
Mefenamic acid (MA) formed solvates with 2-picoline (2PIC), 3-picoline (3PIC), 4-picoline (4PIC) and 3-chloropyridine (3CLPYR). The solvates crystallized in the space group
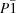
with the carboxylic acid of MA hydrogen-bonded to the nitrogen of the substituted pyridine. Tolfenamic acid (TFA) formed solvates with 2PIC and 3PIC, the crystal structures successfully solved in the space groups
P2
1/
n and
Pbca, respectively. The fenamate conformation varied depending on the acid and the included solvent. Similarities were observed in the structures involving MA. The two solvate structures of TFA had different packing arrangements. Grinding and slurry experiments were also successful for the preparation of all of the compounds except MA·2PIC. Recrystallization, grinding and slurry investigations of MA and 2PIC yielded a polymorph; the structure was successfully solved in
P2
1/
n. Additionally, the thermal stability of the solvates was determined. Desolvation experiments were also performed and the resultant powders were analysed using powder X-ray diffraction.
Supporting information
CCDC references: 1479756; 1479757; 1479758; 1479759; 1479760; 1479761; 1479762
Data collection: APEX2 (Bruker, 2005) for ma2picformI, ma3pic, ma3clpyr, ma4pic, tfa2pic, tfa3pic; COLLECT (2000) for ma2picformII. Cell refinement: SAINT-Plus (Bruker, 2004) for ma2picformI, ma3pic, ma3clpyr, ma4pic, tfa2pic, tfa3pic; DENZO-SMN (Otwinowski & Minor, 1997) for ma2picformII. Data reduction: SAINT-Plus and XPREP (Bruker 2004) for ma2picformI, ma3pic, ma3clpyr, ma4pic, tfa2pic, tfa3pic; DENZO-SMN (Otwinowski & Minor, 1997) for ma2picformII. For all compounds, program(s) used to solve structure: SHELXS97 (Sheldrick, 1990); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: X-SEED (Barbour, 1999); software used to prepare material for publication: SHELXL97 (Sheldrick, 1997).
(ma2picformI) 2-(2,3-dimethylphenyl)aminobenzoic acid 2-methylpyridine solvate form I
top
Crystal data top
| C21H22N2O2 | Z = 2 |
| Mr = 334.41 | F(000) = 356 |
| Triclinic, P1 | Dx = 1.250 Mg m−3 |
| a = 7.6021 (15) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 8.4538 (17) Å | Cell parameters from 22483 reflections |
| c = 15.375 (3) Å | θ = 1.3–28.4° |
| α = 88.99 (3)° | µ = 0.08 mm−1 |
| β = 84.05 (3)° | T = 173 K |
| γ = 64.75 (3)° | Block, colourless |
| V = 888.5 (3) Å3 | 0.48 × 0.27 × 0.14 mm |
Data collection top
Bruker Kappa Duo Apex II
diffractometer | 4474 independent reflections |
| Radiation source: fine-focus sealed tube | 3761 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.028 |
| 1.2° φ scans and ω scans | θmax = 28.4°, θmin = 1.3° |
Absorption correction: multi-scan
SADABS | h = −10→10 |
| Tmin = 0.933, Tmax = 0.989 | k = −11→11 |
| 22483 measured reflections | l = −20→20 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.043 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.123 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0574P)2 + 0.2396P]
where P = (Fo2 + 2Fc2)/3 |
| 4474 reflections | (Δ/σ)max = 0.002 |
| 234 parameters | Δρmax = 0.29 e Å−3 |
| 0 restraints | Δρmin = −0.23 e Å−3 |
Special details top
Experimental. 'SADABS (Sheldrick, 1996)' |
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and
goodness of fit S are based on F2, conventional R-factors R are based
on F, with F set to zero for negative F2. The threshold expression of
F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is
not relevant to the choice of reflections for refinement. R-factors based
on F2 are statistically about twice as large as those based on F, and R-
factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.93288 (15) | 0.85226 (13) | 0.23162 (6) | 0.0416 (2) | |
| N1 | 0.77690 (16) | 0.63686 (14) | 0.19267 (7) | 0.0338 (2) | |
| C9 | 0.74740 (15) | 0.55465 (15) | 0.04623 (7) | 0.0273 (2) | |
| O2 | 0.88541 (16) | 0.94907 (14) | 0.36922 (6) | 0.0462 (3) | |
| H2 | 0.9614 | 0.9926 | 0.3491 | 0.055* | |
| C2 | 0.72635 (16) | 0.77680 (14) | 0.33511 (7) | 0.0272 (2) | |
| N2 | 1.12083 (15) | 1.09333 (13) | 0.31868 (7) | 0.0325 (2) | |
| H7 | 0.846 (2) | 0.698 (2) | 0.1775 (11) | 0.048 (4)* | |
| C10 | 0.75967 (15) | 0.42546 (16) | −0.01340 (8) | 0.0305 (2) | |
| C8 | 0.78127 (16) | 0.50911 (15) | 0.13294 (7) | 0.0278 (2) | |
| C7 | 0.68887 (16) | 0.66913 (14) | 0.27698 (7) | 0.0271 (2) | |
| C6 | 0.54976 (18) | 0.60615 (15) | 0.30710 (8) | 0.0327 (3) | |
| H6 | 0.5212 | 0.5347 | 0.2692 | 0.039* | |
| C13 | 0.82653 (17) | 0.33916 (16) | 0.15942 (8) | 0.0327 (3) | |
| H13 | 0.8506 | 0.3094 | 0.2183 | 0.039* | |
| C1 | 0.85811 (17) | 0.86047 (15) | 0.30668 (8) | 0.0303 (2) | |
| C3 | 0.62827 (18) | 0.81340 (16) | 0.41939 (8) | 0.0333 (3) | |
| H3 | 0.6550 | 0.8846 | 0.4583 | 0.040* | |
| C11 | 0.80384 (18) | 0.25699 (16) | 0.01462 (9) | 0.0360 (3) | |
| H11 | 0.8117 | 0.1701 | −0.0259 | 0.043* | |
| C5 | 0.45488 (19) | 0.64628 (17) | 0.39020 (9) | 0.0381 (3) | |
| H5 | 0.3610 | 0.6028 | 0.4086 | 0.046* | |
| C12 | 0.83660 (18) | 0.21329 (16) | 0.10028 (9) | 0.0366 (3) | |
| H12 | 0.8659 | 0.0976 | 0.1185 | 0.044* | |
| C14 | 0.69770 (19) | 0.73804 (16) | 0.01700 (8) | 0.0368 (3) | |
| H14A | 0.6753 | 0.8146 | 0.0681 | 0.055* | |
| H14C | 0.5791 | 0.7801 | −0.0131 | 0.055* | |
| H14B | 0.8062 | 0.7390 | −0.0230 | 0.055* | |
| C16 | 1.11804 (18) | 1.22465 (16) | 0.36674 (8) | 0.0335 (3) | |
| C4 | 0.4936 (2) | 0.74934 (17) | 0.44798 (9) | 0.0391 (3) | |
| H4 | 0.4289 | 0.7750 | 0.5057 | 0.047* | |
| C15 | 0.72063 (19) | 0.4705 (2) | −0.10663 (8) | 0.0408 (3) | |
| H15B | 0.7449 | 0.3642 | −0.1401 | 0.061* | |
| H15C | 0.8074 | 0.5214 | −0.1325 | 0.061* | |
| H15A | 0.5841 | 0.5552 | −0.1081 | 0.061* | |
| C20 | 1.2549 (2) | 1.02909 (17) | 0.24974 (9) | 0.0388 (3) | |
| H20 | 1.2546 | 0.9368 | 0.2154 | 0.047* | |
| C17 | 1.2552 (2) | 1.29112 (19) | 0.34653 (10) | 0.0471 (3) | |
| H17 | 1.2531 | 1.3838 | 0.3814 | 0.057* | |
| C19 | 1.3933 (2) | 1.09044 (18) | 0.22616 (10) | 0.0445 (3) | |
| H19 | 1.4862 | 1.0423 | 0.1765 | 0.053* | |
| C21 | 0.9619 (2) | 1.2946 (2) | 0.44185 (10) | 0.0472 (3) | |
| H21C | 0.8340 | 1.3529 | 0.4196 | 0.071* | |
| H21A | 0.9850 | 1.3787 | 0.4765 | 0.071* | |
| H21B | 0.9644 | 1.1980 | 0.4787 | 0.071* | |
| C18 | 1.3940 (2) | 1.2228 (2) | 0.27613 (11) | 0.0499 (4) | |
| H18 | 1.4894 | 1.2666 | 0.2622 | 0.060* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0530 (6) | 0.0476 (5) | 0.0372 (5) | −0.0360 (5) | 0.0063 (4) | −0.0081 (4) |
| N1 | 0.0444 (6) | 0.0409 (6) | 0.0277 (5) | −0.0294 (5) | −0.0015 (4) | −0.0040 (4) |
| C9 | 0.0230 (5) | 0.0309 (5) | 0.0276 (5) | −0.0115 (4) | −0.0005 (4) | −0.0023 (4) |
| O2 | 0.0628 (6) | 0.0614 (6) | 0.0374 (5) | −0.0493 (5) | −0.0001 (4) | −0.0086 (4) |
| C2 | 0.0287 (5) | 0.0265 (5) | 0.0296 (5) | −0.0145 (4) | −0.0044 (4) | −0.0006 (4) |
| N2 | 0.0345 (5) | 0.0309 (5) | 0.0373 (5) | −0.0176 (4) | −0.0103 (4) | 0.0019 (4) |
| C10 | 0.0218 (5) | 0.0385 (6) | 0.0291 (6) | −0.0113 (4) | 0.0005 (4) | −0.0075 (5) |
| C8 | 0.0265 (5) | 0.0316 (5) | 0.0280 (5) | −0.0148 (4) | −0.0029 (4) | −0.0032 (4) |
| C7 | 0.0289 (5) | 0.0269 (5) | 0.0278 (5) | −0.0135 (4) | −0.0059 (4) | 0.0003 (4) |
| C6 | 0.0358 (6) | 0.0322 (6) | 0.0366 (6) | −0.0208 (5) | −0.0040 (5) | −0.0037 (5) |
| C13 | 0.0306 (6) | 0.0339 (6) | 0.0330 (6) | −0.0127 (5) | −0.0060 (5) | 0.0025 (5) |
| C1 | 0.0326 (6) | 0.0294 (5) | 0.0335 (6) | −0.0173 (5) | −0.0042 (5) | −0.0019 (4) |
| C3 | 0.0392 (6) | 0.0329 (6) | 0.0324 (6) | −0.0200 (5) | −0.0010 (5) | −0.0063 (5) |
| C11 | 0.0303 (6) | 0.0353 (6) | 0.0415 (7) | −0.0138 (5) | 0.0025 (5) | −0.0150 (5) |
| C5 | 0.0399 (6) | 0.0380 (6) | 0.0437 (7) | −0.0254 (5) | 0.0051 (5) | −0.0050 (5) |
| C12 | 0.0324 (6) | 0.0270 (5) | 0.0476 (7) | −0.0109 (5) | 0.0001 (5) | −0.0019 (5) |
| C14 | 0.0410 (7) | 0.0356 (6) | 0.0331 (6) | −0.0162 (5) | −0.0026 (5) | 0.0030 (5) |
| C16 | 0.0387 (6) | 0.0315 (6) | 0.0357 (6) | −0.0190 (5) | −0.0090 (5) | 0.0015 (5) |
| C4 | 0.0454 (7) | 0.0416 (7) | 0.0347 (6) | −0.0254 (6) | 0.0085 (5) | −0.0086 (5) |
| C15 | 0.0339 (6) | 0.0569 (8) | 0.0283 (6) | −0.0167 (6) | 0.0000 (5) | −0.0092 (5) |
| C20 | 0.0414 (7) | 0.0312 (6) | 0.0421 (7) | −0.0126 (5) | −0.0091 (5) | −0.0015 (5) |
| C17 | 0.0577 (9) | 0.0417 (7) | 0.0556 (9) | −0.0347 (7) | −0.0031 (7) | −0.0035 (6) |
| C19 | 0.0406 (7) | 0.0416 (7) | 0.0450 (8) | −0.0128 (6) | 0.0007 (6) | 0.0055 (6) |
| C21 | 0.0505 (8) | 0.0538 (8) | 0.0425 (8) | −0.0282 (7) | 0.0007 (6) | −0.0097 (6) |
| C18 | 0.0491 (8) | 0.0467 (8) | 0.0633 (10) | −0.0308 (7) | −0.0009 (7) | 0.0096 (7) |
Geometric parameters (Å, º) top
| O1—C1 | 1.2218 (15) | C11—C12 | 1.380 (2) |
| N1—C7 | 1.3709 (15) | C11—H11 | 0.9500 |
| N1—C8 | 1.4174 (15) | C5—C4 | 1.3910 (18) |
| N1—H7 | 0.894 (17) | C5—H5 | 0.9500 |
| C9—C8 | 1.4002 (16) | C12—H12 | 0.9500 |
| C9—C10 | 1.4045 (16) | C14—H14A | 0.9800 |
| C9—C14 | 1.5031 (17) | C14—H14C | 0.9800 |
| O2—C1 | 1.3145 (14) | C14—H14B | 0.9800 |
| O2—H2 | 0.8400 | C16—C17 | 1.3881 (18) |
| C2—C3 | 1.3956 (17) | C16—C21 | 1.494 (2) |
| C2—C7 | 1.4182 (15) | C4—H4 | 0.9500 |
| C2—C1 | 1.4825 (15) | C15—H15B | 0.9800 |
| N2—C20 | 1.3347 (18) | C15—H15C | 0.9800 |
| N2—C16 | 1.3358 (15) | C15—H15A | 0.9800 |
| C10—C11 | 1.3882 (18) | C20—C19 | 1.374 (2) |
| C10—C15 | 1.5034 (17) | C20—H20 | 0.9500 |
| C8—C13 | 1.3911 (17) | C17—C18 | 1.373 (2) |
| C7—C6 | 1.4094 (16) | C17—H17 | 0.9500 |
| C6—C5 | 1.3724 (18) | C19—C18 | 1.370 (2) |
| C6—H6 | 0.9500 | C19—H19 | 0.9500 |
| C13—C12 | 1.3839 (18) | C21—H21C | 0.9800 |
| C13—H13 | 0.9500 | C21—H21A | 0.9800 |
| C3—C4 | 1.3805 (17) | C21—H21B | 0.9800 |
| C3—H3 | 0.9500 | C18—H18 | 0.9500 |
| | | |
| C7—N1—C8 | 126.51 (10) | C11—C12—H12 | 120.2 |
| C7—N1—H7 | 114.2 (10) | C13—C12—H12 | 120.2 |
| C8—N1—H7 | 119.0 (11) | C9—C14—H14A | 109.5 |
| C8—C9—C10 | 118.69 (11) | C9—C14—H14C | 109.5 |
| C8—C9—C14 | 121.06 (11) | H14A—C14—H14C | 109.5 |
| C10—C9—C14 | 120.25 (11) | C9—C14—H14B | 109.5 |
| C1—O2—H2 | 109.5 | H14A—C14—H14B | 109.5 |
| C3—C2—C7 | 119.35 (10) | H14C—C14—H14B | 109.5 |
| C3—C2—C1 | 119.07 (10) | N2—C16—C17 | 120.55 (12) |
| C7—C2—C1 | 121.46 (10) | N2—C16—C21 | 116.94 (11) |
| C20—N2—C16 | 119.12 (11) | C17—C16—C21 | 122.50 (12) |
| C11—C10—C9 | 119.69 (11) | C3—C4—C5 | 118.41 (11) |
| C11—C10—C15 | 120.17 (11) | C3—C4—H4 | 120.8 |
| C9—C10—C15 | 120.13 (11) | C5—C4—H4 | 120.8 |
| C13—C8—C9 | 120.63 (11) | C10—C15—H15B | 109.5 |
| C13—C8—N1 | 120.29 (11) | C10—C15—H15C | 109.5 |
| C9—C8—N1 | 119.03 (10) | H15B—C15—H15C | 109.5 |
| N1—C7—C6 | 121.44 (10) | C10—C15—H15A | 109.5 |
| N1—C7—C2 | 120.64 (10) | H15B—C15—H15A | 109.5 |
| C6—C7—C2 | 117.80 (11) | H15C—C15—H15A | 109.5 |
| C5—C6—C7 | 121.17 (11) | N2—C20—C19 | 122.92 (12) |
| C5—C6—H6 | 119.4 | N2—C20—H20 | 118.5 |
| C7—C6—H6 | 119.4 | C19—C20—H20 | 118.5 |
| C12—C13—C8 | 120.18 (11) | C18—C17—C16 | 119.90 (13) |
| C12—C13—H13 | 119.9 | C18—C17—H17 | 120.1 |
| C8—C13—H13 | 119.9 | C16—C17—H17 | 120.1 |
| O1—C1—O2 | 122.07 (11) | C18—C19—C20 | 118.36 (13) |
| O1—C1—C2 | 123.79 (10) | C18—C19—H19 | 120.8 |
| O2—C1—C2 | 114.13 (10) | C20—C19—H19 | 120.8 |
| C4—C3—C2 | 122.01 (11) | C16—C21—H21C | 109.5 |
| C4—C3—H3 | 119.0 | C16—C21—H21A | 109.5 |
| C2—C3—H3 | 119.0 | H21C—C21—H21A | 109.5 |
| C12—C11—C10 | 121.25 (11) | C16—C21—H21B | 109.5 |
| C12—C11—H11 | 119.4 | H21C—C21—H21B | 109.5 |
| C10—C11—H11 | 119.4 | H21A—C21—H21B | 109.5 |
| C6—C5—C4 | 121.25 (11) | C19—C18—C17 | 119.13 (13) |
| C6—C5—H5 | 119.4 | C19—C18—H18 | 120.4 |
| C4—C5—H5 | 119.4 | C17—C18—H18 | 120.4 |
| C11—C12—C13 | 119.56 (11) | | |
| | | |
| C8—C9—C10—C11 | 0.29 (16) | C7—C2—C1—O1 | 4.62 (18) |
| C14—C9—C10—C11 | −179.38 (10) | C3—C2—C1—O2 | 7.04 (17) |
| C8—C9—C10—C15 | 178.90 (10) | C7—C2—C1—O2 | −176.82 (11) |
| C14—C9—C10—C15 | −0.77 (16) | C7—C2—C3—C4 | −0.68 (19) |
| C10—C9—C8—C13 | 0.12 (16) | C1—C2—C3—C4 | 175.55 (12) |
| C14—C9—C8—C13 | 179.78 (10) | C9—C10—C11—C12 | −0.15 (17) |
| C10—C9—C8—N1 | 177.49 (10) | C15—C10—C11—C12 | −178.76 (11) |
| C14—C9—C8—N1 | −2.84 (16) | C7—C6—C5—C4 | −0.5 (2) |
| C7—N1—C8—C13 | −46.26 (17) | C10—C11—C12—C13 | −0.39 (18) |
| C7—N1—C8—C9 | 136.35 (12) | C8—C13—C12—C11 | 0.80 (18) |
| C8—N1—C7—C6 | −16.14 (18) | C20—N2—C16—C17 | −1.39 (19) |
| C8—N1—C7—C2 | 167.88 (11) | C20—N2—C16—C21 | 178.53 (12) |
| C3—C2—C7—N1 | 177.29 (11) | C2—C3—C4—C5 | −0.4 (2) |
| C1—C2—C7—N1 | 1.15 (17) | C6—C5—C4—C3 | 1.0 (2) |
| C3—C2—C7—C6 | 1.16 (16) | C16—N2—C20—C19 | 0.89 (19) |
| C1—C2—C7—C6 | −174.98 (11) | N2—C16—C17—C18 | 0.6 (2) |
| N1—C7—C6—C5 | −176.68 (12) | C21—C16—C17—C18 | −179.31 (14) |
| C2—C7—C6—C5 | −0.58 (18) | N2—C20—C19—C18 | 0.4 (2) |
| C9—C8—C13—C12 | −0.67 (17) | C20—C19—C18—C17 | −1.2 (2) |
| N1—C8—C13—C12 | −178.01 (11) | C16—C17—C18—C19 | 0.7 (2) |
| C3—C2—C1—O1 | −171.53 (12) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···N2 | 0.84 | 1.78 | 2.6142 (15) | 174 |
| N1—H7···O1 | 0.894 (17) | 1.928 (17) | 2.6661 (14) | 138.7 (14) |
| C12—H12···O1i | 0.95 | 2.60 | 3.480 (2) | 155 |
| C15—H15B···O1ii | 0.98 | 2.62 | 3.329 (2) | 129 |
| Symmetry codes: (i) x, y−1, z; (ii) −x+2, −y+1, −z. |
(ma2picformII) 2-(2,3-dimethylphenyl)aminobenzoic acid 2methylpyridine solvate form II
top
Crystal data top
| C21H22N2O2 | F(000) = 712 |
| Mr = 334.41 | Dx = 1.256 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.7857 (16) Å | Cell parameters from 7473 reflections |
| b = 8.1859 (16) Å | θ = 2.6–27.1° |
| c = 27.953 (6) Å | µ = 0.08 mm−1 |
| β = 96.87 (3)° | T = 173 K |
| V = 1768.7 (6) Å3 | Block, colourless |
| Z = 4 | 0.34 × 0.19 × 0.11 mm |
Data collection top
Nonius KappaCCD area-detector
diffractometer | 2906 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.020 |
| Graphite monochromator | θmax = 27.1°, θmin = 3.8° |
| 1° φ scans and ω scans | h = −9→9 |
| 7453 measured reflections | k = −10→10 |
| 3879 independent reflections | l = −35→35 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.045 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.131 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0708P)2 + 0.3517P]
where P = (Fo2 + 2Fc2)/3 |
| 3879 reflections | (Δ/σ)max = 0.009 |
| 237 parameters | Δρmax = 0.28 e Å−3 |
| 0 restraints | Δρmin = −0.35 e Å−3 |
Special details top
Experimental. none |
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and
goodness of fit S are based on F2, conventional R-factors R are based
on F, with F set to zero for negative F2. The threshold expression of
F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is
not relevant to the choice of reflections for refinement. R-factors based
on F2 are statistically about twice as large as those based on F, and R-
factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.50682 (12) | 0.84893 (14) | 0.06316 (4) | 0.0420 (3) | |
| O2 | 0.67821 (14) | 0.80923 (15) | 0.00585 (4) | 0.0433 (3) | |
| N2 | 0.40271 (15) | 0.66874 (15) | −0.03983 (4) | 0.0368 (3) | |
| H2 | 0.569 (3) | 0.751 (3) | −0.0100 (8) | 0.089 (7)* | |
| H7 | 0.554 (2) | 0.949 (2) | 0.1254 (6) | 0.041 (4)* | |
| C10 | 0.45991 (16) | 1.05292 (18) | 0.25931 (5) | 0.0315 (3) | |
| N1 | 0.63459 (16) | 1.00338 (18) | 0.14251 (4) | 0.0400 (3) | |
| C2 | 0.80224 (17) | 0.93327 (17) | 0.07796 (5) | 0.0306 (3) | |
| C7 | 0.79050 (17) | 1.00271 (17) | 0.12387 (5) | 0.0301 (3) | |
| C5 | 1.09864 (18) | 1.05909 (18) | 0.13103 (5) | 0.0369 (3) | |
| H5 | 1.1994 | 1.1017 | 0.1493 | 0.044* | |
| C9 | 0.50040 (16) | 0.98256 (17) | 0.21616 (5) | 0.0297 (3) | |
| C6 | 0.94329 (17) | 1.06320 (18) | 0.14995 (5) | 0.0333 (3) | |
| H6 | 0.9391 | 1.1077 | 0.1812 | 0.040* | |
| C13 | 0.65067 (18) | 1.23202 (18) | 0.19910 (5) | 0.0369 (3) | |
| H13 | 0.7138 | 1.2940 | 0.1784 | 0.044* | |
| C8 | 0.59697 (16) | 1.07363 (18) | 0.18626 (5) | 0.0313 (3) | |
| C1 | 0.64933 (18) | 0.86171 (17) | 0.04862 (5) | 0.0330 (3) | |
| C3 | 0.96283 (18) | 0.93070 (18) | 0.05994 (5) | 0.0370 (3) | |
| H3 | 0.9703 | 0.8843 | 0.0291 | 0.044* | |
| C11 | 0.51849 (18) | 1.20889 (19) | 0.27175 (5) | 0.0377 (3) | |
| H11 | 0.4939 | 1.2551 | 0.3014 | 0.045* | |
| C16 | 0.41120 (19) | 0.56900 (18) | −0.07752 (5) | 0.0365 (3) | |
| C19 | 0.0992 (2) | 0.6203 (2) | −0.04358 (6) | 0.0425 (4) | |
| H19 | −0.0074 | 0.6412 | −0.0314 | 0.051* | |
| C14 | 0.44577 (19) | 0.81117 (19) | 0.20204 (6) | 0.0399 (4) | |
| H14C | 0.3969 | 0.7585 | 0.2288 | 0.060* | |
| H14A | 0.5465 | 0.7490 | 0.1943 | 0.060* | |
| H14B | 0.3584 | 0.8145 | 0.1737 | 0.060* | |
| C12 | 0.61206 (18) | 1.29841 (19) | 0.24184 (6) | 0.0405 (4) | |
| H12 | 0.6497 | 1.4057 | 0.2508 | 0.049* | |
| C18 | 0.1070 (2) | 0.5160 (2) | −0.08204 (6) | 0.0439 (4) | |
| H18 | 0.0057 | 0.4623 | −0.0965 | 0.053* | |
| C15 | 0.3563 (2) | 0.9609 (2) | 0.29288 (6) | 0.0437 (4) | |
| H15A | 0.3317 | 1.0330 | 0.3192 | 0.066* | |
| H15C | 0.4228 | 0.8662 | 0.3062 | 0.066* | |
| H15B | 0.2473 | 0.9237 | 0.2751 | 0.066* | |
| C20 | 0.24944 (19) | 0.6929 (2) | −0.02345 (5) | 0.0409 (4) | |
| H20 | 0.2448 | 0.7637 | 0.0033 | 0.049* | |
| C17 | 0.2637 (2) | 0.4906 (2) | −0.09922 (6) | 0.0418 (4) | |
| H17 | 0.2708 | 0.4198 | −0.1258 | 0.050* | |
| C4 | 1.11037 (18) | 0.9937 (2) | 0.08569 (6) | 0.0413 (4) | |
| H4 | 1.2176 | 0.9923 | 0.0727 | 0.050* | |
| C21 | 0.5852 (2) | 0.5465 (2) | −0.09444 (6) | 0.0490 (4) | |
| H21C | 0.6635 | 0.6329 | −0.0809 | 0.073* | |
| H21A | 0.5735 | 0.5520 | −0.1297 | 0.073* | |
| H21B | 0.6323 | 0.4398 | −0.0838 | 0.073* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0292 (5) | 0.0559 (7) | 0.0417 (6) | −0.0035 (5) | 0.0083 (4) | −0.0117 (5) |
| O2 | 0.0381 (6) | 0.0591 (7) | 0.0337 (6) | −0.0048 (5) | 0.0091 (4) | −0.0092 (5) |
| N2 | 0.0367 (7) | 0.0398 (7) | 0.0339 (6) | 0.0014 (5) | 0.0041 (5) | 0.0000 (5) |
| C10 | 0.0237 (6) | 0.0403 (8) | 0.0305 (7) | 0.0048 (6) | 0.0031 (5) | 0.0012 (6) |
| N1 | 0.0299 (6) | 0.0587 (8) | 0.0330 (7) | −0.0122 (6) | 0.0097 (5) | −0.0105 (6) |
| C2 | 0.0292 (7) | 0.0292 (7) | 0.0340 (7) | 0.0007 (5) | 0.0067 (5) | 0.0030 (6) |
| C7 | 0.0297 (7) | 0.0301 (7) | 0.0314 (7) | −0.0005 (5) | 0.0071 (5) | 0.0046 (6) |
| C5 | 0.0276 (7) | 0.0361 (8) | 0.0464 (9) | −0.0010 (6) | 0.0023 (6) | 0.0022 (6) |
| C9 | 0.0223 (6) | 0.0346 (7) | 0.0321 (7) | 0.0002 (5) | 0.0030 (5) | −0.0010 (6) |
| C6 | 0.0323 (7) | 0.0338 (7) | 0.0338 (7) | −0.0017 (6) | 0.0041 (6) | 0.0021 (6) |
| C13 | 0.0297 (7) | 0.0382 (8) | 0.0429 (8) | −0.0036 (6) | 0.0048 (6) | 0.0035 (7) |
| C8 | 0.0241 (6) | 0.0398 (8) | 0.0301 (7) | −0.0015 (6) | 0.0032 (5) | −0.0016 (6) |
| C1 | 0.0330 (7) | 0.0344 (7) | 0.0321 (7) | 0.0030 (6) | 0.0066 (6) | 0.0002 (6) |
| C3 | 0.0352 (8) | 0.0375 (8) | 0.0400 (8) | 0.0026 (6) | 0.0115 (6) | −0.0026 (6) |
| C11 | 0.0301 (7) | 0.0470 (9) | 0.0356 (8) | 0.0035 (6) | 0.0022 (6) | −0.0099 (7) |
| C16 | 0.0406 (8) | 0.0365 (8) | 0.0325 (7) | 0.0035 (6) | 0.0052 (6) | 0.0038 (6) |
| C19 | 0.0360 (8) | 0.0475 (9) | 0.0444 (8) | 0.0046 (7) | 0.0064 (6) | 0.0076 (7) |
| C14 | 0.0373 (8) | 0.0383 (8) | 0.0458 (8) | −0.0054 (6) | 0.0115 (6) | −0.0048 (7) |
| C12 | 0.0297 (7) | 0.0358 (8) | 0.0553 (9) | −0.0021 (6) | 0.0024 (6) | −0.0101 (7) |
| C18 | 0.0407 (8) | 0.0449 (9) | 0.0442 (9) | −0.0061 (7) | −0.0027 (7) | 0.0077 (7) |
| C15 | 0.0426 (8) | 0.0527 (10) | 0.0379 (8) | 0.0048 (7) | 0.0138 (6) | 0.0046 (7) |
| C20 | 0.0407 (8) | 0.0437 (8) | 0.0387 (8) | 0.0043 (7) | 0.0066 (6) | −0.0009 (7) |
| C17 | 0.0489 (9) | 0.0396 (8) | 0.0361 (8) | −0.0022 (7) | 0.0023 (7) | −0.0017 (7) |
| C4 | 0.0281 (7) | 0.0434 (9) | 0.0544 (9) | 0.0024 (6) | 0.0135 (6) | 0.0002 (7) |
| C21 | 0.0444 (9) | 0.0577 (11) | 0.0463 (9) | −0.0001 (8) | 0.0117 (7) | −0.0066 (8) |
Geometric parameters (Å, º) top
| O1—C1 | 1.2308 (17) | C5—C6 | 1.3772 (19) |
| O2—C1 | 1.3150 (17) | C5—C4 | 1.389 (2) |
| N2—C16 | 1.3408 (19) | C9—C8 | 1.4036 (19) |
| N2—C20 | 1.3430 (19) | C9—C14 | 1.505 (2) |
| C10—C11 | 1.386 (2) | C13—C12 | 1.378 (2) |
| C10—C9 | 1.4059 (19) | C13—C8 | 1.396 (2) |
| C10—C15 | 1.510 (2) | C3—C4 | 1.380 (2) |
| N1—C7 | 1.3771 (17) | C11—C12 | 1.383 (2) |
| N1—C8 | 1.4134 (18) | C16—C17 | 1.390 (2) |
| C2—C3 | 1.4029 (19) | C16—C21 | 1.499 (2) |
| C2—C7 | 1.4165 (19) | C19—C20 | 1.372 (2) |
| C2—C1 | 1.483 (2) | C19—C18 | 1.380 (2) |
| C7—C6 | 1.409 (2) | C18—C17 | 1.379 (2) |
| | | |
| C16—N2—C20 | 119.14 (13) | C13—C8—C9 | 120.36 (13) |
| C11—C10—C9 | 119.46 (13) | C13—C8—N1 | 120.91 (13) |
| C11—C10—C15 | 119.19 (13) | C9—C8—N1 | 118.71 (13) |
| C9—C10—C15 | 121.34 (13) | O1—C1—O2 | 122.10 (13) |
| C7—N1—C8 | 127.75 (12) | O1—C1—C2 | 123.14 (12) |
| C3—C2—C7 | 119.22 (13) | O2—C1—C2 | 114.75 (12) |
| C3—C2—C1 | 119.11 (12) | C4—C3—C2 | 121.93 (14) |
| C7—C2—C1 | 121.67 (12) | C12—C11—C10 | 121.18 (13) |
| N1—C7—C6 | 122.07 (13) | N2—C16—C17 | 120.55 (13) |
| N1—C7—C2 | 119.97 (12) | N2—C16—C21 | 117.02 (13) |
| C6—C7—C2 | 117.90 (12) | C17—C16—C21 | 122.43 (14) |
| C6—C5—C4 | 121.09 (13) | C20—C19—C18 | 118.27 (14) |
| C8—C9—C10 | 118.97 (13) | C13—C12—C11 | 120.04 (14) |
| C8—C9—C14 | 119.58 (12) | C17—C18—C19 | 119.16 (15) |
| C10—C9—C14 | 121.44 (12) | N2—C20—C19 | 123.02 (14) |
| C5—C6—C7 | 121.25 (13) | C18—C17—C16 | 119.85 (15) |
| C12—C13—C8 | 119.97 (14) | C3—C4—C5 | 118.58 (13) |
| | | |
| C8—N1—C7—C6 | 6.1 (2) | C3—C2—C1—O1 | 174.85 (14) |
| C8—N1—C7—C2 | −176.69 (14) | C7—C2—C1—O1 | −4.6 (2) |
| C3—C2—C7—N1 | −178.74 (13) | C3—C2—C1—O2 | −3.80 (19) |
| C1—C2—C7—N1 | 0.7 (2) | C7—C2—C1—O2 | 176.79 (12) |
| C3—C2—C7—C6 | −1.4 (2) | C7—C2—C3—C4 | 0.1 (2) |
| C1—C2—C7—C6 | 178.00 (12) | C1—C2—C3—C4 | −179.28 (14) |
| C11—C10—C9—C8 | −1.12 (19) | C9—C10—C11—C12 | 1.8 (2) |
| C15—C10—C9—C8 | 179.95 (12) | C15—C10—C11—C12 | −179.23 (13) |
| C11—C10—C9—C14 | 177.57 (12) | C20—N2—C16—C17 | −0.4 (2) |
| C15—C10—C9—C14 | −1.4 (2) | C20—N2—C16—C21 | 179.87 (14) |
| C4—C5—C6—C7 | −0.6 (2) | C8—C13—C12—C11 | −0.7 (2) |
| N1—C7—C6—C5 | 178.93 (13) | C10—C11—C12—C13 | −0.9 (2) |
| C2—C7—C6—C5 | 1.7 (2) | C20—C19—C18—C17 | −1.0 (2) |
| C12—C13—C8—C9 | 1.4 (2) | C16—N2—C20—C19 | −0.1 (2) |
| C12—C13—C8—N1 | 179.75 (13) | C18—C19—C20—N2 | 0.8 (2) |
| C10—C9—C8—C13 | −0.46 (19) | C19—C18—C17—C16 | 0.5 (2) |
| C14—C9—C8—C13 | −179.17 (12) | N2—C16—C17—C18 | 0.3 (2) |
| C10—C9—C8—N1 | −178.86 (12) | C21—C16—C17—C18 | 179.94 (15) |
| C14—C9—C8—N1 | 2.43 (19) | C2—C3—C4—C5 | 0.9 (2) |
| C7—N1—C8—C13 | 46.8 (2) | C6—C5—C4—C3 | −0.7 (2) |
| C7—N1—C8—C9 | −134.76 (15) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···N2 | 1.03 (2) | 1.60 (2) | 2.6265 (17) | 174 (2) |
| N1—H7···O1 | 0.863 (17) | 1.919 (17) | 2.6421 (17) | 140.4 (15) |
| C4—H4···O1i | 0.95 | 2.58 | 3.4341 (19) | 150 |
| Symmetry code: (i) x+1, y, z. |
(ma3pic) 2-(2,3-dimethylphenyl)aminobenzoic acid 3-methylpyridine solvate
top
Crystal data top
| C21H22N2O2 | Z = 2 |
| Mr = 334.41 | F(000) = 356 |
| Triclinic, P1 | Dx = 1.288 Mg m−3 |
| a = 7.6738 (6) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 7.7598 (6) Å | Cell parameters from 20017 reflections |
| c = 16.2725 (12) Å | θ = 2.6–28.4° |
| α = 78.957 (1)° | µ = 0.08 mm−1 |
| β = 83.673 (2)° | T = 173 K |
| γ = 65.094 (1)° | Block, orange |
| V = 862.12 (11) Å3 | 0.36 × 0.30 × 0.29 mm |
Data collection top
Bruker Kappa Duo Apex II
diffractometer | 4332 independent reflections |
| Radiation source: fine-focus sealed tube | 3671 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.026 |
| 1.2° φ scans and ω scans | θmax = 28.4°, θmin = 2.6° |
Absorption correction: multi-scan
SADABS | h = −10→10 |
| Tmin = 0.926, Tmax = 0.976 | k = −10→10 |
| 20017 measured reflections | l = −21→21 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.117 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.059P)2 + 0.2156P]
where P = (Fo2 + 2Fc2)/3 |
| 4332 reflections | (Δ/σ)max = 0.002 |
| 237 parameters | Δρmax = 0.34 e Å−3 |
| 0 restraints | Δρmin = −0.22 e Å−3 |
Special details top
Experimental. 'SADABS (Sheldrick, 1996)' |
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and
goodness of fit S are based on F2, conventional R-factors R are based
on F, with F set to zero for negative F2. The threshold expression of
F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is
not relevant to the choice of reflections for refinement. R-factors based
on F2 are statistically about twice as large as those based on F, and R-
factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.07469 (13) | 0.79435 (12) | 0.14251 (5) | 0.0330 (2) | |
| O2 | 0.22929 (13) | 0.97608 (13) | 0.08350 (5) | 0.0327 (2) | |
| C2 | 0.04471 (14) | 1.05798 (14) | 0.20703 (6) | 0.0201 (2) | |
| N2 | 0.31639 (13) | 0.77296 (13) | −0.03672 (6) | 0.0256 (2) | |
| H7 | −0.090 (2) | 0.811 (2) | 0.2390 (11) | 0.052 (5)* | |
| H2 | 0.259 (3) | 0.896 (3) | 0.0361 (13) | 0.076 (6)* | |
| N1 | −0.11771 (14) | 0.86658 (13) | 0.28649 (6) | 0.0265 (2) | |
| C7 | −0.07473 (15) | 1.02547 (14) | 0.27549 (6) | 0.0206 (2) | |
| C3 | 0.08860 (16) | 1.21816 (15) | 0.19842 (7) | 0.0241 (2) | |
| H3 | 0.1695 | 1.2391 | 0.1527 | 0.029* | |
| C9 | −0.22886 (15) | 0.78590 (14) | 0.43067 (7) | 0.0220 (2) | |
| C5 | −0.10301 (17) | 1.31722 (15) | 0.32046 (7) | 0.0267 (2) | |
| H5 | −0.1553 | 1.4065 | 0.3586 | 0.032* | |
| C8 | −0.25752 (15) | 0.83326 (14) | 0.34426 (7) | 0.0227 (2) | |
| C1 | 0.11681 (15) | 0.93041 (15) | 0.14213 (7) | 0.0225 (2) | |
| C6 | −0.14809 (16) | 1.15946 (15) | 0.33131 (7) | 0.0241 (2) | |
| H6 | −0.2299 | 1.1412 | 0.3772 | 0.029* | |
| C18 | 0.42441 (16) | 0.53182 (17) | −0.15498 (7) | 0.0272 (2) | |
| H18 | 0.4625 | 0.4491 | −0.1958 | 0.033* | |
| C16 | 0.26442 (16) | 0.62605 (16) | −0.02772 (7) | 0.0252 (2) | |
| H16 | 0.1885 | 0.6070 | 0.0204 | 0.030* | |
| C10 | −0.36719 (15) | 0.74252 (15) | 0.48415 (7) | 0.0236 (2) | |
| C13 | −0.41997 (17) | 0.83661 (16) | 0.31176 (7) | 0.0268 (2) | |
| H13 | −0.4387 | 0.8702 | 0.2531 | 0.032* | |
| C20 | 0.42201 (17) | 0.80132 (16) | −0.10473 (7) | 0.0274 (2) | |
| H20 | 0.4597 | 0.9053 | −0.1115 | 0.033* | |
| C17 | 0.31535 (16) | 0.49975 (15) | −0.08496 (7) | 0.0251 (2) | |
| C11 | −0.52731 (16) | 0.74461 (16) | 0.45027 (7) | 0.0277 (2) | |
| H11 | −0.6196 | 0.7136 | 0.4864 | 0.033* | |
| C19 | 0.47803 (17) | 0.68418 (17) | −0.16552 (7) | 0.0290 (2) | |
| H19 | 0.5519 | 0.7079 | −0.2136 | 0.035* | |
| C4 | 0.01763 (17) | 1.34698 (16) | 0.25454 (7) | 0.0277 (2) | |
| H4 | 0.0508 | 1.4539 | 0.2481 | 0.033* | |
| C14 | −0.05255 (16) | 0.77641 (17) | 0.46698 (7) | 0.0288 (2) | |
| H14B | 0.0373 | 0.7917 | 0.4215 | 0.043* | |
| H14C | 0.0094 | 0.6516 | 0.5028 | 0.043* | |
| H14A | −0.0894 | 0.8798 | 0.5004 | 0.043* | |
| C15 | −0.34218 (18) | 0.69274 (18) | 0.57753 (7) | 0.0310 (3) | |
| H15B | −0.3589 | 0.8077 | 0.5998 | 0.047* | |
| H15A | −0.2131 | 0.5925 | 0.5895 | 0.047* | |
| H15C | −0.4384 | 0.6457 | 0.6040 | 0.047* | |
| C21 | 0.2557 (2) | 0.33491 (19) | −0.06983 (9) | 0.0361 (3) | |
| H21B | 0.1759 | 0.3476 | −0.1156 | 0.054* | |
| H21A | 0.1820 | 0.3363 | −0.0166 | 0.054* | |
| H21C | 0.3703 | 0.2133 | −0.0674 | 0.054* | |
| C12 | −0.55404 (17) | 0.79110 (17) | 0.36474 (8) | 0.0301 (3) | |
| H12 | −0.6638 | 0.7918 | 0.3425 | 0.036* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0433 (5) | 0.0337 (4) | 0.0328 (5) | −0.0255 (4) | 0.0158 (4) | −0.0168 (4) |
| O2 | 0.0431 (5) | 0.0366 (5) | 0.0285 (4) | −0.0265 (4) | 0.0162 (4) | −0.0140 (4) |
| C2 | 0.0203 (5) | 0.0203 (5) | 0.0203 (5) | −0.0088 (4) | −0.0005 (4) | −0.0039 (4) |
| N2 | 0.0272 (5) | 0.0261 (4) | 0.0236 (5) | −0.0107 (4) | 0.0012 (4) | −0.0059 (4) |
| N1 | 0.0345 (5) | 0.0256 (5) | 0.0252 (5) | −0.0183 (4) | 0.0112 (4) | −0.0100 (4) |
| C7 | 0.0220 (5) | 0.0195 (5) | 0.0205 (5) | −0.0089 (4) | −0.0004 (4) | −0.0031 (4) |
| C3 | 0.0259 (5) | 0.0255 (5) | 0.0239 (5) | −0.0141 (4) | −0.0008 (4) | −0.0026 (4) |
| C9 | 0.0218 (5) | 0.0192 (5) | 0.0250 (5) | −0.0084 (4) | 0.0022 (4) | −0.0048 (4) |
| C5 | 0.0335 (6) | 0.0225 (5) | 0.0241 (5) | −0.0095 (4) | −0.0041 (4) | −0.0072 (4) |
| C8 | 0.0253 (5) | 0.0194 (5) | 0.0247 (5) | −0.0110 (4) | 0.0055 (4) | −0.0055 (4) |
| C1 | 0.0236 (5) | 0.0242 (5) | 0.0213 (5) | −0.0114 (4) | 0.0028 (4) | −0.0051 (4) |
| C6 | 0.0272 (5) | 0.0239 (5) | 0.0202 (5) | −0.0096 (4) | 0.0022 (4) | −0.0051 (4) |
| C18 | 0.0274 (5) | 0.0306 (6) | 0.0240 (5) | −0.0101 (4) | −0.0021 (4) | −0.0088 (4) |
| C16 | 0.0235 (5) | 0.0286 (5) | 0.0237 (5) | −0.0115 (4) | 0.0002 (4) | −0.0033 (4) |
| C10 | 0.0241 (5) | 0.0201 (5) | 0.0246 (5) | −0.0077 (4) | 0.0038 (4) | −0.0043 (4) |
| C13 | 0.0316 (6) | 0.0273 (5) | 0.0235 (5) | −0.0143 (5) | −0.0014 (4) | −0.0029 (4) |
| C20 | 0.0323 (6) | 0.0275 (5) | 0.0253 (5) | −0.0156 (5) | −0.0001 (4) | −0.0030 (4) |
| C17 | 0.0242 (5) | 0.0246 (5) | 0.0273 (5) | −0.0101 (4) | −0.0056 (4) | −0.0028 (4) |
| C11 | 0.0246 (5) | 0.0289 (5) | 0.0304 (6) | −0.0136 (4) | 0.0056 (4) | −0.0038 (4) |
| C19 | 0.0306 (6) | 0.0353 (6) | 0.0227 (5) | −0.0159 (5) | 0.0029 (4) | −0.0046 (4) |
| C4 | 0.0355 (6) | 0.0237 (5) | 0.0294 (6) | −0.0165 (5) | −0.0050 (5) | −0.0041 (4) |
| C14 | 0.0260 (5) | 0.0312 (6) | 0.0310 (6) | −0.0140 (5) | −0.0016 (4) | −0.0033 (4) |
| C15 | 0.0333 (6) | 0.0336 (6) | 0.0238 (5) | −0.0130 (5) | 0.0040 (4) | −0.0035 (4) |
| C21 | 0.0428 (7) | 0.0344 (6) | 0.0398 (7) | −0.0235 (6) | −0.0029 (6) | −0.0064 (5) |
| C12 | 0.0266 (6) | 0.0340 (6) | 0.0328 (6) | −0.0159 (5) | −0.0018 (5) | −0.0040 (5) |
Geometric parameters (Å, º) top
| O1—C1 | 1.2262 (13) | C9—C14 | 1.5035 (15) |
| O2—C1 | 1.3143 (12) | C5—C6 | 1.3820 (15) |
| C2—C3 | 1.3978 (14) | C5—C4 | 1.3883 (16) |
| C2—C7 | 1.4165 (14) | C8—C13 | 1.3955 (15) |
| C2—C1 | 1.4872 (14) | C18—C19 | 1.3831 (16) |
| N2—C16 | 1.3365 (14) | C18—C17 | 1.3846 (16) |
| N2—C20 | 1.3376 (14) | C16—C17 | 1.3884 (15) |
| N1—C7 | 1.3806 (13) | C10—C11 | 1.3940 (16) |
| N1—C8 | 1.4212 (13) | C10—C15 | 1.5073 (15) |
| C7—C6 | 1.4068 (14) | C13—C12 | 1.3857 (16) |
| C3—C4 | 1.3825 (15) | C20—C19 | 1.3825 (16) |
| C9—C8 | 1.4010 (15) | C17—C21 | 1.5012 (15) |
| C9—C10 | 1.4080 (14) | C11—C12 | 1.3848 (17) |
| | | |
| C3—C2—C7 | 119.22 (9) | O1—C1—C2 | 123.21 (9) |
| C3—C2—C1 | 119.37 (9) | O2—C1—C2 | 114.41 (9) |
| C7—C2—C1 | 121.35 (9) | C5—C6—C7 | 121.03 (10) |
| C16—N2—C20 | 118.48 (9) | C19—C18—C17 | 120.09 (10) |
| C7—N1—C8 | 125.80 (9) | N2—C16—C17 | 123.54 (10) |
| N1—C7—C6 | 121.29 (9) | C11—C10—C9 | 119.65 (10) |
| N1—C7—C2 | 120.53 (9) | C11—C10—C15 | 120.00 (10) |
| C6—C7—C2 | 118.18 (9) | C9—C10—C15 | 120.35 (10) |
| C4—C3—C2 | 121.88 (10) | C12—C13—C8 | 120.22 (10) |
| C8—C9—C10 | 118.85 (10) | N2—C20—C19 | 122.11 (10) |
| C8—C9—C14 | 121.53 (9) | C18—C17—C16 | 117.06 (10) |
| C10—C9—C14 | 119.61 (10) | C18—C17—C21 | 122.26 (10) |
| C6—C5—C4 | 120.89 (10) | C16—C17—C21 | 120.67 (10) |
| C13—C8—C9 | 120.55 (9) | C12—C11—C10 | 121.05 (10) |
| C13—C8—N1 | 117.70 (10) | C20—C19—C18 | 118.72 (10) |
| C9—C8—N1 | 121.61 (10) | C3—C4—C5 | 118.77 (10) |
| O1—C1—O2 | 122.38 (9) | C13—C12—C11 | 119.68 (10) |
| | | |
| C8—N1—C7—C6 | 10.05 (17) | C20—N2—C16—C17 | 0.51 (16) |
| C8—N1—C7—C2 | −169.75 (10) | C8—C9—C10—C11 | 0.98 (15) |
| C3—C2—C7—N1 | −178.81 (10) | C14—C9—C10—C11 | −177.73 (10) |
| C1—C2—C7—N1 | 4.04 (15) | C8—C9—C10—C15 | −179.42 (9) |
| C3—C2—C7—C6 | 1.39 (15) | C14—C9—C10—C15 | 1.87 (15) |
| C1—C2—C7—C6 | −175.76 (9) | C9—C8—C13—C12 | −0.64 (16) |
| C7—C2—C3—C4 | −0.47 (16) | N1—C8—C13—C12 | 175.23 (10) |
| C1—C2—C3—C4 | 176.74 (10) | C16—N2—C20—C19 | 0.15 (17) |
| C10—C9—C8—C13 | −0.26 (15) | C19—C18—C17—C16 | 0.07 (16) |
| C14—C9—C8—C13 | 178.42 (10) | C19—C18—C17—C21 | −179.09 (11) |
| C10—C9—C8—N1 | −175.96 (9) | N2—C16—C17—C18 | −0.61 (17) |
| C14—C9—C8—N1 | 2.72 (15) | N2—C16—C17—C21 | 178.56 (10) |
| C7—N1—C8—C13 | 112.29 (12) | C9—C10—C11—C12 | −0.82 (16) |
| C7—N1—C8—C9 | −71.89 (15) | C15—C10—C11—C12 | 179.58 (10) |
| C3—C2—C1—O1 | −175.36 (10) | N2—C20—C19—C18 | −0.65 (18) |
| C7—C2—C1—O1 | 1.79 (17) | C17—C18—C19—C20 | 0.53 (17) |
| C3—C2—C1—O2 | 4.24 (15) | C2—C3—C4—C5 | −1.02 (17) |
| C7—C2—C1—O2 | −178.62 (9) | C6—C5—C4—C3 | 1.57 (17) |
| C4—C5—C6—C7 | −0.64 (17) | C8—C13—C12—C11 | 0.81 (17) |
| N1—C7—C6—C5 | 179.34 (10) | C10—C11—C12—C13 | −0.08 (17) |
| C2—C7—C6—C5 | −0.86 (16) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H7···O1 | 0.918 (18) | 1.892 (17) | 2.6508 (12) | 138.6 (15) |
| O2—H2···N2 | 1.03 (2) | 1.57 (2) | 2.6006 (12) | 176.7 (18) |
| C16—H16···O1 | 0.95 | 2.54 | 3.2241 (14) | 129 |
(ma3clpyr) 2-(2,3-dimethylphenyl)aminobenzoic acid 3-chloropyridine solvate
top
Crystal data top
| C20H19ClN2O2 | Z = 2 |
| Mr = 354.82 | F(000) = 372 |
| Triclinic, P1 | Dx = 1.379 Mg m−3 |
| a = 7.6718 (15) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 7.7441 (15) Å | Cell parameters from 20017 reflections |
| c = 16.056 (3) Å | θ = 1.3–28.5° |
| α = 80.06 (3)° | µ = 0.24 mm−1 |
| β = 84.88 (3)° | T = 173 K |
| γ = 65.50 (3)° | Block, orange |
| V = 854.8 (4) Å3 | 0.15 × 0.14 × 0.13 mm |
Data collection top
Bruker Kappa Duo Apex II
diffractometer | 4290 independent reflections |
| Radiation source: fine-focus sealed tube | 3370 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.028 |
| 1.2° φ scans and ω scans | θmax = 28.5°, θmin = 1.3° |
Absorption correction: multi-scan
SADABS | h = −10→10 |
| Tmin = 0.891, Tmax = 0.969 | k = −10→10 |
| 13863 measured reflections | l = −21→21 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.122 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0691P)2 + 0.0888P]
where P = (Fo2 + 2Fc2)/3 |
| 4290 reflections | (Δ/σ)max < 0.001 |
| 236 parameters | Δρmax = 0.32 e Å−3 |
| 0 restraints | Δρmin = −0.23 e Å−3 |
Special details top
Experimental. 'SADABS (Sheldrick, 1996)' |
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and
goodness of fit S are based on F2, conventional R-factors R are based
on F, with F set to zero for negative F2. The threshold expression of
F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is
not relevant to the choice of reflections for refinement. R-factors based
on F2 are statistically about twice as large as those based on F, and R-
factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Cl1 | 0.23755 (6) | 0.32298 (5) | 0.42612 (3) | 0.03577 (14) | |
| O1 | 0.07259 (17) | 0.80218 (16) | 0.63918 (7) | 0.0343 (3) | |
| N1 | −0.12767 (19) | 0.87330 (17) | 0.78256 (8) | 0.0262 (3) | |
| O2 | 0.22928 (18) | 0.98357 (17) | 0.58659 (7) | 0.0336 (3) | |
| N2 | 0.32388 (18) | 0.77170 (17) | 0.46329 (8) | 0.0245 (3) | |
| H7 | −0.100 (3) | 0.819 (3) | 0.7356 (13) | 0.048 (6)* | |
| H2 | 0.265 (4) | 0.912 (4) | 0.5493 (16) | 0.075 (8)* | |
| C2 | 0.0346 (2) | 1.06653 (19) | 0.70739 (9) | 0.0198 (3) | |
| C7 | −0.0851 (2) | 1.03223 (18) | 0.77437 (9) | 0.0204 (3) | |
| C10 | −0.3683 (2) | 0.74311 (19) | 0.98100 (9) | 0.0230 (3) | |
| C1 | 0.1122 (2) | 0.9384 (2) | 0.64203 (9) | 0.0222 (3) | |
| C17 | 0.3176 (2) | 0.5047 (2) | 0.41166 (9) | 0.0234 (3) | |
| C3 | 0.0751 (2) | 1.2292 (2) | 0.70079 (9) | 0.0235 (3) | |
| H3 | 0.1551 | 1.2522 | 0.6555 | 0.028* | |
| C8 | −0.2646 (2) | 0.83698 (19) | 0.84015 (9) | 0.0224 (3) | |
| C20 | 0.4374 (2) | 0.7914 (2) | 0.39738 (9) | 0.0260 (3) | |
| H20 | 0.4807 | 0.8913 | 0.3924 | 0.031* | |
| C5 | −0.1173 (2) | 1.3237 (2) | 0.82320 (9) | 0.0264 (3) | |
| H5 | −0.1706 | 1.4117 | 0.8626 | 0.032* | |
| C6 | −0.1599 (2) | 1.1646 (2) | 0.83154 (9) | 0.0238 (3) | |
| H6 | −0.2412 | 1.1446 | 0.8768 | 0.029* | |
| C16 | 0.2637 (2) | 0.6305 (2) | 0.47012 (10) | 0.0245 (3) | |
| H16 | 0.1817 | 0.6163 | 0.5163 | 0.029* | |
| C13 | −0.4278 (2) | 0.8397 (2) | 0.80699 (10) | 0.0260 (3) | |
| H13 | −0.4488 | 0.8736 | 0.7479 | 0.031* | |
| C18 | 0.4339 (2) | 0.5242 (2) | 0.34379 (10) | 0.0267 (3) | |
| H18 | 0.4720 | 0.4388 | 0.3032 | 0.032* | |
| C4 | 0.0017 (2) | 1.3567 (2) | 0.75833 (10) | 0.0270 (3) | |
| H4 | 0.0324 | 1.4650 | 0.7535 | 0.032* | |
| C12 | −0.5592 (2) | 0.7929 (2) | 0.85997 (10) | 0.0288 (3) | |
| H12 | −0.6697 | 0.7936 | 0.8373 | 0.035* | |
| C19 | 0.4939 (2) | 0.6725 (2) | 0.33637 (10) | 0.0274 (3) | |
| H19 | 0.5729 | 0.6918 | 0.2899 | 0.033* | |
| C9 | −0.2324 (2) | 0.78832 (18) | 0.92743 (9) | 0.0218 (3) | |
| C11 | −0.5291 (2) | 0.7452 (2) | 0.94626 (10) | 0.0268 (3) | |
| H11 | −0.6199 | 0.7134 | 0.9824 | 0.032* | |
| C14 | −0.0550 (2) | 0.7795 (2) | 0.96435 (10) | 0.0284 (3) | |
| H14A | 0.0325 | 0.7964 | 0.9185 | 0.043* | |
| H14C | 0.0084 | 0.6546 | 0.9994 | 0.043* | |
| H14B | −0.0903 | 0.8817 | 0.9991 | 0.043* | |
| C15 | −0.3403 (2) | 0.6938 (2) | 1.07514 (10) | 0.0302 (3) | |
| H15B | −0.3627 | 0.8101 | 1.0987 | 0.045* | |
| H15A | −0.2090 | 0.5993 | 1.0869 | 0.045* | |
| H15C | −0.4310 | 0.6400 | 1.1012 | 0.045* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cl1 | 0.0409 (3) | 0.0307 (2) | 0.0437 (3) | −0.02279 (18) | 0.00057 (19) | −0.00538 (17) |
| O1 | 0.0440 (7) | 0.0346 (6) | 0.0371 (7) | −0.0272 (5) | 0.0196 (5) | −0.0192 (5) |
| N1 | 0.0338 (7) | 0.0235 (6) | 0.0264 (7) | −0.0169 (5) | 0.0132 (6) | −0.0100 (5) |
| O2 | 0.0448 (7) | 0.0364 (6) | 0.0312 (6) | −0.0277 (6) | 0.0195 (5) | −0.0156 (5) |
| N2 | 0.0252 (7) | 0.0254 (6) | 0.0237 (6) | −0.0111 (5) | 0.0012 (5) | −0.0042 (5) |
| C2 | 0.0196 (7) | 0.0190 (6) | 0.0208 (7) | −0.0075 (5) | −0.0005 (5) | −0.0035 (5) |
| C7 | 0.0220 (7) | 0.0183 (6) | 0.0208 (7) | −0.0081 (5) | −0.0007 (6) | −0.0024 (5) |
| C10 | 0.0232 (7) | 0.0181 (6) | 0.0255 (7) | −0.0070 (5) | 0.0043 (6) | −0.0044 (5) |
| C1 | 0.0224 (7) | 0.0235 (6) | 0.0218 (7) | −0.0109 (6) | 0.0029 (6) | −0.0045 (5) |
| C17 | 0.0227 (7) | 0.0226 (6) | 0.0264 (8) | −0.0110 (6) | −0.0045 (6) | −0.0007 (5) |
| C3 | 0.0246 (7) | 0.0227 (6) | 0.0257 (8) | −0.0124 (6) | −0.0011 (6) | −0.0027 (6) |
| C8 | 0.0245 (7) | 0.0180 (6) | 0.0253 (7) | −0.0096 (5) | 0.0064 (6) | −0.0056 (5) |
| C20 | 0.0288 (8) | 0.0281 (7) | 0.0244 (8) | −0.0157 (6) | 0.0002 (6) | −0.0021 (6) |
| C5 | 0.0322 (8) | 0.0208 (7) | 0.0248 (8) | −0.0074 (6) | −0.0042 (6) | −0.0074 (6) |
| C6 | 0.0251 (8) | 0.0235 (7) | 0.0210 (7) | −0.0083 (6) | 0.0027 (6) | −0.0046 (5) |
| C16 | 0.0224 (7) | 0.0273 (7) | 0.0236 (7) | −0.0107 (6) | 0.0003 (6) | −0.0024 (6) |
| C13 | 0.0313 (8) | 0.0257 (7) | 0.0225 (7) | −0.0134 (6) | −0.0003 (6) | −0.0030 (6) |
| C18 | 0.0261 (8) | 0.0299 (7) | 0.0241 (8) | −0.0098 (6) | −0.0002 (6) | −0.0086 (6) |
| C4 | 0.0327 (8) | 0.0214 (7) | 0.0311 (8) | −0.0139 (6) | −0.0053 (7) | −0.0049 (6) |
| C12 | 0.0258 (8) | 0.0304 (7) | 0.0326 (9) | −0.0140 (6) | −0.0013 (6) | −0.0036 (6) |
| C19 | 0.0280 (8) | 0.0351 (8) | 0.0213 (7) | −0.0159 (7) | 0.0036 (6) | −0.0036 (6) |
| C9 | 0.0208 (7) | 0.0172 (6) | 0.0267 (8) | −0.0070 (5) | 0.0022 (6) | −0.0043 (5) |
| C11 | 0.0239 (8) | 0.0265 (7) | 0.0302 (8) | −0.0116 (6) | 0.0058 (6) | −0.0043 (6) |
| C14 | 0.0254 (8) | 0.0295 (7) | 0.0314 (8) | −0.0128 (6) | −0.0025 (6) | −0.0022 (6) |
| C15 | 0.0322 (9) | 0.0306 (8) | 0.0252 (8) | −0.0111 (7) | 0.0034 (7) | −0.0039 (6) |
Geometric parameters (Å, º) top
| Cl1—C17 | 1.7293 (14) | C10—C15 | 1.506 (2) |
| O1—C1 | 1.2215 (17) | C17—C18 | 1.374 (2) |
| N1—C7 | 1.3816 (17) | C17—C16 | 1.383 (2) |
| N1—C8 | 1.4212 (19) | C3—C4 | 1.381 (2) |
| O2—C1 | 1.3159 (18) | C8—C13 | 1.394 (2) |
| N2—C16 | 1.3379 (18) | C8—C9 | 1.403 (2) |
| N2—C20 | 1.339 (2) | C20—C19 | 1.378 (2) |
| C2—C3 | 1.4022 (18) | C5—C6 | 1.3828 (19) |
| C2—C7 | 1.413 (2) | C5—C4 | 1.384 (2) |
| C2—C1 | 1.483 (2) | C13—C12 | 1.384 (2) |
| C7—C6 | 1.402 (2) | C18—C19 | 1.389 (2) |
| C10—C11 | 1.391 (2) | C12—C11 | 1.385 (2) |
| C10—C9 | 1.410 (2) | C9—C14 | 1.503 (2) |
| | | |
| C7—N1—C8 | 125.61 (12) | C4—C3—C2 | 121.72 (14) |
| C16—N2—C20 | 118.50 (13) | C13—C8—C9 | 120.55 (13) |
| C3—C2—C7 | 119.31 (13) | C13—C8—N1 | 117.98 (13) |
| C3—C2—C1 | 119.23 (13) | C9—C8—N1 | 121.36 (13) |
| C7—C2—C1 | 121.41 (12) | N2—C20—C19 | 122.62 (13) |
| N1—C7—C6 | 121.24 (13) | C6—C5—C4 | 120.96 (13) |
| N1—C7—C2 | 120.68 (12) | C5—C6—C7 | 121.19 (14) |
| C6—C7—C2 | 118.08 (12) | N2—C16—C17 | 121.67 (14) |
| C11—C10—C9 | 119.50 (14) | C12—C13—C8 | 120.18 (14) |
| C11—C10—C15 | 120.20 (13) | C17—C18—C19 | 117.96 (14) |
| C9—C10—C15 | 120.30 (14) | C3—C4—C5 | 118.73 (13) |
| O1—C1—O2 | 122.29 (13) | C11—C12—C13 | 119.76 (14) |
| O1—C1—C2 | 123.64 (13) | C20—C19—C18 | 119.08 (14) |
| O2—C1—C2 | 114.06 (12) | C8—C9—C10 | 118.83 (13) |
| C18—C17—C16 | 120.16 (13) | C8—C9—C14 | 121.47 (13) |
| C18—C17—Cl1 | 120.99 (11) | C10—C9—C14 | 119.68 (13) |
| C16—C17—Cl1 | 118.86 (12) | C12—C11—C10 | 121.18 (14) |
| | | |
| C8—N1—C7—C6 | 8.2 (2) | C9—C8—C13—C12 | −0.4 (2) |
| C8—N1—C7—C2 | −171.39 (14) | N1—C8—C13—C12 | 175.79 (13) |
| C3—C2—C7—N1 | 179.98 (13) | C16—C17—C18—C19 | 0.0 (2) |
| C1—C2—C7—N1 | 2.4 (2) | Cl1—C17—C18—C19 | −179.57 (11) |
| C3—C2—C7—C6 | 0.4 (2) | C2—C3—C4—C5 | −1.1 (2) |
| C1—C2—C7—C6 | −177.16 (13) | C6—C5—C4—C3 | 1.1 (2) |
| C3—C2—C1—O1 | −174.28 (14) | C8—C13—C12—C11 | 0.6 (2) |
| C7—C2—C1—O1 | 3.3 (2) | N2—C20—C19—C18 | −1.0 (2) |
| C3—C2—C1—O2 | 5.8 (2) | C17—C18—C19—C20 | 0.9 (2) |
| C7—C2—C1—O2 | −176.57 (13) | C13—C8—C9—C10 | −0.3 (2) |
| C7—C2—C3—C4 | 0.3 (2) | N1—C8—C9—C10 | −176.40 (12) |
| C1—C2—C3—C4 | 177.94 (13) | C13—C8—C9—C14 | 178.38 (13) |
| C7—N1—C8—C13 | 112.88 (17) | N1—C8—C9—C14 | 2.3 (2) |
| C7—N1—C8—C9 | −70.9 (2) | C11—C10—C9—C8 | 0.8 (2) |
| C16—N2—C20—C19 | 0.2 (2) | C15—C10—C9—C8 | −178.86 (12) |
| C4—C5—C6—C7 | −0.4 (2) | C11—C10—C9—C14 | −177.87 (12) |
| N1—C7—C6—C5 | −179.91 (13) | C15—C10—C9—C14 | 2.4 (2) |
| C2—C7—C6—C5 | −0.4 (2) | C13—C12—C11—C10 | −0.1 (2) |
| C20—N2—C16—C17 | 0.8 (2) | C9—C10—C11—C12 | −0.7 (2) |
| C18—C17—C16—N2 | −0.9 (2) | C15—C10—C11—C12 | 179.04 (14) |
| Cl1—C17—C16—N2 | 178.71 (11) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C16—H16···O1 | 0.95 | 2.52 | 3.194 (2) | 128 |
| O2—H2···N2 | 0.84 (3) | 1.81 (3) | 2.6487 (19) | 175 (3) |
| N1—H7···O1 | 0.89 (2) | 1.93 (2) | 2.6586 (19) | 137.6 (17) |
(ma4pic) 2-(2,3-dimethylphenyl)aminobenzoic acid 4-methylpyridine solvate
top
Crystal data top
| C21H22N2O2 | Z = 2 |
| Mr = 334.41 | F(000) = 356 |
| Triclinic, P1 | Dx = 1.271 Mg m−3 |
| a = 7.5271 (15) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 7.5702 (15) Å | Cell parameters from 8595 reflections |
| c = 15.664 (3) Å | θ = 1.3–27.9° |
| α = 88.11 (3)° | µ = 0.08 mm−1 |
| β = 78.41 (3)° | T = 173 K |
| γ = 89.20 (3)° | Block, colorless |
| V = 873.9 (3) Å3 | 0.57 × 0.46 × 0.08 mm |
Data collection top
Bruker Kappa Duo Apex II
diffractometer | 4147 independent reflections |
| Radiation source: fine-focus sealed tube | 3128 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.020 |
| 1.2° φ scans and ω scans | θmax = 27.9°, θmin = 1.3° |
Absorption correction: multi-scan
SADABS | h = −9→9 |
| Tmin = 0.922, Tmax = 0.993 | k = −9→9 |
| 8595 measured reflections | l = −20→19 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.046 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.135 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.070P)2 + 0.1127P]
where P = (Fo2 + 2Fc2)/3 |
| 4147 reflections | (Δ/σ)max = 0.006 |
| 237 parameters | Δρmax = 0.27 e Å−3 |
| 0 restraints | Δρmin = −0.26 e Å−3 |
Special details top
Experimental. 'SADABS (Sheldrick, 1996)' |
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and
goodness of fit S are based on F2, conventional R-factors R are based
on F, with F set to zero for negative F2. The threshold expression of
F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is
not relevant to the choice of reflections for refinement. R-factors based
on F2 are statistically about twice as large as those based on F, and R-
factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O2 | 0.45990 (15) | 0.67963 (14) | 0.40671 (7) | 0.0387 (3) | |
| O1 | 0.66682 (14) | 0.47895 (15) | 0.35169 (7) | 0.0387 (3) | |
| C2 | 0.39780 (17) | 0.50945 (17) | 0.29351 (8) | 0.0241 (3) | |
| N1 | 0.61076 (17) | 0.29489 (16) | 0.21643 (8) | 0.0315 (3) | |
| H7 | 0.674 (3) | 0.308 (2) | 0.2562 (12) | 0.046 (5)* | |
| H2 | 0.552 (3) | 0.707 (3) | 0.4441 (14) | 0.066 (6)* | |
| C1 | 0.52078 (18) | 0.55275 (18) | 0.35244 (9) | 0.0268 (3) | |
| C7 | 0.44662 (18) | 0.38226 (17) | 0.22829 (8) | 0.0258 (3) | |
| C9 | 0.71015 (18) | 0.18245 (18) | 0.06917 (9) | 0.0280 (3) | |
| C10 | 0.75189 (18) | 0.03796 (18) | 0.01443 (9) | 0.0280 (3) | |
| C3 | 0.22802 (19) | 0.59221 (18) | 0.30341 (9) | 0.0289 (3) | |
| H3 | 0.1958 | 0.6776 | 0.3471 | 0.035* | |
| C8 | 0.65690 (18) | 0.15059 (18) | 0.15901 (9) | 0.0276 (3) | |
| C5 | 0.1546 (2) | 0.4295 (2) | 0.18786 (9) | 0.0336 (3) | |
| H5 | 0.0718 | 0.4013 | 0.1520 | 0.040* | |
| C6 | 0.32081 (19) | 0.34641 (19) | 0.17562 (9) | 0.0306 (3) | |
| H6 | 0.3515 | 0.2632 | 0.1308 | 0.037* | |
| C11 | 0.74201 (19) | −0.13180 (19) | 0.05060 (9) | 0.0317 (3) | |
| H11 | 0.7700 | −0.2294 | 0.0135 | 0.038* | |
| C13 | 0.64906 (19) | −0.02063 (19) | 0.19399 (9) | 0.0302 (3) | |
| H13 | 0.6140 | −0.0406 | 0.2552 | 0.036* | |
| C12 | 0.6921 (2) | −0.1622 (2) | 0.13999 (10) | 0.0338 (3) | |
| H12 | 0.6875 | −0.2794 | 0.1639 | 0.041* | |
| C4 | 0.10550 (19) | 0.5539 (2) | 0.25182 (9) | 0.0336 (3) | |
| H4 | −0.0095 | 0.6113 | 0.2599 | 0.040* | |
| C15 | 0.8059 (2) | 0.0672 (2) | −0.08305 (9) | 0.0361 (4) | |
| H15B | 0.8607 | −0.0413 | −0.1095 | 0.054* | |
| H15A | 0.8938 | 0.1635 | −0.0962 | 0.054* | |
| H15C | 0.6983 | 0.0984 | −0.1068 | 0.054* | |
| C14 | 0.7237 (2) | 0.3677 (2) | 0.03110 (11) | 0.0419 (4) | |
| H14A | 0.6908 | 0.4518 | 0.0780 | 0.063* | |
| H14C | 0.6410 | 0.3825 | −0.0096 | 0.063* | |
| H14B | 0.8484 | 0.3898 | 0.0000 | 0.063* | |
| N2 | 0.68427 (16) | 0.76449 (15) | 0.50589 (7) | 0.0299 (3) | |
| C18 | 0.93135 (19) | 0.86481 (18) | 0.60673 (9) | 0.0286 (3) | |
| C19 | 0.75445 (19) | 0.92713 (18) | 0.62262 (9) | 0.0289 (3) | |
| H19 | 0.7142 | 1.0053 | 0.6689 | 0.035* | |
| C20 | 0.63648 (19) | 0.87580 (19) | 0.57129 (9) | 0.0293 (3) | |
| H20 | 0.5161 | 0.9215 | 0.5829 | 0.035* | |
| C17 | 0.9810 (2) | 0.75022 (19) | 0.53821 (10) | 0.0335 (3) | |
| H17 | 1.1012 | 0.7045 | 0.5244 | 0.040* | |
| C16 | 0.8550 (2) | 0.7033 (2) | 0.49045 (10) | 0.0342 (3) | |
| H16 | 0.8910 | 0.6238 | 0.4444 | 0.041* | |
| C21 | 1.0629 (2) | 0.9182 (2) | 0.66117 (11) | 0.0414 (4) | |
| H21A | 1.0249 | 0.8685 | 0.7205 | 0.062* | |
| H21C | 1.0656 | 1.0474 | 0.6629 | 0.062* | |
| H21B | 1.1841 | 0.8735 | 0.6357 | 0.062* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O2 | 0.0428 (6) | 0.0389 (6) | 0.0382 (6) | 0.0116 (5) | −0.0154 (5) | −0.0192 (5) |
| O1 | 0.0301 (5) | 0.0477 (7) | 0.0415 (6) | 0.0073 (5) | −0.0118 (4) | −0.0203 (5) |
| C2 | 0.0273 (7) | 0.0224 (6) | 0.0215 (6) | −0.0013 (5) | −0.0023 (5) | 0.0000 (5) |
| N1 | 0.0312 (6) | 0.0354 (7) | 0.0306 (6) | 0.0076 (5) | −0.0110 (5) | −0.0142 (5) |
| C1 | 0.0288 (7) | 0.0258 (7) | 0.0246 (6) | −0.0003 (5) | −0.0019 (5) | −0.0045 (5) |
| C7 | 0.0277 (7) | 0.0249 (7) | 0.0242 (6) | 0.0003 (5) | −0.0042 (5) | −0.0005 (5) |
| C9 | 0.0277 (7) | 0.0267 (7) | 0.0304 (7) | 0.0009 (5) | −0.0073 (5) | −0.0039 (5) |
| C10 | 0.0253 (7) | 0.0316 (7) | 0.0275 (7) | −0.0007 (5) | −0.0055 (5) | −0.0064 (5) |
| C3 | 0.0302 (7) | 0.0281 (7) | 0.0262 (7) | 0.0032 (5) | −0.0003 (5) | −0.0019 (5) |
| C8 | 0.0256 (7) | 0.0291 (7) | 0.0290 (7) | 0.0031 (5) | −0.0068 (5) | −0.0086 (5) |
| C5 | 0.0318 (7) | 0.0414 (8) | 0.0294 (7) | −0.0019 (6) | −0.0106 (6) | 0.0019 (6) |
| C6 | 0.0354 (8) | 0.0326 (8) | 0.0251 (7) | −0.0001 (6) | −0.0083 (6) | −0.0052 (6) |
| C11 | 0.0332 (7) | 0.0291 (7) | 0.0324 (7) | 0.0004 (6) | −0.0043 (6) | −0.0095 (6) |
| C13 | 0.0299 (7) | 0.0351 (8) | 0.0251 (7) | 0.0012 (6) | −0.0042 (5) | −0.0020 (6) |
| C12 | 0.0382 (8) | 0.0266 (7) | 0.0355 (8) | −0.0013 (6) | −0.0051 (6) | 0.0005 (6) |
| C4 | 0.0280 (7) | 0.0383 (8) | 0.0337 (8) | 0.0044 (6) | −0.0051 (6) | 0.0024 (6) |
| C15 | 0.0389 (8) | 0.0415 (9) | 0.0275 (7) | −0.0028 (6) | −0.0046 (6) | −0.0059 (6) |
| C14 | 0.0551 (10) | 0.0298 (8) | 0.0386 (9) | −0.0002 (7) | −0.0043 (7) | −0.0005 (6) |
| N2 | 0.0330 (6) | 0.0299 (6) | 0.0263 (6) | 0.0014 (5) | −0.0052 (5) | −0.0025 (5) |
| C18 | 0.0305 (7) | 0.0262 (7) | 0.0289 (7) | −0.0029 (5) | −0.0067 (5) | 0.0074 (5) |
| C19 | 0.0318 (7) | 0.0278 (7) | 0.0264 (7) | 0.0006 (5) | −0.0035 (5) | −0.0045 (5) |
| C20 | 0.0275 (7) | 0.0303 (7) | 0.0291 (7) | 0.0036 (5) | −0.0027 (5) | −0.0033 (6) |
| C17 | 0.0289 (7) | 0.0318 (8) | 0.0373 (8) | 0.0053 (6) | −0.0013 (6) | 0.0023 (6) |
| C16 | 0.0390 (8) | 0.0311 (8) | 0.0301 (7) | 0.0056 (6) | −0.0007 (6) | −0.0060 (6) |
| C21 | 0.0371 (9) | 0.0432 (9) | 0.0467 (9) | −0.0054 (7) | −0.0160 (7) | 0.0052 (7) |
Geometric parameters (Å, º) top
| O2—C1 | 1.3192 (16) | C3—C4 | 1.382 (2) |
| O1—C1 | 1.2243 (17) | C8—C13 | 1.388 (2) |
| C2—C3 | 1.3979 (19) | C5—C6 | 1.373 (2) |
| C2—C7 | 1.4177 (18) | C5—C4 | 1.390 (2) |
| C2—C1 | 1.4793 (19) | C11—C12 | 1.387 (2) |
| N1—C7 | 1.3754 (18) | C13—C12 | 1.382 (2) |
| N1—C8 | 1.4322 (17) | N2—C20 | 1.3376 (18) |
| C7—C6 | 1.4095 (19) | N2—C16 | 1.3382 (19) |
| C9—C8 | 1.3975 (19) | C18—C19 | 1.384 (2) |
| C9—C10 | 1.4051 (19) | C18—C17 | 1.391 (2) |
| C9—C14 | 1.503 (2) | C18—C21 | 1.498 (2) |
| C10—C11 | 1.384 (2) | C19—C20 | 1.379 (2) |
| C10—C15 | 1.5089 (19) | C17—C16 | 1.377 (2) |
| | | |
| C3—C2—C7 | 119.08 (12) | C13—C8—C9 | 120.67 (12) |
| C3—C2—C1 | 119.54 (12) | C13—C8—N1 | 119.04 (12) |
| C7—C2—C1 | 121.36 (12) | C9—C8—N1 | 120.29 (13) |
| C7—N1—C8 | 123.49 (12) | C6—C5—C4 | 121.18 (14) |
| O1—C1—O2 | 122.15 (13) | C5—C6—C7 | 121.24 (13) |
| O1—C1—C2 | 123.74 (12) | C10—C11—C12 | 121.26 (13) |
| O2—C1—C2 | 114.11 (12) | C12—C13—C8 | 120.18 (13) |
| N1—C7—C6 | 120.46 (12) | C13—C12—C11 | 119.48 (14) |
| N1—C7—C2 | 121.60 (12) | C3—C4—C5 | 118.36 (13) |
| C6—C7—C2 | 117.94 (12) | C20—N2—C16 | 117.38 (13) |
| C8—C9—C10 | 118.93 (13) | C19—C18—C17 | 116.99 (13) |
| C8—C9—C14 | 120.95 (13) | C19—C18—C21 | 121.35 (13) |
| C10—C9—C14 | 120.12 (13) | C17—C18—C21 | 121.66 (14) |
| C11—C10—C9 | 119.46 (13) | C20—C19—C18 | 120.03 (13) |
| C11—C10—C15 | 120.17 (13) | N2—C20—C19 | 122.82 (13) |
| C9—C10—C15 | 120.37 (13) | C16—C17—C18 | 119.66 (14) |
| C4—C3—C2 | 122.20 (13) | N2—C16—C17 | 123.10 (13) |
| | | |
| C3—C2—C1—O1 | 175.47 (13) | C7—N1—C8—C9 | 78.66 (18) |
| C7—C2—C1—O1 | −3.0 (2) | C4—C5—C6—C7 | 1.0 (2) |
| C3—C2—C1—O2 | −4.67 (18) | N1—C7—C6—C5 | 178.50 (13) |
| C7—C2—C1—O2 | 176.87 (12) | C2—C7—C6—C5 | −1.1 (2) |
| C8—N1—C7—C6 | −7.6 (2) | C9—C10—C11—C12 | −0.2 (2) |
| C8—N1—C7—C2 | 172.05 (13) | C15—C10—C11—C12 | −179.78 (13) |
| C3—C2—C7—N1 | −179.07 (12) | C9—C8—C13—C12 | −0.7 (2) |
| C1—C2—C7—N1 | −0.61 (19) | N1—C8—C13—C12 | 179.71 (13) |
| C3—C2—C7—C6 | 0.56 (19) | C8—C13—C12—C11 | −0.4 (2) |
| C1—C2—C7—C6 | 179.03 (12) | C10—C11—C12—C13 | 0.9 (2) |
| C8—C9—C10—C11 | −0.9 (2) | C2—C3—C4—C5 | −0.4 (2) |
| C14—C9—C10—C11 | 178.80 (13) | C6—C5—C4—C3 | −0.2 (2) |
| C8—C9—C10—C15 | 178.66 (13) | C17—C18—C19—C20 | −0.27 (19) |
| C14—C9—C10—C15 | −1.6 (2) | C21—C18—C19—C20 | 179.92 (13) |
| C7—C2—C3—C4 | 0.2 (2) | C16—N2—C20—C19 | −0.6 (2) |
| C1—C2—C3—C4 | −178.33 (12) | C18—C19—C20—N2 | 0.8 (2) |
| C10—C9—C8—C13 | 1.4 (2) | C19—C18—C17—C16 | −0.5 (2) |
| C14—C9—C8—C13 | −178.35 (13) | C21—C18—C17—C16 | 179.33 (13) |
| C10—C9—C8—N1 | −179.04 (12) | C20—N2—C16—C17 | −0.2 (2) |
| C14—C9—C8—N1 | 1.2 (2) | C18—C17—C16—N2 | 0.8 (2) |
| C7—N1—C8—C13 | −101.76 (17) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H7···O1 | 0.868 (19) | 2.000 (19) | 2.6793 (17) | 134.4 (16) |
| O2—H2···N2 | 1.02 (2) | 1.60 (2) | 2.6132 (18) | 175.0 (19) |
(tfa2pic) 2-[(3-chloro-2-methylphenyl)amino]benzoic acid) 2-methylpyridine solvate
top
Crystal data top
| C20H19ClN2O2 | F(000) = 744 |
| Mr = 354.82 | Dx = 1.353 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.7585 (16) Å | Cell parameters from 24570 reflections |
| b = 8.0699 (16) Å | θ = 2.6–28.4° |
| c = 28.056 (6) Å | µ = 0.24 mm−1 |
| β = 97.29 (3)° | T = 173 K |
| V = 1742.4 (6) Å3 | Block, colorless |
| Z = 4 | 0.24 × 0.23 × 0.22 mm |
Data collection top
Bruker Kappa Duo Apex II
diffractometer | 4345 independent reflections |
| Radiation source: fine-focus sealed tube | 3478 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.038 |
| 1.2° φ scans and ω scans | θmax = 28.4°, θmin = 2.6° |
Absorption correction: multi-scan
SADABS | h = −10→10 |
| Tmin = 0.897, Tmax = 0.950 | k = −10→10 |
| 24570 measured reflections | l = −37→37 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.105 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0509P)2 + 0.5161P]
where P = (Fo2 + 2Fc2)/3 |
| 4345 reflections | (Δ/σ)max = 0.002 |
| 236 parameters | Δρmax = 0.30 e Å−3 |
| 0 restraints | Δρmin = −0.28 e Å−3 |
Special details top
Experimental. 'SADABS (Sheldrick, 1996)' |
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and
goodness of fit S are based on F2, conventional R-factors R are based
on F, with F set to zero for negative F2. The threshold expression of
F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is
not relevant to the choice of reflections for refinement. R-factors based
on F2 are statistically about twice as large as those based on F, and R-
factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Cl1 | 0.66000 (5) | 0.93524 (5) | 0.205423 (13) | 0.03861 (12) | |
| O1 | 0.49311 (12) | 0.84441 (14) | 0.43656 (4) | 0.0345 (2) | |
| O2 | 0.32335 (14) | 0.80501 (15) | 0.49381 (4) | 0.0359 (3) | |
| C2 | 0.19649 (17) | 0.93174 (16) | 0.42215 (5) | 0.0247 (3) | |
| N2 | 0.59942 (15) | 0.66601 (15) | 0.53974 (4) | 0.0301 (3) | |
| H7 | 0.442 (2) | 0.946 (2) | 0.3738 (6) | 0.031 (4)* | |
| H2 | 0.430 (3) | 0.756 (3) | 0.5107 (9) | 0.101 (9)* | |
| C10 | 0.53807 (16) | 1.04602 (18) | 0.24302 (5) | 0.0256 (3) | |
| C6 | 0.05306 (18) | 1.06539 (18) | 0.35047 (5) | 0.0285 (3) | |
| H6 | 0.0564 | 1.1116 | 0.3195 | 0.034* | |
| N1 | 0.36271 (16) | 1.00046 (19) | 0.35755 (4) | 0.0354 (3) | |
| C1 | 0.35091 (17) | 0.85760 (17) | 0.45119 (5) | 0.0269 (3) | |
| C9 | 0.49966 (16) | 0.97326 (17) | 0.28558 (5) | 0.0241 (3) | |
| C7 | 0.20633 (17) | 1.00183 (17) | 0.37628 (5) | 0.0253 (3) | |
| C15 | 0.59130 (19) | 0.57089 (18) | 0.57860 (5) | 0.0293 (3) | |
| C3 | 0.03625 (18) | 0.92933 (18) | 0.44017 (5) | 0.0304 (3) | |
| H3 | 0.0297 | 0.8817 | 0.4709 | 0.036* | |
| C8 | 0.40072 (17) | 1.06929 (18) | 0.31414 (5) | 0.0260 (3) | |
| C5 | −0.10205 (18) | 1.06137 (18) | 0.36973 (5) | 0.0319 (3) | |
| H5 | −0.2037 | 1.1057 | 0.3518 | 0.038* | |
| C13 | 0.34757 (17) | 1.22897 (19) | 0.30006 (5) | 0.0316 (3) | |
| H13 | 0.2828 | 1.2932 | 0.3199 | 0.038* | |
| C12 | 0.38844 (18) | 1.29440 (19) | 0.25739 (6) | 0.0340 (3) | |
| H12 | 0.3510 | 1.4030 | 0.2480 | 0.041* | |
| C20 | 0.4158 (2) | 0.5506 (2) | 0.59506 (6) | 0.0396 (4) | |
| H20A | 0.3574 | 0.4539 | 0.5793 | 0.059* | |
| H20C | 0.3459 | 0.6500 | 0.5867 | 0.059* | |
| H20B | 0.4296 | 0.5348 | 0.6300 | 0.059* | |
| C14 | 0.55494 (19) | 0.80070 (18) | 0.30056 (5) | 0.0324 (3) | |
| H14C | 0.6016 | 0.7448 | 0.2739 | 0.049* | |
| H14B | 0.4546 | 0.7388 | 0.3091 | 0.049* | |
| H14A | 0.6448 | 0.8058 | 0.3284 | 0.049* | |
| C16 | 0.7395 (2) | 0.49485 (19) | 0.60177 (5) | 0.0339 (3) | |
| H16 | 0.7324 | 0.4270 | 0.6291 | 0.041* | |
| C4 | −0.11259 (18) | 0.9940 (2) | 0.41473 (6) | 0.0349 (3) | |
| H4 | −0.2198 | 0.9925 | 0.4278 | 0.042* | |
| C17 | 0.8974 (2) | 0.5184 (2) | 0.58481 (5) | 0.0360 (3) | |
| H17 | 0.9995 | 0.4669 | 0.6004 | 0.043* | |
| C19 | 0.75305 (19) | 0.6887 (2) | 0.52354 (5) | 0.0343 (3) | |
| H19 | 0.7573 | 0.7567 | 0.4961 | 0.041* | |
| C11 | 0.48339 (17) | 1.20283 (19) | 0.22832 (5) | 0.0312 (3) | |
| H11 | 0.5106 | 1.2469 | 0.1988 | 0.037* | |
| C18 | 0.90534 (19) | 0.6176 (2) | 0.54500 (6) | 0.0355 (3) | |
| H18 | 1.0124 | 0.6364 | 0.5328 | 0.043* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cl1 | 0.0416 (2) | 0.0442 (2) | 0.03322 (19) | −0.00067 (16) | 0.01696 (15) | −0.00518 (15) |
| O1 | 0.0244 (5) | 0.0467 (6) | 0.0331 (5) | 0.0033 (4) | 0.0064 (4) | 0.0103 (5) |
| O2 | 0.0314 (5) | 0.0503 (7) | 0.0271 (5) | 0.0048 (5) | 0.0077 (4) | 0.0090 (5) |
| C2 | 0.0242 (6) | 0.0236 (6) | 0.0268 (6) | −0.0008 (5) | 0.0053 (5) | −0.0030 (5) |
| N2 | 0.0299 (6) | 0.0321 (7) | 0.0279 (6) | −0.0014 (5) | 0.0025 (5) | 0.0000 (5) |
| C10 | 0.0199 (6) | 0.0330 (7) | 0.0242 (6) | −0.0029 (5) | 0.0042 (5) | −0.0018 (5) |
| C6 | 0.0277 (7) | 0.0303 (7) | 0.0273 (6) | 0.0024 (6) | 0.0029 (5) | −0.0017 (5) |
| N1 | 0.0262 (6) | 0.0539 (8) | 0.0274 (6) | 0.0142 (6) | 0.0086 (5) | 0.0115 (6) |
| C1 | 0.0280 (7) | 0.0276 (7) | 0.0256 (6) | −0.0018 (5) | 0.0053 (5) | 0.0002 (5) |
| C9 | 0.0193 (6) | 0.0274 (7) | 0.0252 (6) | −0.0005 (5) | 0.0017 (5) | 0.0009 (5) |
| C7 | 0.0239 (6) | 0.0258 (7) | 0.0267 (6) | 0.0014 (5) | 0.0055 (5) | −0.0035 (5) |
| C15 | 0.0323 (7) | 0.0286 (7) | 0.0269 (6) | −0.0016 (6) | 0.0038 (5) | −0.0037 (5) |
| C3 | 0.0276 (7) | 0.0307 (7) | 0.0345 (7) | −0.0026 (6) | 0.0105 (6) | 0.0013 (6) |
| C8 | 0.0209 (6) | 0.0333 (7) | 0.0237 (6) | 0.0013 (5) | 0.0024 (5) | 0.0007 (5) |
| C5 | 0.0230 (7) | 0.0306 (7) | 0.0411 (8) | 0.0012 (5) | 0.0009 (6) | −0.0017 (6) |
| C13 | 0.0249 (7) | 0.0335 (8) | 0.0365 (7) | 0.0046 (6) | 0.0047 (5) | −0.0040 (6) |
| C12 | 0.0252 (7) | 0.0301 (8) | 0.0457 (8) | 0.0020 (6) | 0.0011 (6) | 0.0091 (6) |
| C20 | 0.0353 (8) | 0.0457 (9) | 0.0395 (8) | 0.0018 (7) | 0.0111 (6) | 0.0070 (7) |
| C14 | 0.0302 (7) | 0.0306 (8) | 0.0376 (8) | 0.0044 (6) | 0.0090 (6) | 0.0055 (6) |
| C16 | 0.0403 (8) | 0.0309 (8) | 0.0299 (7) | 0.0024 (6) | 0.0024 (6) | 0.0006 (6) |
| C4 | 0.0239 (7) | 0.0365 (8) | 0.0462 (9) | −0.0023 (6) | 0.0116 (6) | 0.0006 (7) |
| C17 | 0.0330 (8) | 0.0362 (8) | 0.0369 (8) | 0.0054 (6) | −0.0024 (6) | −0.0062 (6) |
| C19 | 0.0339 (8) | 0.0372 (8) | 0.0322 (7) | −0.0044 (6) | 0.0058 (6) | 0.0005 (6) |
| C11 | 0.0256 (7) | 0.0373 (8) | 0.0305 (7) | −0.0023 (6) | 0.0033 (5) | 0.0088 (6) |
| C18 | 0.0290 (7) | 0.0399 (9) | 0.0380 (8) | −0.0041 (6) | 0.0053 (6) | −0.0069 (7) |
Geometric parameters (Å, º) top
| Cl1—C10 | 1.7494 (14) | N1—C8 | 1.4036 (18) |
| O1—C1 | 1.2296 (17) | C9—C8 | 1.4098 (19) |
| O2—C1 | 1.3116 (16) | C9—C14 | 1.5014 (19) |
| C2—C3 | 1.4004 (19) | C15—C16 | 1.390 (2) |
| C2—C7 | 1.4166 (19) | C15—C20 | 1.501 (2) |
| C2—C1 | 1.4865 (19) | C3—C4 | 1.381 (2) |
| N2—C19 | 1.3411 (19) | C8—C13 | 1.395 (2) |
| N2—C15 | 1.3415 (18) | C5—C4 | 1.386 (2) |
| C10—C11 | 1.381 (2) | C13—C12 | 1.382 (2) |
| C10—C9 | 1.3963 (18) | C12—C11 | 1.381 (2) |
| C6—C5 | 1.381 (2) | C16—C17 | 1.382 (2) |
| C6—C7 | 1.4083 (19) | C17—C18 | 1.382 (2) |
| N1—C7 | 1.3820 (17) | C19—C18 | 1.381 (2) |
| | | |
| C3—C2—C7 | 119.09 (13) | C6—C7—C2 | 118.25 (12) |
| C3—C2—C1 | 119.15 (12) | N2—C15—C16 | 120.75 (13) |
| C7—C2—C1 | 121.74 (12) | N2—C15—C20 | 116.77 (13) |
| C19—N2—C15 | 119.28 (13) | C16—C15—C20 | 122.48 (13) |
| C11—C10—C9 | 123.36 (13) | C4—C3—C2 | 122.04 (13) |
| C11—C10—Cl1 | 117.21 (10) | C13—C8—N1 | 121.62 (13) |
| C9—C10—Cl1 | 119.42 (11) | C13—C8—C9 | 120.63 (12) |
| C5—C6—C7 | 120.75 (13) | N1—C8—C9 | 117.72 (12) |
| C7—N1—C8 | 128.17 (12) | C6—C5—C4 | 121.37 (13) |
| O1—C1—O2 | 122.23 (13) | C12—C13—C8 | 120.43 (13) |
| O1—C1—C2 | 123.14 (12) | C11—C12—C13 | 120.37 (14) |
| O2—C1—C2 | 114.62 (12) | C17—C16—C15 | 119.68 (14) |
| C10—C9—C8 | 116.46 (12) | C3—C4—C5 | 118.49 (13) |
| C10—C9—C14 | 123.02 (12) | C18—C17—C16 | 119.34 (14) |
| C8—C9—C14 | 120.50 (12) | N2—C19—C18 | 122.92 (14) |
| N1—C7—C6 | 122.15 (12) | C12—C11—C10 | 118.73 (13) |
| N1—C7—C2 | 119.56 (12) | C19—C18—C17 | 118.03 (14) |
| | | |
| C3—C2—C1—O1 | 174.80 (14) | C7—N1—C8—C9 | −135.99 (15) |
| C7—C2—C1—O1 | −3.9 (2) | C10—C9—C8—C13 | −0.76 (19) |
| C3—C2—C1—O2 | −4.43 (19) | C14—C9—C8—C13 | −179.30 (12) |
| C7—C2—C1—O2 | 176.86 (12) | C10—C9—C8—N1 | −178.91 (12) |
| C11—C10—C9—C8 | −0.44 (19) | C14—C9—C8—N1 | 2.56 (19) |
| Cl1—C10—C9—C8 | −179.90 (9) | C7—C6—C5—C4 | −0.4 (2) |
| C11—C10—C9—C14 | 178.05 (13) | N1—C8—C13—C12 | 179.26 (13) |
| Cl1—C10—C9—C14 | −1.41 (18) | C9—C8—C13—C12 | 1.2 (2) |
| C8—N1—C7—C6 | 5.5 (2) | C8—C13—C12—C11 | −0.4 (2) |
| C8—N1—C7—C2 | −176.98 (14) | N2—C15—C16—C17 | 0.5 (2) |
| C5—C6—C7—N1 | 178.44 (14) | C20—C15—C16—C17 | 179.98 (14) |
| C5—C6—C7—C2 | 0.9 (2) | C2—C3—C4—C5 | 0.8 (2) |
| C3—C2—C7—N1 | −178.11 (13) | C6—C5—C4—C3 | −0.4 (2) |
| C1—C2—C7—N1 | 0.6 (2) | C15—C16—C17—C18 | 0.0 (2) |
| C3—C2—C7—C6 | −0.5 (2) | C15—N2—C19—C18 | 0.3 (2) |
| C1—C2—C7—C6 | 178.24 (13) | C13—C12—C11—C10 | −0.8 (2) |
| C19—N2—C15—C16 | −0.7 (2) | C9—C10—C11—C12 | 1.2 (2) |
| C19—N2—C15—C20 | 179.83 (14) | Cl1—C10—C11—C12 | −179.32 (11) |
| C7—C2—C3—C4 | −0.4 (2) | N2—C19—C18—C17 | 0.2 (2) |
| C1—C2—C3—C4 | −179.11 (14) | C16—C17—C18—C19 | −0.4 (2) |
| C7—N1—C8—C13 | 45.9 (2) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H7···O1 | 0.842 (17) | 1.938 (17) | 2.6370 (17) | 139.6 (15) |
| O2—H2···N2 | 0.98 (3) | 1.63 (3) | 2.6088 (17) | 177 (3) |
(tfa3pic) 2-[(3-chloro-2-methylphenyl)amino]benzoic acid) 3-methylpyridine solvate
top
Crystal data top
| C20H19ClN2O2 | Dx = 1.341 Mg m−3 |
| Mr = 354.82 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pbca | Cell parameters from 51960 reflections |
| a = 7.8069 (16) Å | θ = 1.3–28.4° |
| b = 13.987 (3) Å | µ = 0.23 mm−1 |
| c = 32.185 (6) Å | T = 173 K |
| V = 3514.5 (12) Å3 | Platelet, colourless |
| Z = 8 | 0.24 × 0.12 × 0.08 mm |
| F(000) = 1488 | |
Data collection top
Bruker Kappa Duo Apex II
diffractometer | 4397 independent reflections |
| Radiation source: fine-focus sealed tube | 3281 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.063 |
| 1.2° φ scans and ω scans | θmax = 28.4°, θmin = 1.3° |
Absorption correction: multi-scan
SADABS | h = −10→10 |
| Tmin = 0.898, Tmax = 0.982 | k = −18→18 |
| 51960 measured reflections | l = −42→42 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.108 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.046P)2 + 1.5661P]
where P = (Fo2 + 2Fc2)/3 |
| 4397 reflections | (Δ/σ)max = 0.001 |
| 236 parameters | Δρmax = 0.28 e Å−3 |
| 0 restraints | Δρmin = −0.30 e Å−3 |
Special details top
Experimental. 'SADABS (Sheldrick, 1996)' |
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and
goodness of fit S are based on F2, conventional R-factors R are based
on F, with F set to zero for negative F2. The threshold expression of
F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is
not relevant to the choice of reflections for refinement. R-factors based
on F2 are statistically about twice as large as those based on F, and R-
factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Cl1 | 0.01395 (6) | 0.85841 (3) | 0.449816 (12) | 0.03870 (13) | |
| O1 | −0.44507 (16) | 0.82314 (10) | 0.62910 (3) | 0.0384 (3) | |
| N2 | −0.78777 (17) | 0.74218 (9) | 0.68041 (4) | 0.0283 (3) | |
| N1 | −0.15088 (18) | 0.90005 (10) | 0.60302 (4) | 0.0289 (3) | |
| H7 | −0.247 (2) | 0.8671 (13) | 0.5987 (6) | 0.034 (5)* | |
| H2 | −0.604 (4) | 0.7922 (19) | 0.6894 (8) | 0.082 (8)* | |
| C10 | 0.0616 (2) | 0.86761 (11) | 0.50270 (5) | 0.0271 (3) | |
| C3 | −0.2470 (2) | 0.95402 (11) | 0.71286 (4) | 0.0264 (3) | |
| H3 | −0.3302 | 0.9382 | 0.7333 | 0.032* | |
| O2 | −0.50295 (16) | 0.83080 (10) | 0.69654 (4) | 0.0383 (3) | |
| C9 | −0.0688 (2) | 0.88339 (10) | 0.53137 (5) | 0.0243 (3) | |
| C2 | −0.26218 (19) | 0.91527 (11) | 0.67308 (4) | 0.0237 (3) | |
| C14 | −0.2531 (2) | 0.89472 (13) | 0.51910 (5) | 0.0338 (4) | |
| H14A | −0.2601 | 0.9100 | 0.4894 | 0.051* | |
| H14C | −0.3048 | 0.9466 | 0.5353 | 0.051* | |
| H14B | −0.3147 | 0.8350 | 0.5246 | 0.051* | |
| C13 | 0.1510 (2) | 0.87984 (11) | 0.58479 (5) | 0.0294 (3) | |
| H13 | 0.1827 | 0.8837 | 0.6132 | 0.035* | |
| C7 | −0.1378 (2) | 0.93778 (11) | 0.64259 (5) | 0.0247 (3) | |
| C8 | −0.02002 (19) | 0.88924 (11) | 0.57355 (4) | 0.0246 (3) | |
| C1 | −0.4105 (2) | 0.85241 (12) | 0.66386 (5) | 0.0269 (3) | |
| C15 | −0.8131 (2) | 0.69616 (11) | 0.64465 (5) | 0.0281 (3) | |
| H15 | −0.7203 | 0.6920 | 0.6256 | 0.034* | |
| C19 | −0.9170 (2) | 0.74782 (12) | 0.70734 (5) | 0.0306 (3) | |
| H19 | −0.9000 | 0.7808 | 0.7328 | 0.037* | |
| C17 | −1.0992 (2) | 0.66056 (12) | 0.66217 (5) | 0.0339 (4) | |
| H17 | −1.2073 | 0.6327 | 0.6561 | 0.041* | |
| C11 | 0.2319 (2) | 0.85857 (12) | 0.51363 (5) | 0.0323 (4) | |
| H11 | 0.3173 | 0.8482 | 0.4931 | 0.039* | |
| C5 | 0.0035 (2) | 1.03899 (12) | 0.69313 (5) | 0.0317 (3) | |
| H5 | 0.0937 | 1.0819 | 0.6998 | 0.038* | |
| C4 | −0.1148 (2) | 1.01470 (12) | 0.72337 (5) | 0.0302 (3) | |
| H4 | −0.1050 | 1.0393 | 0.7508 | 0.036* | |
| C12 | 0.2751 (2) | 0.86491 (12) | 0.55505 (5) | 0.0332 (4) | |
| H12 | 0.3916 | 0.8589 | 0.5632 | 0.040* | |
| C6 | −0.0072 (2) | 1.00201 (11) | 0.65347 (5) | 0.0294 (3) | |
| H6 | 0.0750 | 1.0203 | 0.6332 | 0.035* | |
| C18 | −1.0740 (2) | 0.70742 (12) | 0.69938 (5) | 0.0341 (4) | |
| H18 | −1.1639 | 0.7117 | 0.7192 | 0.041* | |
| C16 | −0.9669 (2) | 0.65417 (12) | 0.63372 (5) | 0.0306 (3) | |
| C20 | −0.9879 (3) | 0.60588 (17) | 0.59246 (7) | 0.0565 (6) | |
| H20C | −0.9046 | 0.5538 | 0.5900 | 0.085* | |
| H20B | −0.9687 | 0.6524 | 0.5702 | 0.085* | |
| H20A | −1.1042 | 0.5799 | 0.5902 | 0.085* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cl1 | 0.0478 (3) | 0.0468 (3) | 0.02147 (19) | −0.0049 (2) | 0.00098 (17) | −0.00275 (16) |
| O1 | 0.0327 (6) | 0.0588 (8) | 0.0238 (6) | −0.0145 (6) | −0.0012 (5) | −0.0074 (5) |
| N2 | 0.0252 (7) | 0.0317 (7) | 0.0280 (7) | −0.0020 (5) | −0.0025 (5) | 0.0019 (5) |
| N1 | 0.0257 (7) | 0.0389 (8) | 0.0222 (6) | −0.0053 (6) | 0.0018 (5) | −0.0032 (5) |
| C10 | 0.0349 (8) | 0.0256 (8) | 0.0207 (7) | −0.0029 (6) | 0.0011 (6) | 0.0006 (6) |
| C3 | 0.0261 (8) | 0.0302 (8) | 0.0230 (7) | 0.0006 (6) | −0.0010 (6) | 0.0011 (6) |
| O2 | 0.0337 (6) | 0.0579 (8) | 0.0234 (5) | −0.0198 (6) | 0.0008 (5) | −0.0004 (5) |
| C9 | 0.0270 (8) | 0.0231 (7) | 0.0227 (7) | −0.0009 (6) | −0.0007 (6) | 0.0008 (6) |
| C2 | 0.0222 (7) | 0.0266 (7) | 0.0224 (7) | −0.0002 (6) | −0.0017 (6) | 0.0015 (6) |
| C14 | 0.0287 (8) | 0.0420 (10) | 0.0307 (8) | 0.0011 (7) | −0.0045 (7) | −0.0044 (7) |
| C13 | 0.0316 (8) | 0.0314 (8) | 0.0251 (8) | 0.0027 (7) | −0.0026 (6) | 0.0019 (6) |
| C7 | 0.0247 (7) | 0.0266 (7) | 0.0228 (7) | 0.0009 (6) | −0.0005 (6) | 0.0014 (6) |
| C8 | 0.0277 (8) | 0.0241 (7) | 0.0218 (7) | −0.0008 (6) | 0.0016 (6) | 0.0021 (5) |
| C1 | 0.0237 (7) | 0.0331 (8) | 0.0240 (7) | −0.0004 (6) | −0.0025 (6) | 0.0013 (6) |
| C15 | 0.0269 (8) | 0.0316 (8) | 0.0257 (7) | −0.0005 (6) | 0.0022 (6) | 0.0020 (6) |
| C19 | 0.0362 (9) | 0.0305 (8) | 0.0249 (8) | 0.0009 (7) | 0.0004 (6) | −0.0005 (6) |
| C17 | 0.0258 (8) | 0.0308 (9) | 0.0451 (10) | −0.0051 (7) | −0.0021 (7) | 0.0022 (7) |
| C11 | 0.0292 (8) | 0.0362 (9) | 0.0315 (8) | 0.0001 (7) | 0.0077 (7) | −0.0017 (7) |
| C5 | 0.0293 (8) | 0.0310 (8) | 0.0348 (8) | −0.0061 (7) | −0.0037 (7) | −0.0019 (7) |
| C4 | 0.0324 (8) | 0.0337 (9) | 0.0245 (7) | −0.0007 (7) | −0.0047 (6) | −0.0037 (6) |
| C12 | 0.0255 (8) | 0.0380 (9) | 0.0360 (9) | 0.0034 (7) | −0.0006 (7) | 0.0013 (7) |
| C6 | 0.0279 (8) | 0.0307 (8) | 0.0297 (8) | −0.0038 (7) | 0.0014 (6) | 0.0023 (6) |
| C18 | 0.0313 (9) | 0.0346 (9) | 0.0365 (9) | 0.0017 (7) | 0.0097 (7) | 0.0024 (7) |
| C16 | 0.0335 (8) | 0.0292 (8) | 0.0291 (8) | −0.0012 (7) | −0.0031 (7) | −0.0002 (6) |
| C20 | 0.0583 (14) | 0.0687 (15) | 0.0425 (11) | −0.0088 (11) | −0.0065 (10) | −0.0210 (10) |
Geometric parameters (Å, º) top
| Cl1—C10 | 1.7470 (16) | C2—C7 | 1.416 (2) |
| O1—C1 | 1.2214 (18) | C2—C1 | 1.484 (2) |
| N2—C19 | 1.332 (2) | C13—C12 | 1.378 (2) |
| N2—C15 | 1.334 (2) | C13—C8 | 1.390 (2) |
| N1—C7 | 1.3822 (19) | C7—C6 | 1.403 (2) |
| N1—C8 | 1.402 (2) | C15—C16 | 1.382 (2) |
| C10—C11 | 1.381 (2) | C19—C18 | 1.374 (2) |
| C10—C9 | 1.392 (2) | C17—C18 | 1.380 (2) |
| C3—C4 | 1.379 (2) | C17—C16 | 1.383 (2) |
| C3—C2 | 1.395 (2) | C11—C12 | 1.378 (2) |
| O2—C1 | 1.3106 (19) | C5—C6 | 1.380 (2) |
| C9—C8 | 1.412 (2) | C5—C4 | 1.384 (2) |
| C9—C14 | 1.500 (2) | C16—C20 | 1.499 (2) |
| | | |
| C19—N2—C15 | 118.53 (14) | C13—C8—C9 | 120.25 (14) |
| C7—N1—C8 | 127.62 (14) | N1—C8—C9 | 117.39 (14) |
| C11—C10—C9 | 123.36 (15) | O1—C1—O2 | 122.42 (15) |
| C11—C10—Cl1 | 116.52 (12) | O1—C1—C2 | 123.65 (14) |
| C9—C10—Cl1 | 120.12 (13) | O2—C1—C2 | 113.94 (13) |
| C4—C3—C2 | 121.84 (14) | N2—C15—C16 | 123.61 (15) |
| C10—C9—C8 | 116.70 (14) | N2—C19—C18 | 122.00 (15) |
| C10—C9—C14 | 122.93 (14) | C18—C17—C16 | 119.95 (16) |
| C8—C9—C14 | 120.37 (14) | C12—C11—C10 | 118.44 (15) |
| C3—C2—C7 | 119.43 (14) | C6—C5—C4 | 121.21 (15) |
| C3—C2—C1 | 118.67 (13) | C3—C4—C5 | 118.57 (14) |
| C7—C2—C1 | 121.89 (13) | C13—C12—C11 | 120.64 (16) |
| C12—C13—C8 | 120.61 (15) | C5—C6—C7 | 120.99 (15) |
| N1—C7—C6 | 121.89 (14) | C19—C18—C17 | 118.97 (15) |
| N1—C7—C2 | 120.18 (14) | C15—C16—C17 | 116.93 (15) |
| C6—C7—C2 | 117.88 (14) | C15—C16—C20 | 120.78 (16) |
| C13—C8—N1 | 122.29 (14) | C17—C16—C20 | 122.28 (16) |
| | | |
| C11—C10—C9—C8 | −0.6 (2) | C7—C2—C1—O1 | −7.0 (2) |
| Cl1—C10—C9—C8 | 179.53 (11) | C3—C2—C1—O2 | −7.7 (2) |
| C11—C10—C9—C14 | 178.96 (16) | C7—C2—C1—O2 | 173.14 (14) |
| Cl1—C10—C9—C14 | −0.9 (2) | C19—N2—C15—C16 | 0.4 (2) |
| C4—C3—C2—C7 | 0.7 (2) | C15—N2—C19—C18 | 0.3 (2) |
| C4—C3—C2—C1 | −178.52 (14) | C9—C10—C11—C12 | 0.4 (3) |
| C8—N1—C7—C6 | 24.1 (2) | Cl1—C10—C11—C12 | −179.69 (13) |
| C8—N1—C7—C2 | −158.62 (15) | C2—C3—C4—C5 | 1.4 (2) |
| C3—C2—C7—N1 | −179.96 (14) | C6—C5—C4—C3 | −1.5 (3) |
| C1—C2—C7—N1 | −0.8 (2) | C8—C13—C12—C11 | −0.3 (3) |
| C3—C2—C7—C6 | −2.6 (2) | C10—C11—C12—C13 | 0.1 (3) |
| C1—C2—C7—C6 | 176.52 (14) | C4—C5—C6—C7 | −0.6 (3) |
| C12—C13—C8—N1 | 176.98 (15) | N1—C7—C6—C5 | 179.90 (15) |
| C12—C13—C8—C9 | 0.1 (2) | C2—C7—C6—C5 | 2.6 (2) |
| C7—N1—C8—C13 | 26.2 (2) | N2—C19—C18—C17 | −0.8 (3) |
| C7—N1—C8—C9 | −156.88 (15) | C16—C17—C18—C19 | 0.5 (3) |
| C10—C9—C8—C13 | 0.3 (2) | N2—C15—C16—C17 | −0.6 (2) |
| C14—C9—C8—C13 | −179.24 (15) | N2—C15—C16—C20 | 178.28 (17) |
| C10—C9—C8—N1 | −176.71 (14) | C18—C17—C16—C15 | 0.1 (2) |
| C14—C9—C8—N1 | 3.7 (2) | C18—C17—C16—C20 | −178.74 (18) |
| C3—C2—C1—O1 | 172.11 (16) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H7···O1 | 0.889 (19) | 1.933 (19) | 2.6715 (19) | 139.4 (16) |
| O2—H2···N2 | 0.98 (3) | 1.62 (3) | 2.5981 (18) | 171 (2) |
| C12—H12···O1i | 0.95 | 2.53 | 3.285 (2) | 137 |
| C19—H19···O2ii | 0.95 | 2.51 | 3.372 (2) | 151 |
| Symmetry codes: (i) x+1, y, z; (ii) x−1/2, y, −z+3/2. |

 journal menu
journal menu











