
This work reports a new acetonitrile (ACN)-solvated cocrystal of piroxicam (PRX) and succinic acid (SA), 2C
15H
13N
3O
4S·0.5C
4H
6O
4·C
2H
3N or PRX:SA:ACN (4:1:2), which adopts the triclinic space group
P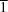
. The outcome of crystallization from ACN solution can be controlled by varying only the PRX:SA ratio, with a higher PRX:SA ratio in solution unexpectedly favouring a lower stoichiometric ratio in the solid product. In the new solvate, zwitterionic (Z) and non-ionized (NI) PRX molecules co-exist in the asymmetric unit. In contrast, the nonsolvated PRX–SA cocrystal contains only NI-type PRX molecules. The ACN molecule entrapped in PRX–SA·ACN does not form any hydrogen bonds with the surrounding molecules. In the solvated cocrystal, Z-type molecules form dimers linked by intermolecular N—H

O hydrogen bonds, whereas every pair of NI-type molecules is linked to SA
via N—H

O and O—H

N hydrogen bonds. Thermogravimetry and differential scanning calorimetry suggest that thermal desolvation of the solvate sample occurs at 148 °C, and is followed by recrystallization, presumably of a multicomponent PRX–SA structure. Vibrational spectra (IR and Raman spectroscopy) of PRX–SA·ACN and PRX–SA are also used to demonstrate the ability of spectroscopic techniques to distinguish between NI- and Z-type PRX molecules in the solid state. Hence, vibrational spectroscopy can be used to distinguish the PRX–SA cocrystal and its ACN solvate.
Supporting information
CCDC reference: 1860488
Data collection: CrysAlis PRO (Rigaku OD, 2018); cell refinement: CrysAlis PRO (Rigaku OD, 2018); data reduction: CrysAlis PRO (Rigaku OD, 2018); program(s) used to solve structure: SHELXT (Sheldrick, 2015a); program(s) used to refine structure: SHELXL2014 (Sheldrick, 2015b); molecular graphics: OLEX2 (Dolomanov et al., 2009); software used to prepare material for publication: OLEX2 (Dolomanov et al., 2009).
4-Hydroxy-2-methyl-N-(pyridin-2-yl)-2H-1,2-benzothiazine-3-\
carboxamide 1,1-dioxide–succinic acid–acetonitrile (4/1/2)
top
Crystal data top
| 4C15H13N3O4S·C4H6O4·2C2H3N | Z = 1 |
| Mr = 1525.57 | F(000) = 794 |
| Triclinic, P1 | Dx = 1.499 Mg m−3 |
| a = 9.1561 (3) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 12.7660 (5) Å | Cell parameters from 11463 reflections |
| c = 15.1022 (5) Å | θ = 4.0–29.3° |
| α = 83.030 (3)° | µ = 0.23 mm−1 |
| β = 75.313 (3)° | T = 120 K |
| γ = 83.972 (3)° | Block, clear yellowish yellow |
| V = 1689.87 (11) Å3 | 0.44 × 0.16 × 0.09 mm |
Data collection top
Agilent SuperNova Dual Source
diffractometer with an Atlas detector | 6505 reflections with I > 2σ(I) |
| Radiation source: micro-focus sealed X-ray tube, SuperNova (Mo) X-ray Source | Rint = 0.047 |
| Mirror monochromator | θmax = 29.8°, θmin = 2.9° |
| Detector resolution: 5.1574 pixels mm-1 | h = −12→11 |
| ω scans | k = −16→17 |
| 30897 measured reflections | l = −20→19 |
| 8512 independent reflections | |
Refinement top
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.043 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.111 | w = 1/[σ2(Fo2) + (0.0488P)2 + 0.6069P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max = 0.001 |
| 8512 reflections | Δρmax = 0.43 e Å−3 |
| 501 parameters | Δρmin = −0.42 e Å−3 |
| 0 restraints | |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| S1 | 0.56249 (5) | 0.51097 (4) | 0.81566 (3) | 0.01718 (11) | |
| S1A | 0.51996 (5) | 0.95151 (4) | 0.20324 (3) | 0.01738 (11) | |
| O2A | 0.35755 (14) | 0.89787 (10) | 0.49328 (8) | 0.0189 (3) | |
| O1 | 0.85023 (13) | 0.54236 (10) | 0.55856 (8) | 0.0178 (3) | |
| O1A | 0.63606 (13) | 0.84018 (10) | 0.49432 (8) | 0.0191 (3) | |
| O3A | 0.52506 (14) | 0.83890 (10) | 0.20667 (9) | 0.0218 (3) | |
| O4 | 0.61816 (14) | 0.52241 (11) | 0.89421 (9) | 0.0246 (3) | |
| O2 | 0.38987 (13) | 0.63539 (10) | 0.58349 (8) | 0.0204 (3) | |
| O3 | 0.55616 (14) | 0.40694 (10) | 0.79172 (9) | 0.0228 (3) | |
| O5 | 0.91844 (14) | 0.95196 (11) | 0.16737 (9) | 0.0246 (3) | |
| O6 | 1.11296 (16) | 0.83233 (12) | 0.12959 (10) | 0.0294 (3) | |
| O4A | 0.56602 (14) | 1.01347 (11) | 0.11726 (9) | 0.0259 (3) | |
| N1A | 1.06678 (15) | 0.79626 (11) | 0.31394 (10) | 0.0166 (3) | |
| N2 | 0.66103 (17) | 0.59141 (12) | 0.48374 (10) | 0.0169 (3) | |
| N1 | 0.89245 (16) | 0.55523 (12) | 0.37868 (10) | 0.0157 (3) | |
| N3A | 0.61791 (15) | 0.97913 (12) | 0.27301 (10) | 0.0161 (3) | |
| N3 | 0.66282 (16) | 0.57882 (12) | 0.72692 (10) | 0.0165 (3) | |
| N2A | 0.82028 (15) | 0.86478 (12) | 0.35951 (11) | 0.0156 (3) | |
| C6 | 0.34274 (18) | 0.62285 (13) | 0.74572 (12) | 0.0154 (3) | |
| C10A | 0.94686 (18) | 0.80958 (13) | 0.38521 (12) | 0.0142 (3) | |
| C7 | 0.44819 (19) | 0.61706 (13) | 0.65212 (11) | 0.0154 (3) | |
| C10 | 0.74555 (19) | 0.59175 (14) | 0.39554 (12) | 0.0160 (3) | |
| C9A | 0.67602 (18) | 0.87595 (13) | 0.41232 (12) | 0.0149 (3) | |
| C8 | 0.60287 (19) | 0.58837 (14) | 0.64623 (11) | 0.0154 (3) | |
| C6A | 0.30083 (18) | 0.98616 (14) | 0.35675 (12) | 0.0158 (3) | |
| C7A | 0.41397 (18) | 0.93752 (13) | 0.40644 (12) | 0.0151 (3) | |
| C8A | 0.56444 (18) | 0.93173 (13) | 0.36512 (12) | 0.0148 (3) | |
| C14A | 1.19510 (19) | 0.74678 (14) | 0.33108 (13) | 0.0199 (4) | |
| H14A | 1.2788 | 0.7381 | 0.2819 | 0.024* | |
| C9 | 0.71416 (19) | 0.57116 (13) | 0.56295 (12) | 0.0154 (3) | |
| C3A | 0.0904 (2) | 1.08212 (14) | 0.26020 (14) | 0.0214 (4) | |
| H3A | 0.0199 | 1.1141 | 0.2286 | 0.026* | |
| C5 | 0.19658 (19) | 0.67127 (14) | 0.75407 (12) | 0.0182 (4) | |
| H5 | 0.1665 | 0.7012 | 0.7016 | 0.022* | |
| C11A | 0.95027 (19) | 0.77222 (14) | 0.47520 (12) | 0.0166 (3) | |
| H11A | 0.8654 | 0.7814 | 0.5236 | 0.020* | |
| C1A | 0.33696 (18) | 0.99931 (14) | 0.26045 (12) | 0.0167 (3) | |
| C1 | 0.38137 (19) | 0.57745 (14) | 0.82679 (12) | 0.0171 (4) | |
| C16 | 1.01926 (19) | 0.91257 (15) | 0.10927 (12) | 0.0203 (4) | |
| C12 | 0.7771 (2) | 0.63371 (15) | 0.23344 (13) | 0.0215 (4) | |
| H12 | 0.7380 | 0.6610 | 0.1836 | 0.026* | |
| C12A | 1.0841 (2) | 0.72088 (14) | 0.49062 (13) | 0.0196 (4) | |
| H12A | 1.0898 | 0.6949 | 0.5500 | 0.023* | |
| C4A | 0.05204 (19) | 1.06884 (14) | 0.35496 (14) | 0.0207 (4) | |
| H4A | −0.0445 | 1.0922 | 0.3868 | 0.025* | |
| C4 | 0.0957 (2) | 0.67538 (15) | 0.83937 (13) | 0.0216 (4) | |
| H4 | −0.0009 | 0.7086 | 0.8436 | 0.026* | |
| C2A | 0.2346 (2) | 1.04773 (14) | 0.21159 (13) | 0.0202 (4) | |
| H2AC | 0.2617 | 1.0571 | 0.1477 | 0.024* | |
| C11 | 0.6857 (2) | 0.63191 (15) | 0.32054 (12) | 0.0192 (4) | |
| H11 | 0.5845 | 0.6572 | 0.3299 | 0.023* | |
| C13 | 0.9296 (2) | 0.59466 (15) | 0.21870 (12) | 0.0213 (4) | |
| H13 | 0.9920 | 0.5955 | 0.1595 | 0.026* | |
| C14 | 0.9839 (2) | 0.55563 (15) | 0.29280 (12) | 0.0194 (4) | |
| H14 | 1.0845 | 0.5290 | 0.2845 | 0.023* | |
| C5A | 0.15555 (19) | 1.02107 (14) | 0.40357 (13) | 0.0185 (4) | |
| H5A | 0.1278 | 1.0124 | 0.4675 | 0.022* | |
| C17 | 1.0507 (2) | 0.95073 (16) | 0.00934 (13) | 0.0239 (4) | |
| H17A | 1.0347 | 0.8950 | −0.0247 | 0.029* | |
| H17B | 1.1558 | 0.9666 | −0.0124 | 0.029* | |
| C2 | 0.2816 (2) | 0.58051 (16) | 0.91262 (13) | 0.0231 (4) | |
| H2A | 0.3106 | 0.5498 | 0.9652 | 0.028* | |
| C15 | 0.7219 (2) | 0.67557 (16) | 0.74421 (14) | 0.0258 (4) | |
| H15A | 0.6406 | 0.7298 | 0.7568 | 0.039* | |
| H15B | 0.7976 | 0.6994 | 0.6910 | 0.039* | |
| H15C | 0.7659 | 0.6605 | 0.7961 | 0.039* | |
| C13A | 1.2085 (2) | 0.70835 (14) | 0.41802 (14) | 0.0217 (4) | |
| H13A | 1.2992 | 0.6748 | 0.4275 | 0.026* | |
| C15A | 0.6571 (2) | 1.08983 (15) | 0.26579 (14) | 0.0219 (4) | |
| H15D | 0.6980 | 1.1138 | 0.2024 | 0.033* | |
| H15E | 0.7311 | 1.0936 | 0.3002 | 0.033* | |
| H15F | 0.5678 | 1.1339 | 0.2901 | 0.033* | |
| C3 | 0.1373 (2) | 0.63041 (16) | 0.91858 (13) | 0.0252 (4) | |
| H3 | 0.0688 | 0.6336 | 0.9756 | 0.030* | |
| C18 | 0.6546 (3) | 0.75781 (19) | 0.00166 (14) | 0.0364 (5) | |
| C19 | 0.4929 (3) | 0.76025 (19) | 0.01312 (15) | 0.0348 (5) | |
| H19A | 0.4702 | 0.7040 | −0.0169 | 0.052* | |
| H19B | 0.4571 | 0.8270 | −0.0136 | 0.052* | |
| H19C | 0.4439 | 0.7514 | 0.0775 | 0.052* | |
| N4 | 0.7818 (3) | 0.7562 (2) | −0.00821 (16) | 0.0619 (7) | |
| H6 | 1.093 (3) | 0.819 (2) | 0.191 (2) | 0.049 (8)* | |
| H2AB | 0.837 (2) | 0.8879 (17) | 0.3023 (16) | 0.022 (5)* | |
| H1 | 0.928 (2) | 0.5362 (16) | 0.4253 (15) | 0.020 (5)* | |
| H2 | 0.565 (2) | 0.6139 (16) | 0.4959 (14) | 0.020 (5)* | |
| H2AA | 0.438 (3) | 0.872 (2) | 0.5126 (18) | 0.050 (8)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| S1 | 0.0155 (2) | 0.0214 (2) | 0.0137 (2) | 0.00247 (16) | −0.00492 (16) | 0.00131 (16) |
| S1A | 0.0146 (2) | 0.0221 (2) | 0.0146 (2) | −0.00002 (16) | −0.00349 (16) | 0.00032 (17) |
| O2A | 0.0148 (6) | 0.0251 (7) | 0.0152 (6) | −0.0006 (5) | −0.0022 (5) | 0.0012 (5) |
| O1 | 0.0151 (6) | 0.0217 (7) | 0.0156 (6) | 0.0001 (5) | −0.0039 (5) | 0.0008 (5) |
| O1A | 0.0170 (6) | 0.0231 (7) | 0.0155 (6) | −0.0003 (5) | −0.0035 (5) | 0.0021 (5) |
| O3A | 0.0218 (6) | 0.0227 (7) | 0.0212 (7) | 0.0012 (5) | −0.0051 (5) | −0.0056 (5) |
| O4 | 0.0213 (6) | 0.0384 (8) | 0.0142 (6) | 0.0031 (6) | −0.0083 (5) | 0.0007 (6) |
| O2 | 0.0183 (6) | 0.0282 (7) | 0.0159 (6) | 0.0005 (5) | −0.0085 (5) | −0.0001 (5) |
| O3 | 0.0243 (6) | 0.0185 (7) | 0.0237 (7) | 0.0020 (5) | −0.0058 (5) | 0.0027 (5) |
| O5 | 0.0193 (6) | 0.0352 (8) | 0.0149 (6) | 0.0046 (5) | −0.0008 (5) | 0.0013 (6) |
| O6 | 0.0316 (7) | 0.0323 (8) | 0.0175 (7) | 0.0106 (6) | −0.0010 (6) | 0.0023 (6) |
| O4A | 0.0198 (6) | 0.0375 (8) | 0.0177 (7) | −0.0016 (6) | −0.0041 (5) | 0.0059 (6) |
| N1A | 0.0142 (7) | 0.0159 (7) | 0.0192 (7) | −0.0003 (5) | −0.0039 (6) | −0.0006 (6) |
| N2 | 0.0156 (7) | 0.0212 (8) | 0.0142 (7) | −0.0005 (6) | −0.0049 (6) | −0.0013 (6) |
| N1 | 0.0168 (7) | 0.0173 (8) | 0.0135 (7) | −0.0002 (6) | −0.0059 (6) | 0.0000 (6) |
| N3A | 0.0129 (7) | 0.0184 (8) | 0.0162 (7) | −0.0020 (5) | −0.0043 (6) | 0.0035 (6) |
| N3 | 0.0159 (7) | 0.0207 (8) | 0.0134 (7) | −0.0013 (6) | −0.0055 (6) | −0.0003 (6) |
| N2A | 0.0143 (7) | 0.0176 (8) | 0.0143 (8) | 0.0002 (5) | −0.0044 (6) | 0.0018 (6) |
| C6 | 0.0161 (8) | 0.0153 (9) | 0.0155 (8) | −0.0010 (6) | −0.0055 (7) | −0.0012 (7) |
| C10A | 0.0130 (7) | 0.0130 (8) | 0.0174 (8) | −0.0018 (6) | −0.0051 (6) | −0.0011 (6) |
| C7 | 0.0187 (8) | 0.0147 (8) | 0.0131 (8) | −0.0019 (6) | −0.0055 (7) | 0.0009 (6) |
| C10 | 0.0179 (8) | 0.0145 (9) | 0.0162 (8) | −0.0031 (6) | −0.0047 (7) | −0.0008 (6) |
| C9A | 0.0143 (8) | 0.0138 (8) | 0.0172 (8) | −0.0018 (6) | −0.0049 (7) | −0.0012 (6) |
| C8 | 0.0162 (8) | 0.0176 (9) | 0.0132 (8) | −0.0016 (6) | −0.0062 (7) | 0.0014 (6) |
| C6A | 0.0123 (8) | 0.0151 (8) | 0.0201 (9) | −0.0016 (6) | −0.0039 (7) | −0.0018 (7) |
| C7A | 0.0161 (8) | 0.0146 (8) | 0.0148 (8) | −0.0017 (6) | −0.0035 (7) | −0.0021 (6) |
| C8A | 0.0145 (8) | 0.0154 (9) | 0.0150 (8) | −0.0008 (6) | −0.0050 (6) | −0.0003 (6) |
| C14A | 0.0148 (8) | 0.0174 (9) | 0.0279 (10) | 0.0000 (7) | −0.0067 (7) | −0.0020 (7) |
| C9 | 0.0177 (8) | 0.0120 (8) | 0.0169 (8) | −0.0025 (6) | −0.0054 (7) | 0.0003 (6) |
| C3A | 0.0165 (8) | 0.0181 (9) | 0.0315 (11) | −0.0004 (7) | −0.0111 (8) | 0.0002 (8) |
| C5 | 0.0177 (8) | 0.0176 (9) | 0.0197 (9) | 0.0008 (7) | −0.0079 (7) | 0.0010 (7) |
| C11A | 0.0181 (8) | 0.0167 (9) | 0.0155 (8) | −0.0019 (7) | −0.0047 (7) | −0.0013 (7) |
| C1A | 0.0142 (8) | 0.0167 (9) | 0.0196 (9) | −0.0021 (6) | −0.0051 (7) | −0.0008 (7) |
| C1 | 0.0155 (8) | 0.0174 (9) | 0.0183 (9) | 0.0006 (6) | −0.0054 (7) | −0.0003 (7) |
| C16 | 0.0164 (8) | 0.0249 (10) | 0.0185 (9) | −0.0025 (7) | −0.0029 (7) | 0.0001 (7) |
| C12 | 0.0254 (9) | 0.0234 (10) | 0.0178 (9) | −0.0027 (7) | −0.0103 (8) | 0.0019 (7) |
| C12A | 0.0229 (9) | 0.0167 (9) | 0.0220 (9) | −0.0015 (7) | −0.0121 (7) | 0.0012 (7) |
| C4A | 0.0130 (8) | 0.0187 (9) | 0.0314 (11) | −0.0003 (7) | −0.0063 (7) | −0.0051 (8) |
| C4 | 0.0154 (8) | 0.0237 (10) | 0.0249 (10) | 0.0033 (7) | −0.0045 (7) | −0.0041 (8) |
| C2A | 0.0207 (9) | 0.0198 (9) | 0.0211 (9) | −0.0030 (7) | −0.0085 (7) | 0.0022 (7) |
| C11 | 0.0190 (8) | 0.0210 (9) | 0.0191 (9) | −0.0012 (7) | −0.0081 (7) | −0.0010 (7) |
| C13 | 0.0230 (9) | 0.0260 (10) | 0.0141 (9) | −0.0054 (7) | −0.0019 (7) | −0.0015 (7) |
| C14 | 0.0179 (8) | 0.0214 (9) | 0.0178 (9) | −0.0019 (7) | −0.0027 (7) | −0.0011 (7) |
| C5A | 0.0146 (8) | 0.0191 (9) | 0.0213 (9) | −0.0022 (7) | −0.0025 (7) | −0.0038 (7) |
| C17 | 0.0224 (9) | 0.0298 (11) | 0.0157 (9) | 0.0022 (8) | −0.0007 (7) | 0.0011 (8) |
| C2 | 0.0206 (9) | 0.0324 (11) | 0.0151 (9) | 0.0034 (8) | −0.0050 (7) | −0.0010 (8) |
| C15 | 0.0303 (10) | 0.0261 (11) | 0.0257 (10) | −0.0075 (8) | −0.0133 (8) | −0.0018 (8) |
| C13A | 0.0176 (8) | 0.0172 (9) | 0.0328 (11) | 0.0018 (7) | −0.0130 (8) | −0.0009 (8) |
| C15A | 0.0186 (8) | 0.0202 (10) | 0.0265 (10) | −0.0045 (7) | −0.0064 (8) | 0.0034 (8) |
| C3 | 0.0193 (9) | 0.0345 (12) | 0.0183 (9) | 0.0035 (8) | −0.0003 (7) | −0.0022 (8) |
| C18 | 0.0418 (13) | 0.0415 (14) | 0.0211 (11) | 0.0069 (10) | −0.0023 (9) | −0.0051 (9) |
| C19 | 0.0411 (12) | 0.0364 (13) | 0.0269 (11) | −0.0051 (10) | −0.0098 (10) | 0.0016 (9) |
| N4 | 0.0391 (13) | 0.093 (2) | 0.0449 (14) | 0.0120 (12) | 0.0023 (10) | −0.0163 (13) |
Geometric parameters (Å, º) top
| S1—O4 | 1.4327 (13) | C6—C1 | 1.402 (2) |
| S1—O3 | 1.4287 (14) | C10A—C11A | 1.392 (2) |
| S1—N3 | 1.6283 (15) | C7—C8 | 1.408 (2) |
| S1—C1 | 1.7597 (17) | C10—C11 | 1.400 (2) |
| S1A—O3A | 1.4285 (14) | C9A—C8A | 1.467 (2) |
| S1A—O4A | 1.4258 (13) | C8—C9 | 1.428 (2) |
| S1A—N3A | 1.6354 (15) | C6A—C7A | 1.472 (2) |
| S1A—C1A | 1.7622 (18) | C6A—C1A | 1.400 (2) |
| O2A—C7A | 1.339 (2) | C6A—C5A | 1.394 (2) |
| O1—C9 | 1.249 (2) | C7A—C8A | 1.359 (2) |
| O1A—C9A | 1.243 (2) | C14A—C13A | 1.376 (3) |
| O2—C7 | 1.269 (2) | C3A—C4A | 1.378 (3) |
| O5—C16 | 1.214 (2) | C3A—C2A | 1.393 (3) |
| O6—C16 | 1.324 (2) | C5—C4 | 1.385 (3) |
| N1A—C10A | 1.341 (2) | C11A—C12A | 1.387 (2) |
| N1A—C14A | 1.344 (2) | C1A—C2A | 1.387 (2) |
| N2—C10 | 1.360 (2) | C1—C2 | 1.386 (3) |
| N2—C9 | 1.389 (2) | C16—C17 | 1.493 (3) |
| N1—C10 | 1.347 (2) | C12—C11 | 1.368 (3) |
| N1—C14 | 1.354 (2) | C12—C13 | 1.404 (3) |
| N3A—C8A | 1.433 (2) | C12A—C13A | 1.377 (3) |
| N3A—C15A | 1.479 (2) | C4A—C5A | 1.390 (2) |
| N3—C8 | 1.446 (2) | C4—C3 | 1.387 (3) |
| N3—C15 | 1.475 (2) | C13—C14 | 1.357 (2) |
| N2A—C10A | 1.407 (2) | C17—C17i | 1.524 (4) |
| N2A—C9A | 1.362 (2) | C2—C3 | 1.392 (2) |
| C6—C7 | 1.501 (2) | C18—C19 | 1.444 (3) |
| C6—C5 | 1.396 (2) | C18—N4 | 1.135 (3) |
| | | |
| O4—S1—N3 | 108.25 (8) | C1A—C6A—C7A | 120.63 (15) |
| O4—S1—C1 | 109.80 (8) | C5A—C6A—C7A | 121.38 (16) |
| O3—S1—O4 | 118.80 (8) | C5A—C6A—C1A | 117.99 (15) |
| O3—S1—N3 | 108.52 (8) | O2A—C7A—C6A | 115.32 (14) |
| O3—S1—C1 | 108.08 (8) | O2A—C7A—C8A | 123.16 (15) |
| N3—S1—C1 | 102.09 (8) | C8A—C7A—C6A | 121.50 (15) |
| O3A—S1A—N3A | 107.47 (8) | N3A—C8A—C9A | 118.19 (14) |
| O3A—S1A—C1A | 108.85 (8) | C7A—C8A—N3A | 120.38 (14) |
| O4A—S1A—O3A | 120.15 (8) | C7A—C8A—C9A | 121.42 (15) |
| O4A—S1A—N3A | 108.97 (8) | N1A—C14A—C13A | 123.07 (17) |
| O4A—S1A—C1A | 109.22 (8) | O1—C9—N2 | 120.79 (16) |
| N3A—S1A—C1A | 100.36 (8) | O1—C9—C8 | 124.47 (15) |
| C10A—N1A—C14A | 118.27 (15) | N2—C9—C8 | 114.73 (14) |
| C10—N2—C9 | 126.60 (15) | C4A—C3A—C2A | 120.16 (16) |
| C10—N1—C14 | 122.92 (15) | C4—C5—C6 | 120.97 (16) |
| C8A—N3A—S1A | 112.74 (11) | C12A—C11A—C10A | 118.00 (16) |
| C8A—N3A—C15A | 114.87 (14) | C6A—C1A—S1A | 116.90 (12) |
| C15A—N3A—S1A | 117.03 (11) | C2A—C1A—S1A | 121.03 (14) |
| C8—N3—S1 | 113.83 (11) | C2A—C1A—C6A | 122.07 (16) |
| C8—N3—C15 | 115.29 (14) | C6—C1—S1 | 117.16 (13) |
| C15—N3—S1 | 116.95 (12) | C2—C1—S1 | 120.23 (13) |
| C9A—N2A—C10A | 127.38 (15) | C2—C1—C6 | 122.57 (15) |
| C5—C6—C7 | 119.74 (15) | O5—C16—O6 | 122.55 (17) |
| C5—C6—C1 | 117.19 (16) | O5—C16—C17 | 123.48 (16) |
| C1—C6—C7 | 123.01 (15) | O6—C16—C17 | 113.97 (16) |
| N1A—C10A—N2A | 113.35 (15) | C11—C12—C13 | 120.56 (16) |
| N1A—C10A—C11A | 122.36 (15) | C13A—C12A—C11A | 120.02 (16) |
| C11A—C10A—N2A | 124.29 (16) | C3A—C4A—C5A | 120.92 (17) |
| O2—C7—C6 | 117.07 (14) | C5—C4—C3 | 120.63 (16) |
| O2—C7—C8 | 124.47 (16) | C1A—C2A—C3A | 118.66 (17) |
| C8—C7—C6 | 118.43 (14) | C12—C11—C10 | 119.53 (16) |
| N2—C10—C11 | 121.94 (15) | C14—C13—C12 | 118.47 (17) |
| N1—C10—N2 | 119.85 (15) | N1—C14—C13 | 120.33 (16) |
| N1—C10—C11 | 118.19 (16) | C4A—C5A—C6A | 120.18 (17) |
| O1A—C9A—N2A | 124.25 (15) | C16—C17—C17i | 111.9 (2) |
| O1A—C9A—C8A | 120.33 (15) | C1—C2—C3 | 118.74 (16) |
| N2A—C9A—C8A | 115.40 (15) | C14A—C13A—C12A | 118.27 (16) |
| C7—C8—N3 | 120.94 (15) | C4—C3—C2 | 119.89 (17) |
| C7—C8—C9 | 124.80 (15) | N4—C18—C19 | 179.3 (3) |
| C9—C8—N3 | 114.20 (14) | | |
| Symmetry code: (i) −x+2, −y+2, −z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C11A—H11A···O1A | 0.93 | 2.29 | 2.869 (2) | 120 |
| C15—H15C···O4 | 0.96 | 2.45 | 2.843 (2) | 105 |
| C15A—H15D···O4A | 0.96 | 2.50 | 2.886 (2) | 104 |
| O6—H6···N1A | 0.89 (3) | 1.81 (3) | 2.697 (2) | 176 (3) |
| N2A—H2AB···O5 | 0.86 (2) | 2.08 (2) | 2.927 (2) | 168.8 (19) |
| N2A—H2AB···N3A | 0.86 (2) | 2.33 (2) | 2.7326 (19) | 109.1 (16) |
| N1—H1···O1 | 0.85 (2) | 1.97 (2) | 2.6322 (19) | 134.5 (18) |
| N1—H1···O1ii | 0.85 (2) | 2.22 (2) | 2.8690 (18) | 132.9 (18) |
| N2—H2···O2 | 0.88 (2) | 1.82 (2) | 2.6009 (19) | 147.1 (19) |
| O2A—H2AA···O1A | 0.88 (3) | 1.78 (3) | 2.5823 (17) | 152 (3) |
| Symmetry code: (ii) −x+2, −y+1, −z+1. |
Crystallographic data for PRX–SA and PRX–SA·ACN at 120 K top| Compound | PRX-SA | PRX-SA·ACN |
| Chemical formula in the asymmetric unit | C15H13N3O4S.0.5C4H6O4 | 2C15H13N3O4S·0.5C4H6O4·2C2H3N |
| Formula weight | 390.39 | 762.16 |
| Crystal system | Triclinic | Triclinic |
| Space group | P1 | P1 |
| a (Å) | 7.6779 (9) | 9.156 (3) |
| b (Å) | 8.4177 (9) | 12.7660 (5) |
| c (Å) | 14.3934 (10) | 15.1022 (5) |
| α (°) | 79.528 (8) | 83.030 (3) |
| β (°) | 74.650 (8) | 75.313 (3) |
| γ (°) | 72.820 (10) | 83.972 (3) |
| V (Å3) | 851.61 (16) | 1689.87 (11) |
| Z | 2 | 2 |
| Density (calc) (Mg m-3) | 1.522 | 1.499 |
| Crystal colour | colourless | yellow |
| Radiation type | Cu Kα | Mo Kα |
| Theta range (°) | 5.5180–74.1820 | 4.0350–29.3170 |
| hkl range | -9 < h < 9, -9 < k < 10, -17 < l <17 | -12 < h < 11, -16 < k < 17, -20 < l < 19 |
| Completeness (%) | 99 | 100 |
| Rint (%) | 5.46 | 4.73 |
| Refinement method | Least-squares minimization | Least-squares minimization |
| R[F2 > 2σ(F2 )] wR(F2 ) | R1 = 5.48%, wR2 = 15.38% | R1 = 4.27%, wR2 = 11.07% |
| | |
| Goodness-of-fit | 1.055 | 1.055 |
 O hydrogen bonds, whereas every pair of NI-type molecules is linked to SA via N—H
O hydrogen bonds, whereas every pair of NI-type molecules is linked to SA via N—H O and O—H
O and O—H N hydrogen bonds. Thermogravimetry and differential scanning calorimetry suggest that thermal desolvation of the solvate sample occurs at 148 °C, and is followed by recrystallization, presumably of a multicomponent PRX–SA structure. Vibrational spectra (IR and Raman spectroscopy) of PRX–SA·ACN and PRX–SA are also used to demonstrate the ability of spectroscopic techniques to distinguish between NI- and Z-type PRX molecules in the solid state. Hence, vibrational spectroscopy can be used to distinguish the PRX–SA cocrystal and its ACN solvate.
N hydrogen bonds. Thermogravimetry and differential scanning calorimetry suggest that thermal desolvation of the solvate sample occurs at 148 °C, and is followed by recrystallization, presumably of a multicomponent PRX–SA structure. Vibrational spectra (IR and Raman spectroscopy) of PRX–SA·ACN and PRX–SA are also used to demonstrate the ability of spectroscopic techniques to distinguish between NI- and Z-type PRX molecules in the solid state. Hence, vibrational spectroscopy can be used to distinguish the PRX–SA cocrystal and its ACN solvate.
 journal menu
journal menu











