—C26—H26 | 120.4 |
| C13—C12—H12 | 120.1 | C27—C26—H26 | 120.4 |
| C12—C13—C14 | 120.6 (7) | C22—C27—C26 | 122.6 (5) |
| C12—C13—H13 | 119.7 | C22—C27—H27 | 118.7 |
| C14—C13—H13 | 119.7 | C26—C27—H27 | 118.7 |
| C9—C14—C13 | 119.7 (6) | | |
| | | |
| O2—S1—N1—C4 | −46.4 (4) | C2—C3—C8—C7 | 178.7 (5) |
| O3—S1—N1—C4 | −175.2 (3) | C11—C10—C9—C14 | 2.2 (9) |
| C9—S1—N1—C4 | 69.0 (4) | C11—C10—C9—S1 | −176.5 (5) |
| O2—S1—N1—C1 | 174.7 (3) | O2—S1—C9—C14 | 27.7 (5) |
| O3—S1—N1—C1 | 45.9 (4) | O3—S1—C9—C14 | 160.7 (4) |
| C9—S1—N1—C1 | −69.8 (4) | N1—S1—C9—C14 | −84.8 (4) |
| O1—P1—O4—C16 | −29.7 (5) | O2—S1—C9—C10 | −153.6 (4) |
| O5—P1—O4—C16 | −154.8 (4) | O3—S1—C9—C10 | −20.6 (5) |
| C15—P1—O4—C16 | 97.8 (4) | N1—S1—C9—C10 | 93.9 (4) |
| O1—P1—O5—C18 | −3.8 (9) | C9—C10—C11—C12 | −0.1 (10) |
| O4—P1—O5—C18 | 121.7 (9) | C10—C11—C12—C13 | −1.5 (11) |
| C15—P1—O5—C18 | −127.9 (9) | C11—C12—C13—C14 | 1.1 (11) |
| C4—N1—C1—C2 | 2.1 (4) | C10—C9—C14—C13 | −2.6 (9) |
| S1—N1—C1—C2 | 145.6 (3) | S1—C9—C14—C13 | 176.1 (5) |
| C4—N1—C1—C15 | 174.5 (4) | C12—C13—C14—C9 | 0.9 (10) |
| S1—N1—C1—C15 | −42.0 (5) | C2—C1—C15—P1 | 85.9 (5) |
| N1—C1—C2—C20 | 177.8 (4) | N1—C1—C15—P1 | −85.0 (4) |
| C15—C1—C2—C20 | 5.9 (7) | O1—P1—C15—C1 | 163.6 (3) |
| N1—C1—C2—C3 | −2.0 (5) | O5—P1—C15—C1 | −70.3 (4) |
| C15—C1—C2—C3 | −173.8 (4) | O4—P1—C15—C1 | 34.0 (4) |
| C1—C2—C3—C4 | 1.1 (5) | P1—O4—C16—C17 | −157.4 (4) |
| C20—C2—C3—C4 | −178.6 (4) | P1—O5—C18—C19 | 165.8 (7) |
| C1—C2—C3—C8 | −178.8 (5) | C1—C2—C20—C21 | −170.5 (5) |
| C20—C2—C3—C8 | 1.5 (8) | C3—C2—C20—C21 | 9.2 (8) |
| C8—C3—C4—C5 | −0.2 (7) | C2—C20—C21—C22 | −178.9 (5) |
| C2—C3—C4—C5 | 179.9 (4) | C20—C21—C22—C23 | −0.4 (9) |
| C8—C3—C4—N1 | −179.9 (4) | C20—C21—C22—C27 | −177.5 (5) |
| C2—C3—C4—N1 | 0.2 (5) | C27—C22—C23—C24 | −1.1 (8) |
| C1—N1—C4—C3 | −1.4 (4) | C21—C22—C23—C24 | −178.3 (5) |
| S1—N1—C4—C3 | −146.1 (3) | C22—C23—C24—C25 | −2.6 (9) |
| C1—N1—C4—C5 | 178.9 (4) | C23—C24—C25—C26 | 4.3 (9) |
| S1—N1—C4—C5 | 34.2 (6) | C23—C24—C25—Br1 | −177.7 (4) |
| C3—C4—C5—C6 | 0.6 (7) | C24—C25—C26—C27 | −2.2 (9) |
| N1—C4—C5—C6 | −179.8 (4) | Br1—C25—C26—C27 | 179.7 (5) |
| C4—C5—C6—C7 | 0.3 (8) | C23—C22—C27—C26 | 3.3 (9) |
| C5—C6—C7—C8 | −1.7 (9) | C21—C22—C27—C26 | −179.4 (6) |
| C6—C7—C8—C3 | 2.0 (8) | C25—C26—C27—C22 | −1.7 (9) |
| C4—C3—C8—C7 | −1.1 (7) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C5—H5···O2 | 0.93 | 2.31 | 2.893 (6) | 121 |
| C15—H15B···O3 | 0.97 | 2.17 | 2.904 (6) | 132 |
| C15—H15A···O1i | 0.97 | 2.35 | 3.301 (5) | 167 |
| C14—H14···Br1ii | 0.93 | 2.88 | 3.660 (7) | 142 |
| Symmetry codes: (i) −x+1/2, y−1/2, −z+1/2; (ii) x−1/2, −y−1/2, z+1/2. |
Experimental details
| | (I) | (II) |
| Crystal data |
| Chemical formula | C27H26Cl2NO5PS | C27H27BrNO5PS |
| Mr | 578.42 | 588.44 |
| Crystal system, space group | Monoclinic, P21/n | Monoclinic, P21/n |
| Temperature (K) | 293 | 293 |
| a, b, c (Å) | 16.608 (2), 8.357 (1), 19.647 (5) | 16.837 (1), 8.323 (1), 19.611 (1) |
| β (°) | 93.30 (1) | 104.11 (1) |
| V (Å3) | 2722.4 (9) | 2665.3 (3) |
| Z | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα |
| µ (mm−1) | 0.41 | 1.72 |
| Crystal size (mm) | 0.36 × 0.36 × 0.21 | 0.3 × 0.2 × 0.2 |
| |
| Data collection |
| Diffractometer | Enraf-Nonius CAD-4
diffractometer | Enraf-Nonius CAD-4
diffractometer |
| Absorption correction | ψ scan
(North et al., 1968) | ψ scan
(North et al., 1968) |
| Tmin, Tmax | 0.865, 0.917 | 0.461, 0.709 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 4904, 4759, 2932 | 4827, 4683, 2474 |
| Rint | 0.030 | 0.060 |
| (sin θ/λ)max (Å−1) | 0.594 | 0.594 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.059, 0.181, 1.01 | 0.050, 0.144, 1.01 |
| No. of reflections | 4759 | 4683 |
| No. of parameters | 336 | 327 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.39, −0.32 | 0.36, −0.28 |
Selected geometric parameters (Å, º) for (I) top| P1—O1 | 1.463 (3) | S1—C9 | 1.754 (4) |
| P1—O4 | 1.566 (3) | Cl1—C25 | 1.739 (4) |
| P1—O5 | 1.564 (3) | Cl2—C27 | 1.735 (4) |
| P1—C15 | 1.798 (4) | N1—C1 | 1.424 (5) |
| S1—O2 | 1.422 (3) | N1—C4 | 1.433 (5) |
| S1—O3 | 1.427 (3) | C18—C19 | 1.395 (9) |
| S1—N1 | 1.670 (3) | | |
| | | |
| O1—P1—O4 | 116.1 (2) | O3—S1—C9 | 108.3 (2) |
| O1—P1—O5 | 116.0 (2) | C16—O4—P1 | 125.4 (3) |
| O1—P1—C15 | 113.2 (2) | C18—O5—P1 | 130.8 (4) |
| O4—P1—C15 | 106.9 (2) | C4—C3—C8 | 118.7 (4) |
| O5—P1—C15 | 103.0 (2) | C3—C4—C5 | 122.2 (4) |
| O2—S1—N1 | 106.5 (2) | C4—C5—C6 | 116.9 (5) |
| O3—S1—N1 | 107.3 (2) | C7—C6—C5 | 121.9 (4) |
| O2—S1—C9 | 109.6 (2) | C8—C7—C6 | 120.7 (4) |
| | | |
| O2—S1—N1—C4 | 38.7 (4) | O3—S1—C9—C10 | 2.0 (4) |
| C9—S1—N1—C4 | −77.6 (3) | N1—S1—C9—C10 | −112.4 (4) |
| O1—P1—O4—C16 | 27.0 (5) | O2—S1—C9—C14 | −45.5 (4) |
| O5—P1—O4—C16 | 152.5 (4) | C2—C1—C15—P1 | −83.8 (5) |
| O4—P1—O5—C18 | −114.8 (8) | O1—P1—C15—C1 | −164.6 (3) |
| S1—N1—C1—C15 | 40.1 (5) | O4—P1—C15—C1 | −35.5 (3) |
| S1—N1—C4—C5 | −35.6 (6) | O5—P1—C15—C1 | 69.4 (3) |
Hydrogen-bond geometry (Å, º) for (I) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C5—H5···O2 | 0.93 | 2.32 | 2.896 (6) | 120 |
| C10—H10···O3 | 0.93 | 2.47 | 2.867 (7) | 106 |
| C15—H15A···O3 | 0.97 | 2.20 | 2.915 (5) | 130 |
| C15—H15B···O1i | 0.97 | 2.40 | 3.347 (5) | 165 |
| C23—H23···O1i | 0.93 | 2.46 | 3.382 (5) | 173 |
| C19—H19B···Cl1ii | 0.96 | 2.87 | 3.826 (7) | 172 |
| Symmetry codes: (i) −x+1/2, y−1/2, −z+1/2; (ii) x, y+1, z. |
Selected geometric parameters (Å, º) for (II) top| Br1—C25 | 1.897 (5) | P1—O5 | 1.564 (4) |
| S1—O2 | 1.412 (3) | P1—O4 | 1.567 (4) |
| S1—O3 | 1.425 (3) | P1—C15 | 1.792 (4) |
| S1—N1 | 1.685 (4) | N1—C4 | 1.420 (5) |
| S1—C9 | 1.753 (5) | N1—C1 | 1.430 (5) |
| P1—O1 | 1.456 (3) | C18—C19 | 1.403 (9) |
| | | |
| O2—S1—N1 | 105.8 (2) | C18—O5—P1 | 127.5 (4) |
| O1—P1—O5 | 116.0 (2) | C4—C3—C8 | 118.9 (4) |
| O1—P1—O4 | 116.5 (2) | C3—C4—C5 | 122.3 (4) |
| O1—P1—C15 | 112.8 (2) | C6—C5—C4 | 117.5 (5) |
| O5—P1—C15 | 103.6 (2) | C5—C6—C7 | 121.3 (5) |
| C16—O4—P1 | 124.3 (3) | C8—C7—C6 | 121.1 (5) |
| | | |
| O2—S1—N1—C4 | −46.4 (4) | O3—S1—C9—C10 | −20.6 (5) |
| O1—P1—O4—C16 | −29.7 (5) | N1—S1—C9—C10 | 93.9 (4) |
| O5—P1—O4—C16 | −154.8 (4) | C2—C1—C15—P1 | 85.9 (5) |
| S1—N1—C1—C15 | −42.0 (5) | O1—P1—C15—C1 | 163.6 (3) |
| S1—N1—C4—C5 | 34.2 (6) | O5—P1—C15—C1 | −70.3 (4) |
| O2—S1—C9—C14 | 27.7 (5) | O4—P1—C15—C1 | 34.0 (4) |
Hydrogen-bond geometry (Å, º) for (II) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C5—H5···O2 | 0.93 | 2.31 | 2.893 (6) | 121 |
| C15—H15B···O3 | 0.97 | 2.17 | 2.904 (6) | 132 |
| C15—H15A···O1i | 0.97 | 2.35 | 3.301 (5) | 167 |
| C14—H14···Br1ii | 0.93 | 2.88 | 3.660 (7) | 142 |
| Symmetry codes: (i) −x+1/2, y−1/2, −z+1/2; (ii) x−1/2, −y−1/2, z+1/2. |
 O and C—H
O and C—H halogen interactions. The intermolecular hydrogen bonds in (I) form cyclic dimers with graph-set descriptors R
halogen interactions. The intermolecular hydrogen bonds in (I) form cyclic dimers with graph-set descriptors R
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2002/08/00/na1565/na1565fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2002/08/00/na1565/na1565fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2002/08/00/na1565/na1565fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/c/issues/2002/08/00/na1565/na1565fig4thm.gif)




Organophosphorus compounds have attracted considerable interest because of their applications as insecticides, bactericides, flame retardants, lubricants, etc. (Ismail, 1975). Phosphate esters are present in many enzymes, nucleic acids, bacteria, vitamin and viruses. Organophosphorous compounds are also reported as effective antitumour compounds.
The indole ring system is present in a number of natural products, many of which are found to possess antibacterial (Okabe & Adachi, 1998), antitumour (Schollmeyer et al., 1995), antidepressant (Papenstasion & Newmeyer, 1972), psychotropic (Grinev et al., 1978), hypertensive (Merk, 1971), antimicrobial (El-Sayed et al., 1986; Gadaginamath & Patil, 1999) and antiinflammatory activities (Rodriguez et al., 1985; Polletto et al., 1974). Indoles also intercalate with DNA (Sivaraman et al., 1996), and this intercalation between the base pairs in DNA has been implicated for their medicinal properties. The indole ring system occurs in plants (Nigovic et al., 2000), for example, indole-3-acetic acid is a naturally occurring plant-growth hormone that controls several growth activities of plants (Moore, 1989; Fargasova, 1994). Indoles have also been proven to display high aldose reductase inhibitory activity (Rajeswaran et al., 1999).
Sulfonamide-containing drugs acts as diuretics and sulthiame, as a carbonic anhydrase inhibitor, has been shown to possess anticonvulsant activity (Crawford & Kennedy, 1959; Camerman & Camerman, 1975; Tanimukai et al., 1965). Sulfonamides inhibit the growth of bacterial organisms and are also useful for treating urinary and gastrointestinal infections.
Both title compounds are excellent intermediates for elaborating the substituent at the 2-position of the indole ring. Compounds (I) and (II) could be converted into a 2-vinyl derivative by the Wittig reaction. These compounds have been converted into analogues of the anticancer alkaloid ellipticine (Srinivasan & Mohanakrishnan, 1995). Similarly, compounds of this type have been shown to undergo a Diels-Alder reaction to give 2-(quinolin-2-yl) indoles (Srinivasan & Elango, 1999). Against this background, and in order to obtain detailed information on their molecular conformation in the solid state, X-ray studies of the title compounds, (I) and (II), have been carried out and the results are presented here. \sch
Figs. 1 and 2 show the molecular structures of (I) and (II), respectively, with the atom-numbering schemes. The mean P—O (P1—O4 and P1—O5) single-bond distance of 1.565 (3) Å for (I) and (II) is in agreement with the reported value of 1.564 (5) Å for the structure of diethyl (1-hydroxy-2-butynyl)phosphonate (Sanders et al., 1996). The P1═O1 double-bond distances in (I) and (II) are comparable with the reported values of 1.464 (2), 1.459 (3) and 1.454 (4) Å [Naidu et al. (1992), Boehlow et al. (1997) and Yokota et al. (1990), respectively Rephrasing OK?]. The P1—C15 single-bond lengths in both compounds are in good agreement with the reported values of 1.791 (2) and 1.806 (6) Å (Weichsel & Lis, 1996; Liu et al., 1995; Perales & García-Blanco, 1977; Howells et al., 1973).
In (I) and (II), the average values of the S1═O2 and S1═O3 distances [1.417 (3) and 1.426 (3) Å, respectively] are comparable with the literature value of 1.427 (4) Å (Datta et al., 1993; Ghosh et al., 1989; Seetharaman & Rajan, 1995). The S1—N1 and S1—C9 bond distances in (I) and (II) compare well with the literature values of 1.642 (24) and 1.758 (18) Å, respectively (Allen et al., 1987).
The relatively large values of the C—N distances in the indole moiety (N1—C1 and N1—C4) are due to the electron-withdrawing character of the phenylsulfonyl group (Govindasamy et al., 1997, 1998). As in similar structures (Hazel & Collin, 1972; Ezra & Collin, 1973; Sanders et al., 1996), short C—C bond distances [C18—C19 in (I) and (II)], along with high thermal motion, are observed for the ethyl C atoms. The Csp2—X bond distances [X is Cl in (I) and Br in (II)] are comparable with the reported values of 1.734 (19) and 1.883 (15) Å (Allen et al., 1987). Selected geometric parameters for (I) and (II) are given in Tables 1 and 3, respectively.
Atom P1 adopts a distorted tetrahedral configuration in both compounds, and the widening of the O1—P1—O4 angles, and the resultant narrowing of the O5—P1—C15 angles, from the ideal tetrahedral value are attributed to Thorpe-Ingold effect (Bassindale, 1984). The P1—C15 bond is (-) synclinal to the C1—C2 bond in (I), whereas it is (+) synclinal to C1—C2 in (II). The conformation about P1—C15 is, as expected, staggered in (I) and (II).
In both compounds, the indole system is not strictly planar, and the dihedral angle formed by the pyrrole and benzo planes is 2.1 (1)° in (I) and 0.9 (2)° in (II). The O2—S1—N1—C4 and O2—S1—C9—C14 torsion angles in both compounds describe the conformation of the phenylsulfonyl with respect to the indole system, which causes the best planes of the indole and phenyl rings to form a dihedral angle of 77.3 (1)° in (I) and 71.6 (2)° in (II), as observed in similar structures (Yokum & Fronzeck, 1997; Sankaranarayanan et al., 2000).
The phenyl rings of the dichlorophenyl in (I) and the bromophenyl in (II) are orthogonal to the phenyl ring of the sulfonyl substituent in each, forming dihedral angles of 82.0 (1)° in (I) and 78.7 (2)° in (II). The dihedral angle formed by the weighted least-squares planes through the pyrrole ring and the halophenyl group is 12.0 (1)° in (I) and 11.4° in (II). Atoms C15 and C20 are out of the indole plane by 0.205 (4) and 0.057 (4) Å, respectively, in (I) and by 0.097 (4) and 0.038 (4) Å, respectively, in (II), both on one side, while atom S1 is out of the plane on the other side, by 0.730 (1) Å in (I) and 0.812 Å in (II).
In the benzene ring of the indole system, the endocyclic angles at C3 and C5 are contracted to 118.7 (4) and 116.9 (5)°, respectively, in (I), and 118.9 (4) and 117.5 (5)°, respectively, in (II), while those at C4, C6 and C7 are expanded to 122.2 (4), 121.9 (4) and 120.7 (3)°, respectively, in (I), and 122.3 (4), 121.3 (5) and 121.1 (5)°, respectively, in (II). This would appear to be a real effect caused by the fusion of the smaller pyrrole ring to the six-membered benzene ring, and the strain is taken up by angular distortion rather than by bond-length distortions (Allen, 1981). A similar effect has also been observed by Varghese et al. (1986) and Sankaranarayanan et al. (2000).
The angular disposition of the bond about atom S1 shows a significant deviation from that of a regular tetrahedron, with the largest deviation in the O—S—O angle. The widening of the O2—S1—O3 angle to 119.1 (2)° in (I) and 119.5 (2)° in (II) from the ideal tetrahedral value is presumably the result of the repulsive interaction between the short S═O bonds, similar to that observed in related structures (Rodriguez et al., 1985; Beddoes et al., 1986). The orientation of the indole substituent is influenced by a weak C5—H5···O2 interaction, defined by the torsion angle C5—C4—N1—S1 in (I) and C4—N1—S1—O2 in (II), while the orientation of the phenyl ring bound to the sulfonyl group is governed by a C10—H10···O3 interaction, defined by the N1—S1—C9—C10 torsion angle in both compounds.
In addition to van der Waals interactions, the molecular structures and packing are stabilized by C—H···O and C—H···halogen interactions (Tables 2 and 4). In compound (I), the hydrogen bonds form cyclic dimers with graph-set descriptors (Bernstein et al., 1995) R21(10) and R22(8) about a 21 axis. This system of rings is joined through the O5—C19—C18 ethoxy chain by a hydrogen bond to the Cl1 atom of a molecule shifted by b in the [010] direction. For R21(10), the ring is C15—H15B···O1i···H23—C23—C22—C21—C20—C2—C1, and for R22(8), the ring is C15—H15B···O1i—P1i—C15i—H15Bi···O1···P1 [symmetry code: (i) Please provide missing symmetry code]. There are three intramolecular rings in (I), namely, S(6) C5—H5···O2—S1—N1—C4, S(6) C15—H15A···O3—S1—N1—C1 and S(5) C10—H10···O3—S1—C9 (Fig. 3). In (II), there is a C22(20) chain, O1i···H15A—C15—C1—N1—S1—C9—C14—H14···Br1ii—C25ii—C24ii—C23ii—C22ii– C21ii—C20ii—C2ii—C1ii—C15ii—H15ii [symmetry code: (ii) Please provide missing symmetry code]. There are also two intramolecular rings in (II), namely, S(6) C5—H5···O2—S1—N1—C4 and S(6) C15—H15A···O3—S1—N1—C1 (Fig. 4).