The structure of the conjugative coupling protein TrwBΔN70 from Escherichia coli plasmid R388 was solved using two crystal forms. This large multimeric membrane protein of 437 residues per monomer is involved in cell-to-cell single-strand DNA transfer. Diffraction data to 2.4 Å were available from trigonal crystals obtained from ammonium sulfate and to 2.5 Å from monoclinic crystals grown from tartrate. A single tantalum bromide (Ta6Br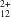 ) derivative of the trigonal form, which presented a protein hexamer with C6 local symmetry in the asymmetric unit, was used in a three-wavelength MAD experiment to achieve 4.5 Å resolution for initial phases. Sixfold averaging and phase extension increased the effective phasing resolution and eventually produced a straightforwardly traceable electron-density map. The monoclinic structure was solved by molecular replacement, i.e. a hexamer of the trigonal form was used as a search model. Two such hexamers are present in the asymmetric unit.
) derivative of the trigonal form, which presented a protein hexamer with C6 local symmetry in the asymmetric unit, was used in a three-wavelength MAD experiment to achieve 4.5 Å resolution for initial phases. Sixfold averaging and phase extension increased the effective phasing resolution and eventually produced a straightforwardly traceable electron-density map. The monoclinic structure was solved by molecular replacement, i.e. a hexamer of the trigonal form was used as a search model. Two such hexamers are present in the asymmetric unit.

 journal menu
journal menu











