
The structure of the new salt 1-(
o-tolyl)biguanidium chloride, C
9H
14N
5+·Cl
−, has been determined by single-crystal X-ray diffraction. The salt crystallizes in the monoclinic space group
C2/
c. In this structure, the chloride and biguanidium hydrophilic ions are mostly connected to each other
via N—H

N and N—H

Cl hydrogen bonds to form layers parallel to the
ab plane around
y =
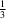
and
y =
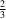
. The 2-methylbenzyl groups form layers between these layers around
y = 0 and
y =
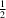
, with the methyl group forming C—H

π interactions with the aromatic ring. Intermolecular interactions on the Hirshfeld surface were investigated in terms of contact enrichment and electrostatic energy, and confirm the role of strong hydrogen bonds along with hydrophobic interactions. A correlation between electrostatic energy and contact enrichment is found only for the strongly attractive (N—H

Cl
−) and repulsive contacts. Electrostatic energies between ions reveal that the interacting biguanidium cation pairs are repulsive and that the crystal is maintained by attractive cation

Cl
− dimers. The vibrational absorption bands were identified by IR spectroscopy.
Supporting information
CCDC reference: 1877569
Data collection: CrysAlis PRO (Rigaku OD, 2015); cell refinement: CrysAlis PRO (Rigaku OD, 2015); data reduction: CrysAlis PRO (Rigaku OD, 2015); program(s) used to solve structure: SHELXT (Sheldrick, 2015a); program(s) used to refine structure: SHELXL (Sheldrick, 2008, 2015b) and
MoPro (Jelsch et al., 2005); molecular graphics: DIAMOND (Brandenburg, 1998).
2-[Amino(iminiumyl)methyl]-1-(2-methylphenyl)guanidine chloride
top
Crystal data top
| C9H14N5+·Cl− | F(000) = 960 |
| Mr = 227.68 | Dx = 1.293 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 8409 reflections |
| a = 18.4640 (7) Å | θ = 3.7–32.4° |
| b = 13.7652 (6) Å | µ = 0.30 mm−1 |
| c = 9.3158 (3) Å | T = 110 K |
| β = 98.847 (4)° | Prism, white |
| V = 2339.54 (16) Å3 | 0.23 × 0.15 × 0.04 mm |
| Z = 8 | |
Data collection top
Agilent SuperNova Dual Source
diffractometer with an Atlas detector | 3970 independent reflections |
| Radiation source: fine-focus sealed tube | 2840 reflections with > 2.0σ(I) |
| Mirror monochromator | Rint = 0.063 |
| ω scans | θmax = 31.9°, θmin = 3.7° |
Absorption correction: multi-scan
(CrysAlis PRO; Rigaku OD, 2015) | h = −27→27 |
| Tmin = 0.850, Tmax = 1.000 | k = −19→20 |
| 29979 measured reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.057 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.065 | All H-atom parameters refined |
| S = 0.97 | w = 1/[2.1*σ2(Fo2)] |
| 3970 reflections | (Δ/σ)max = −0.002 |
| 192 parameters | Δρmax = 0.64 e Å−3 |
| 47 restraints | Δρmin = −0.40 e Å−3 |
Special details top
Refinement. Refinement of F2 against reflections.
The threshold expression of F2 > 2sigma(F2) is
used for calculating R-factors(gt) and is not
relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice
as large as those based on F, and R-factors based on
ALL data will be even larger. A single crystal was carefully selected under a polarizing
microscope in order to perform its structural analysis. X-ray diffraction data
were collected on a SuperNova (Dual, Cu at zero Atlas) diffractometer at 110 K, using graphite-monochromated Mo Kα = 0.71073 Å radiation. The structure
was solved using a direct method with the SHELXT program
The structure was
refined using the full-matrix least-squares procedure using the SHELXL program
(Sheldrick, 2008; Sheldrick 2015).
The drawings were made with Diamond (Brandenburg, 1998). Crystal
data and experimental parameters used for the intensity data collection are
summarized in Table 1. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Cl | 0.414821 (13) | 0.88578 (2) | −0.04433 (3) | 0.01974 (2) | |
| N2 | 0.33610 (4) | 0.78604 (6) | 0.42786 (10) | 0.01757 (7) | |
| N3 | 0.30580 (4) | 0.81756 (7) | 0.65219 (9) | 0.01997 (8) | |
| H1 | 0.3193 (6) | 0.8406 (9) | 0.756 (10) | 0.0244 (2)* | |
| H2 | 0.257 (10) | 0.7853 (8) | 0.6192 (12) | 0.0244 (2)* | |
| N4 | 0.36565 (5) | 0.75392 (7) | 0.20089 (10) | 0.01987 (8) | |
| H4 | 0.3785 (6) | 0.7843 (8) | 0.109 (10) | 0.0244 (2)* | |
| N5 | 0.39543 (5) | 0.90299 (7) | 0.29973 (9) | 0.02277 (8) | |
| H3 | 0.3896 (7) | 0.957 (10) | 0.3710 (9) | 0.0280 (3)* | |
| H5 | 0.4122 (6) | 0.9204 (8) | 0.204 (10) | 0.0279 (3)* | |
| N6 | 0.41948 (5) | 0.86368 (8) | 0.61097 (9) | 0.02504 (8) | |
| H6 | 0.4310 (6) | 0.8816 (9) | 0.718 (10) | 0.0302 (3)* | |
| H7 | 0.460 (10) | 0.8615 (10) | 0.5498 (9) | 0.0302 (3)* | |
| C7 | 0.35465 (5) | 0.82389 (8) | 0.56102 (12) | 0.01720 (8) | |
| C8 | 0.36674 (5) | 0.81496 (8) | 0.31484 (12) | 0.01704 (8) | |
| C9 | 0.35099 (6) | 0.65283 (8) | 0.20189 (13) | 0.02048 (9) | |
| C1 | 0.39317 (6) | 0.59042 (9) | 0.29875 (14) | 0.02626 (11) | |
| C2 | 0.29635 (6) | 0.61761 (8) | 0.09368 (13) | 0.02776 (11) | |
| H8 | 0.2707 (5) | 0.667 (10) | 0.0104 (8) | 0.0335 (3)* | |
| C3 | 0.28159 (7) | 0.51917 (10) | 0.08305 (15) | 0.03686 (13) | |
| H9 | 0.240 (10) | 0.4912 (5) | −0.0010 (9) | 0.0449 (4)* | |
| C4 | 0.37666 (7) | 0.49095 (9) | 0.28582 (15) | 0.03765 (14) | |
| H14 | 0.410 (10) | 0.4425 (4) | 0.3608 (9) | 0.0464 (5)* | |
| C6 | 0.45508 (6) | 0.62439 (8) | 0.40909 (13) | 0.03502 (12) | |
| H10 | 0.4772 (4) | 0.692 (10) | 0.3770 (9) | 0.0523 (5)* | |
| H11 | 0.499 (10) | 0.5723 (4) | 0.4226 (10) | 0.0523 (5)* | |
| H12 | 0.4374 (4) | 0.6349 (7) | 0.513 (10) | 0.0522 (5)* | |
| C5 | 0.32202 (8) | 0.45601 (8) | 0.17956 (18) | 0.04085 (15) | |
| H13 | 0.3110 (6) | 0.379 (10) | 0.1731 (12) | 0.0506 (5)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cl | 0.02111 (12) | 0.02624 (15) | 0.01251 (14) | 0.00064 (10) | 0.00457 (9) | 0.00235 (11) |
| N2 | 0.0193 (4) | 0.0218 (5) | 0.0124 (5) | −0.0053 (4) | 0.0050 (3) | −0.0019 (4) |
| N3 | 0.0189 (4) | 0.0286 (6) | 0.0135 (5) | −0.0043 (4) | 0.0061 (4) | −0.0024 (4) |
| N4 | 0.0276 (5) | 0.0197 (5) | 0.0135 (5) | −0.0037 (4) | 0.0070 (4) | −0.0023 (4) |
| N5 | 0.0334 (5) | 0.0224 (5) | 0.0140 (5) | −0.0096 (4) | 0.0085 (4) | −0.0021 (4) |
| N6 | 0.0196 (4) | 0.0427 (7) | 0.0132 (5) | −0.0091 (4) | 0.0040 (4) | −0.0041 (5) |
| C7 | 0.0177 (5) | 0.0217 (6) | 0.0128 (6) | −0.0026 (4) | 0.0044 (4) | −0.0013 (5) |
| C8 | 0.0193 (5) | 0.0187 (6) | 0.0136 (6) | −0.0030 (4) | 0.0040 (4) | −0.0014 (5) |
| C9 | 0.0228 (5) | 0.0209 (6) | 0.0198 (6) | −0.0025 (4) | 0.0097 (4) | −0.0023 (5) |
| C1 | 0.0272 (6) | 0.0225 (6) | 0.0309 (7) | 0.0004 (5) | 0.0100 (5) | 0.0019 (6) |
| C2 | 0.0296 (6) | 0.0275 (7) | 0.0267 (7) | −0.0071 (5) | 0.0061 (5) | −0.0101 (6) |
| C3 | 0.0390 (7) | 0.0318 (8) | 0.0411 (9) | −0.0097 (6) | 0.0104 (6) | −0.0136 (7) |
| C4 | 0.0437 (8) | 0.0253 (7) | 0.0470 (10) | 0.0036 (6) | 0.0164 (7) | 0.0058 (7) |
| C6 | 0.0337 (7) | 0.0381 (8) | 0.0322 (8) | 0.0062 (6) | 0.0015 (6) | 0.0058 (6) |
| C5 | 0.0487 (8) | 0.0216 (7) | 0.0554 (11) | −0.0071 (6) | 0.0184 (7) | −0.0103 (7) |
Geometric parameters (Å, º) top
| N2—C8 | 1.3306 (13) | C9—C1 | 1.3938 (16) |
| N2—C7 | 1.3405 (14) | C9—C2 | 1.3986 (15) |
| N3—C7 | 1.3335 (12) | C1—C4 | 1.4039 (17) |
| N3—H1 | 1.01 (9) | C1—C6 | 1.4902 (17) |
| N3—H2 | 1.01 (16) | C2—C3 | 1.3826 (16) |
| N4—C8 | 1.3516 (14) | C2—H8 | 1.08 (9) |
| N4—C9 | 1.4179 (14) | C3—C5 | 1.384 (2) |
| N4—H4 | 1.01 (8) | C3—H9 | 1.08 (11) |
| N5—C8 | 1.3385 (13) | C4—C5 | 1.3863 (19) |
| N5—H3 | 1.01 (10) | C4—H14 | 1.08 (9) |
| N5—H5 | 1.01 (9) | C6—H11 | 1.08 (14) |
| N6—C7 | 1.3334 (13) | C6—H10 | 1.08 (12) |
| N6—H7 | 1.02 (15) | C6—H12 | 1.08 (9) |
| N6—H6 | 1.02 (9) | C5—H13 | 1.08 (14) |
| | | |
| C8—N2—C7 | 122.46 (8) | C9—C1—C4 | 116.95 (10) |
| C7—N3—H1 | 120 (4) | C9—C1—C6 | 123.10 (9) |
| C7—N3—H2 | 120 (8) | C4—C1—C6 | 119.91 (10) |
| H1—N3—H2 | 120 (2) | C9—C2—C3 | 120.24 (10) |
| C8—N4—C9 | 125.79 (8) | C9—C2—H8 | 119 (3) |
| C8—N4—H4 | 115 (4) | C3—C2—H8 | 120 (7) |
| C9—N4—H4 | 118.7 (6) | C2—C3—C5 | 119.24 (10) |
| C8—N5—H3 | 121 (7) | C2—C3—H9 | 121 (2) |
| C8—N5—H5 | 119 (1) | C5—C3—H9 | 120 (4) |
| H3—N5—H5 | 119 (4) | C1—C4—C5 | 121.61 (11) |
| C7—N6—H7 | 120 (9) | C1—C4—H14 | 117 (2) |
| C7—N6—H6 | 119 (1) | C5—C4—H14 | 122 (6) |
| H7—N6—H6 | 120 (4) | C1—C6—H11 | 111 (7) |
| N2—C7—N6 | 124.79 (8) | C1—C6—H10 | 111 (2) |
| N2—C7—N3 | 117.54 (8) | C1—C6—H12 | 111 (3) |
| N6—C7—N3 | 117.61 (8) | H11—C6—H10 | 107 (6) |
| N4—C8—N2 | 118.60 (8) | H11—C6—H12 | 108 (4) |
| N4—C8—N5 | 116.01 (8) | H10—C6—H12 | 108 (2) |
| N2—C8—N5 | 125.27 (8) | C3—C5—C4 | 120.43 (11) |
| N4—C9—C1 | 121.39 (9) | C3—C5—H13 | 120 (5) |
| N4—C9—C2 | 116.92 (9) | C4—C5—H13 | 119 (3) |
| C1—C9—C2 | 121.48 (10) | | |
| | | |
| N2—C8—N4—C9 | −16.30 (15) | C8—N4—C9—C2 | 127.35 (15) |
| N2—C8—N4—H4 | 166 (2) | C9—C1—C4—C5 | 1.41 (17) |
| N2—C8—N5—H3 | −8 (4) | C9—C1—C4—H14 | 180 (9) |
| N2—C8—N5—H5 | −173 (3) | C9—C1—C6—H11 | −143 (9) |
| N2—C7—N6—H7 | −5 (4) | C9—C1—C6—H10 | −24 (1) |
| N2—C7—N6—H6 | −171 (2) | C9—C1—C6—H12 | 96 (5) |
| N2—C7—N3—H1 | 175 (2) | C9—C2—C3—C5 | −0.72 (17) |
| N2—C7—N3—H2 | 0 (5) | C9—C2—C3—H9 | −179 (10) |
| N3—C7—N2—C8 | 156.67 (15) | C1—C9—C2—C3 | 1.78 (17) |
| N3—C7—N6—H7 | 172 (4) | C1—C9—C2—H8 | −170 (1) |
| N3—C7—N6—H6 | 6 (2) | C1—C4—C5—C3 | −0.44 (19) |
| H1—N3—C7—N6 | −2 (2) | C1—C4—C5—H13 | 180 (1) |
| H2—N3—C7—N6 | −178 (5) | C2—C9—C1—C4 | −2.07 (16) |
| N4—C8—N2—C7 | 157.09 (14) | C2—C9—C1—C6 | 175.84 (17) |
| N4—C8—N5—H3 | 168 (4) | C2—C3—C5—C4 | 0.07 (19) |
| N4—C8—N5—H5 | 3 (2) | C2—C3—C5—H13 | 180 (1) |
| N4—C9—C1—C4 | −176.71 (15) | H8—C2—C3—C5 | 171 (2) |
| N4—C9—C1—C6 | 1.20 (15) | H8—C2—C3—H9 | −7 (11) |
| N4—C9—C2—C3 | 176.65 (15) | C3—C5—C4—H14 | −179 (10) |
| N4—C9—C2—H8 | 4 (2) | H9—C3—C5—C4 | 179 (13) |
| H4—N4—C8—N5 | −10 (2) | H9—C3—C5—H13 | −2 (13) |
| H4—N4—C9—C1 | 120 (2) | C4—C1—C6—H11 | 35 (9) |
| H4—N4—C9—C2 | −55 (2) | C4—C1—C6—H10 | 154 (1) |
| N5—C8—N4—C9 | 167.49 (15) | C4—C1—C6—H12 | −86 (5) |
| N5—C8—N2—C7 | −27.08 (15) | H14—C4—C1—C6 | 2 (13) |
| N6—C7—N2—C8 | −26.32 (16) | H14—C4—C5—H13 | 2 (13) |
| C8—N4—C9—C1 | −57.78 (16) | C6—C1—C4—C5 | −176.57 (18) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N4—H4···Cl | 1.01 (8) | 2.17 (7) | 3.1584 (9) | 163 (1) |
| N5—H5···Cl | 1.01 (9) | 2.37 (9) | 3.2880 (9) | 150 (6) |
| N6—H7···N5 | 1.02 (15) | 2.52 (6) | 2.9156 (13) | 103 (5) |
| C6—H10···C8 | 1.08 (12) | 2.65 (9) | 3.1420 (15) | 108 (5) |
| N3—H1···Cli | 1.01 (9) | 2.44 (6) | 3.3431 (9) | 148 (4) |
| N6—H6···Cli | 1.02 (9) | 2.28 (9) | 3.2398 (9) | 157 (1) |
| N5—H3···Clii | 1.01 (10) | 2.32 (13) | 3.2454 (9) | 150 (3) |
| N6—H7···Cliii | 1.02 (15) | 2.34 (18) | 3.2284 (9) | 146 (11) |
| N3—H2···N2iv | 1.01 (16) | 1.96 (15) | 2.9754 (12) | 174 (1) |
| Symmetry codes: (i) x, y, z+1; (ii) x, −y+2, z+1/2; (iii) −x+1, y, −z+1/2; (iv) −x+1/2, −y+3/2, −z+1. |
Analysis of contacts on the Hirshfeld surface. Reciprocal contacts
X···Y and Y···X are merged. The second line shows
the chemical content on the surface. The % of contact types between chemical
species are given in the next five lines, followed by their enrichment ratios.
The major contacts, as well as the most enriched ones, are highlighted in bold.
The lower part of the table give the contact enrichment when the N atom with
lone pair Nlp is distinguished from the other N—H/NH2 atoms top| Atom | Hn | C | N | Cl | Hc | |
| Surface % | 27.3 | 19.3 | 8.1 | 15.5 | 29.8 | |
| Hn | 4.4 | | | | | |
| C | 6.4 | 5.0 | | % contacts | | |
| N | 4.9 | 1.1 | 0 | | | |
| Cl | 27.3 | 1.2 | 1.2 | 0.2 | | |
| Hc | 7.8 | 18.7 | 7.1 | 3.8 | 11.0 | |
| Hn | 0.58 | | | | | |
| C | 0.62 | 1.43 | | Enrichment | | |
| N | 1.24 | 0.42 | 0 | | | |
| Cl | 2.92 | 0.19 | 0.48 | 0.08 | | |
| Hc | 0.48 | 1.69 | 1.68 | 0.38 | 1.25 | |
| | | | | | |
| Atom | Hn | C | Nlp | Nh | Cl | Hc |
| Nlp | 2.37 | 0.05 | 0 | 0 | 0 | 0.98 |
| Nh | 0.39 | 0.36 | 0 | 0 | 0.88 | 2.24 |
Comparison between C—N bond distances in 1-(o-tolyl)biguanidium and
metformin chloride (Niranjana et al., 2017) top| 1-(o-Tolyl)biguanidium chloride | Metformin chloride | | |
| N6—C7 | 1.3334 (13) | N5—C4 | 1.3398 (3) |
| N3—C7 | 1.3335 (13) | N4—C4 | 1.3407 (4) |
| N2—C7 | 1.3407 (13) | N3—C4 | 1.3332 (3) |
| N2—C8 | 1.3307 (13) | C4—N3 | 1.3332 (3) |
| N5—C8 | 1.3385 (13) | N2—C3 | 1.3372 (4) |
| N4—C8 | 1.3516 (14) | N1—C3 | 1.3376 (4) |
| N4—C9 | 1.4180 (14) | N1—C1(CH3) | 1.4590 (4) |
| | N1—C2(CH3) | 1.4566 (5) |
Electrostatic energy (kJ mol-1) between the TBG+ cations and the neighbouring
cations in direct contact. The summation was performed by attributing a
coefficient 1/2 to the involutional symmetry operators σ and a unitary
weight to the others top | Symmetry code | Energy | Coefficient |
| TBG+···Cl- | x, y, z | -385 | 1/2 |
| x, y, z+1 | -410 | 1 |
| x, -y+2, z+1/2 | -369 | 1 |
| -x+1, y, -z+1/2 | -369 | 1/2 |
| x, -y+1, z-1/2 | 126 | 1 |
| -x+1, y, -z+1/2 | 216 | 1/2 |
| TBG+···TBG+ | -x+1, y, -z+3/2 | 245 | 1/2 |
| -x+1, -y+1, -z+1 | 135 | 1/2 |
| -x+1/2, y-1/2, -z+1/2 | 138 | 1 |
| -x+1/2, -y+3/2, -z | 141 | 1/2 |
| -x+1/2, -y+3/2, -z+1 | 60 | 1/2 |
| Sum | | -472 | |
Calculated energies of the possible cations obtained by protonation of
1-(o-tolyl)biguanide top| Structure | Absolute energy (Ha) | Relative energy versus structure 6 (kJ mol-1) |
| 1 | -624.898406392 | 230 |
| 2 | -624.903083434 | 216 |
| 3 | -624.916688817 | 182 |
| 4 | -624.954584111 | 83 |
| 5 | -624.966125315 | 52 |
| 6 | -624.986333171 | 0 |
| 7 | -624.964705612 | 55 |
 N and N—H
N and N—H Cl hydrogen bonds to form layers parallel to the ab plane around y =
Cl hydrogen bonds to form layers parallel to the ab plane around y =  π interactions with the aromatic ring. Intermolecular interactions on the Hirshfeld surface were investigated in terms of contact enrichment and electrostatic energy, and confirm the role of strong hydrogen bonds along with hydrophobic interactions. A correlation between electrostatic energy and contact enrichment is found only for the strongly attractive (N—H
π interactions with the aromatic ring. Intermolecular interactions on the Hirshfeld surface were investigated in terms of contact enrichment and electrostatic energy, and confirm the role of strong hydrogen bonds along with hydrophobic interactions. A correlation between electrostatic energy and contact enrichment is found only for the strongly attractive (N—H Cl−) and repulsive contacts. Electrostatic energies between ions reveal that the interacting biguanidium cation pairs are repulsive and that the crystal is maintained by attractive cation
Cl−) and repulsive contacts. Electrostatic energies between ions reveal that the interacting biguanidium cation pairs are repulsive and that the crystal is maintained by attractive cation Cl− dimers. The vibrational absorption bands were identified by IR spectroscopy.
Cl− dimers. The vibrational absorption bands were identified by IR spectroscopy.
 journal menu
journal menu











