
The title compounds, (9-fluoro-4
H-chromeno[4,3-
c]isoxazol-3-yl)methanol, C
11H
8FNO
3, (I), and (9-chloro-4
H-chromeno[4,3-
c]isoxazol-3-yl)methanol, C
11H
8ClNO
3, (II), crystallize in the orthorhombic space group
Pbca with
Z′ = 1 and the triclinic space group
P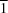
with
Z′ = 6, respectively. The simple replacement of F by Cl in the main molecular scaffold of (I) and (II) results in significant differences in the intermolecular interaction patterns and a corresponding change in the point-group symmetry from
D2h to
Ci =
S2. These striking differences are manifested through the presence of C—H

F and the absence of O—H

O and C—H

O interactions in (I), and the absence of C—H

Cl and the presence of O—H

O and C—H

O interactions in (II). However, the geometry of the synthons formed by the O—H

N and O—H
 X
X (
X = F or Cl) interactions observed in the constitution of the supramolecular networks of both (I) and (II) remains similar. Also, C—H

O interactions are not preferred in the presence of F in (I), while they are much preferred in the presence of Cl in (II).
Supporting information
CCDC references: 848752; 860080
Samples of (I) and (II) were prepared according to the procedure described by
Liaskopoulos et al. (2007), starting from 2-fluoro- or
2-chloro-6-hydroxy-benzaldehyde with 4-chloro-but-2-yn-1-ol in equimolar
amounts. Crystals suitable for single-crystal X-ray diffraction were grown by
slow evaporation of solutions in ethanol.
In both compounds, the H atoms attached to hydroxyl atoms O3 were located in a
difference map and refined freely. All other H atoms were placed geometrically
and treated as riding, with C—H = 0.95–0.99 Å and with Uiso(H) =
1.2Ueq(C). [Please check added text]
For both compounds, data collection: APEX2 (Bruker, 2009); cell refinement: SAINT (Bruker, 2009); data reduction: SAINT (Bruker, 2009); program(s) used to solve structure: SHELXTL (Sheldrick, 2008); program(s) used to refine structure: SHELXTL (Sheldrick, 2008); molecular graphics: SHELXTL (Sheldrick, 2008); software used to prepare material for publication: SHELXTL (Sheldrick, 2008) and PLATON (Spek, 2009).
(I) (9-Fluoro-4
H-chromeno[4,3-
c]isoxazol-3-yl)methanol
top
Crystal data top
| C11H8FNO3 | F(000) = 912 |
| Mr = 221.18 | Dx = 1.618 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 3017 reflections |
| a = 14.1516 (2) Å | θ = 1.0–1.0° |
| b = 7.0064 (1) Å | µ = 0.13 mm−1 |
| c = 18.3134 (2) Å | T = 100 K |
| V = 1815.81 (4) Å3 | Block, colourless |
| Z = 8 | 0.35 × 0.10 × 0.07 mm |
Data collection top
Bruker SMART APEXII CCD area-detector
diffractometer | 3017 independent reflections |
| Radiation source: fine-focus sealed tube | 2279 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.040 |
| ϕ and ω scans | θmax = 31.5°, θmin = 2.2° |
Absorption correction: multi-scan
(SADABS; Bruker, 2009) | h = −20→20 |
| Tmin = 0.955, Tmax = 0.991 | k = −10→10 |
| 18961 measured reflections | l = −26→26 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.044 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.123 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0673P)2 + 0.3939P]
where P = (Fo2 + 2Fc2)/3 |
| 3017 reflections | (Δ/σ)max = 0.001 |
| 149 parameters | Δρmax = 0.54 e Å−3 |
| 0 restraints | Δρmin = −0.24 e Å−3 |
Crystal data top
| C11H8FNO3 | V = 1815.81 (4) Å3 |
| Mr = 221.18 | Z = 8 |
| Orthorhombic, Pbca | Mo Kα radiation |
| a = 14.1516 (2) Å | µ = 0.13 mm−1 |
| b = 7.0064 (1) Å | T = 100 K |
| c = 18.3134 (2) Å | 0.35 × 0.10 × 0.07 mm |
Data collection top
Bruker SMART APEXII CCD area-detector
diffractometer | 3017 independent reflections |
Absorption correction: multi-scan
(SADABS; Bruker, 2009) | 2279 reflections with I > 2σ(I) |
| Tmin = 0.955, Tmax = 0.991 | Rint = 0.040 |
| 18961 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.044 | 0 restraints |
| wR(F2) = 0.123 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | Δρmax = 0.54 e Å−3 |
| 3017 reflections | Δρmin = −0.24 e Å−3 |
| 149 parameters | |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| F1 | 0.41065 (5) | 0.05586 (12) | 0.59370 (4) | 0.01837 (18) | |
| O1 | 0.08873 (6) | 0.14786 (14) | 0.52350 (5) | 0.0155 (2) | |
| O2 | 0.19614 (6) | 0.10219 (14) | 0.75469 (5) | 0.0183 (2) | |
| O3 | −0.05528 (7) | 0.08591 (17) | 0.73037 (5) | 0.0249 (2) | |
| N1 | 0.26517 (7) | 0.10383 (17) | 0.69879 (6) | 0.0169 (2) | |
| C1 | 0.21639 (8) | 0.08882 (17) | 0.63780 (6) | 0.0127 (2) | |
| C2 | 0.25101 (8) | 0.09622 (18) | 0.56288 (6) | 0.0123 (2) | |
| C3 | 0.34492 (8) | 0.08806 (18) | 0.54103 (6) | 0.0134 (2) | |
| C4 | 0.37431 (9) | 0.11263 (19) | 0.47017 (7) | 0.0158 (2) | |
| H4A | 0.4393 | 0.1052 | 0.4575 | 0.019* | |
| C5 | 0.30535 (9) | 0.14883 (19) | 0.41753 (7) | 0.0169 (3) | |
| H5A | 0.3238 | 0.1702 | 0.3683 | 0.020* | |
| C6 | 0.21028 (9) | 0.15418 (19) | 0.43580 (6) | 0.0159 (2) | |
| H6A | 0.1640 | 0.1759 | 0.3991 | 0.019* | |
| C7 | 0.18288 (8) | 0.12752 (18) | 0.50808 (6) | 0.0132 (2) | |
| C8 | 0.05216 (8) | 0.05084 (19) | 0.58765 (6) | 0.0148 (2) | |
| H8A | 0.0450 | −0.0870 | 0.5772 | 0.018* | |
| H8B | −0.0108 | 0.1028 | 0.6002 | 0.018* | |
| C9 | 0.11769 (8) | 0.07771 (18) | 0.65028 (6) | 0.0132 (2) | |
| C10 | 0.10894 (8) | 0.08776 (19) | 0.72365 (6) | 0.0151 (2) | |
| C11 | 0.02658 (9) | 0.0855 (2) | 0.77433 (7) | 0.0188 (3) | |
| H11A | 0.0282 | −0.0301 | 0.8054 | 0.023* | |
| H11B | 0.0276 | 0.1994 | 0.8063 | 0.023* | |
| H3 | −0.0993 (15) | 0.095 (3) | 0.7581 (12) | 0.040 (6)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| F1 | 0.0095 (3) | 0.0282 (5) | 0.0175 (3) | 0.0019 (3) | −0.0015 (3) | 0.0013 (3) |
| O1 | 0.0094 (4) | 0.0243 (5) | 0.0128 (4) | 0.0016 (3) | −0.0005 (3) | 0.0027 (3) |
| O2 | 0.0100 (4) | 0.0326 (6) | 0.0122 (4) | −0.0010 (3) | 0.0004 (3) | 0.0006 (4) |
| O3 | 0.0099 (4) | 0.0485 (7) | 0.0161 (4) | 0.0013 (4) | 0.0013 (3) | 0.0009 (4) |
| N1 | 0.0102 (4) | 0.0277 (6) | 0.0128 (4) | −0.0009 (4) | 0.0011 (3) | 0.0007 (4) |
| C1 | 0.0091 (5) | 0.0155 (6) | 0.0134 (5) | 0.0006 (4) | −0.0005 (4) | 0.0004 (4) |
| C2 | 0.0106 (5) | 0.0140 (5) | 0.0124 (5) | −0.0001 (4) | 0.0000 (4) | 0.0005 (4) |
| C3 | 0.0094 (5) | 0.0155 (6) | 0.0155 (5) | 0.0002 (4) | −0.0014 (4) | 0.0004 (4) |
| C4 | 0.0120 (5) | 0.0176 (6) | 0.0178 (5) | −0.0001 (4) | 0.0029 (4) | 0.0000 (4) |
| C5 | 0.0171 (6) | 0.0192 (6) | 0.0144 (5) | −0.0002 (5) | 0.0030 (4) | 0.0008 (4) |
| C6 | 0.0157 (5) | 0.0186 (6) | 0.0134 (5) | 0.0010 (5) | 0.0000 (4) | 0.0007 (4) |
| C7 | 0.0108 (5) | 0.0153 (6) | 0.0136 (5) | 0.0008 (4) | −0.0007 (4) | −0.0006 (4) |
| C8 | 0.0106 (5) | 0.0206 (6) | 0.0132 (5) | −0.0010 (4) | 0.0000 (4) | 0.0016 (4) |
| C9 | 0.0089 (5) | 0.0174 (6) | 0.0134 (5) | 0.0002 (4) | −0.0005 (4) | 0.0010 (4) |
| C10 | 0.0094 (5) | 0.0215 (6) | 0.0146 (5) | 0.0006 (4) | −0.0005 (4) | 0.0002 (5) |
| C11 | 0.0117 (5) | 0.0307 (8) | 0.0141 (5) | 0.0009 (5) | 0.0023 (4) | 0.0006 (5) |
Geometric parameters (Å, º) top
| F1—C3 | 1.3589 (13) | C4—C5 | 1.3949 (17) |
| O1—C7 | 1.3693 (13) | C4—H4A | 0.9500 |
| O1—C8 | 1.4526 (14) | C5—C6 | 1.3870 (17) |
| O2—C10 | 1.3624 (14) | C5—H5A | 0.9500 |
| O2—N1 | 1.4151 (13) | C6—C7 | 1.3919 (16) |
| O3—C11 | 1.4107 (15) | C6—H6A | 0.9500 |
| O3—H3 | 0.81 (2) | C8—C9 | 1.4870 (16) |
| N1—C1 | 1.3172 (15) | C8—H8A | 0.9900 |
| C1—C9 | 1.4175 (16) | C8—H8B | 0.9900 |
| C1—C2 | 1.4577 (16) | C9—C10 | 1.3512 (16) |
| C2—C3 | 1.3891 (16) | C10—C11 | 1.4901 (17) |
| C2—C7 | 1.4089 (16) | C11—H11A | 0.9900 |
| C3—C4 | 1.3736 (16) | C11—H11B | 0.9900 |
| | | |
| C7—O1—C8 | 117.70 (9) | O1—C7—C6 | 116.95 (10) |
| C10—O2—N1 | 108.91 (9) | O1—C7—C2 | 122.36 (10) |
| C11—O3—H3 | 105.9 (15) | C6—C7—C2 | 120.51 (11) |
| C1—N1—O2 | 104.54 (9) | O1—C8—C9 | 110.02 (9) |
| N1—C1—C9 | 112.59 (10) | O1—C8—H8A | 109.7 |
| N1—C1—C2 | 128.25 (11) | C9—C8—H8A | 109.7 |
| C9—C1—C2 | 119.01 (10) | O1—C8—H8B | 109.7 |
| C3—C2—C7 | 117.13 (10) | C9—C8—H8B | 109.7 |
| C3—C2—C1 | 126.25 (11) | H8A—C8—H8B | 108.2 |
| C7—C2—C1 | 116.48 (10) | C10—C9—C1 | 104.35 (10) |
| F1—C3—C4 | 118.96 (10) | C10—C9—C8 | 135.76 (11) |
| F1—C3—C2 | 117.21 (10) | C1—C9—C8 | 119.81 (10) |
| C4—C3—C2 | 123.83 (11) | C9—C10—O2 | 109.61 (10) |
| C3—C4—C5 | 117.64 (11) | C9—C10—C11 | 133.68 (11) |
| C3—C4—H4A | 121.2 | O2—C10—C11 | 116.70 (10) |
| C5—C4—H4A | 121.2 | O3—C11—C10 | 106.66 (10) |
| C6—C5—C4 | 121.12 (11) | O3—C11—H11A | 110.4 |
| C6—C5—H5A | 119.4 | C10—C11—H11A | 110.4 |
| C4—C5—H5A | 119.4 | O3—C11—H11B | 110.4 |
| C5—C6—C7 | 119.74 (11) | C10—C11—H11B | 110.4 |
| C5—C6—H6A | 120.1 | H11A—C11—H11B | 108.6 |
| C7—C6—H6A | 120.1 | | |
| | | |
| C10—O2—N1—C1 | −0.52 (13) | C3—C2—C7—O1 | −176.63 (11) |
| O2—N1—C1—C9 | 0.16 (14) | C1—C2—C7—O1 | −0.77 (18) |
| O2—N1—C1—C2 | 175.44 (12) | C3—C2—C7—C6 | −1.77 (18) |
| N1—C1—C2—C3 | 11.9 (2) | C1—C2—C7—C6 | 174.09 (12) |
| C9—C1—C2—C3 | −173.10 (12) | C7—O1—C8—C9 | 44.47 (14) |
| N1—C1—C2—C7 | −163.55 (13) | N1—C1—C9—C10 | 0.24 (15) |
| C9—C1—C2—C7 | 11.46 (17) | C2—C1—C9—C10 | −175.52 (11) |
| C7—C2—C3—F1 | −179.32 (11) | N1—C1—C9—C8 | −177.07 (11) |
| C1—C2—C3—F1 | 5.27 (19) | C2—C1—C9—C8 | 7.17 (17) |
| C7—C2—C3—C4 | 1.43 (19) | O1—C8—C9—C10 | 149.93 (15) |
| C1—C2—C3—C4 | −173.97 (12) | O1—C8—C9—C1 | −33.80 (16) |
| F1—C3—C4—C5 | −178.88 (11) | C1—C9—C10—O2 | −0.57 (14) |
| C2—C3—C4—C5 | 0.35 (19) | C8—C9—C10—O2 | 176.10 (13) |
| C3—C4—C5—C6 | −1.86 (19) | C1—C9—C10—C11 | 179.68 (14) |
| C4—C5—C6—C7 | 1.5 (2) | C8—C9—C10—C11 | −3.7 (3) |
| C8—O1—C7—C6 | 155.91 (11) | N1—O2—C10—C9 | 0.69 (14) |
| C8—O1—C7—C2 | −29.06 (17) | N1—O2—C10—C11 | −179.51 (11) |
| C5—C6—C7—O1 | 175.49 (12) | C9—C10—C11—O3 | −5.0 (2) |
| C5—C6—C7—C2 | 0.36 (19) | O2—C10—C11—O3 | 175.24 (11) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3···N1i | 0.81 (2) | 2.07 (2) | 2.8557 (14) | 163 (2) |
| O3—H3···F1i | 0.81 (2) | 2.73 (2) | 3.2644 (12) | 125.3 (18) |
| C8—H8A···F1ii | 0.99 | 2.60 | 3.5095 (16) | 153 |
| C4—H4A···F1iii | 0.95 | 2.58 | 3.4674 (14) | 155 |
| Symmetry codes: (i) x−1/2, y, −z+3/2; (ii) −x+1/2, y−1/2, z; (iii) −x+1, −y, −z+1. |
(II) (9-Chloro-4
H-chromeno[4,3-
c]isoxazol-3-yl)methanol
top
Crystal data top
| C11H8ClNO3 | Z = 12 |
| Mr = 237.63 | F(000) = 1464 |
| Triclinic, P1 | Dx = 1.608 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.4306 (1) Å | Cell parameters from 17629 reflections |
| b = 14.8892 (2) Å | θ = 2–32.7° |
| c = 21.4949 (2) Å | µ = 0.38 mm−1 |
| α = 98.764 (1)° | T = 100 K |
| β = 97.610 (1)° | Block, colourless |
| γ = 94.013 (1)° | 0.32 × 0.32 × 0.17 mm |
| V = 2944.12 (6) Å3 | |
Data collection top
Bruker SMART APEXII CCD area-detector
diffractometer | 21416 independent reflections |
| Radiation source: fine-focus sealed tube | 17626 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.023 |
| ϕ and ω scans | θmax = 32.7°, θmin = 1.0° |
Absorption correction: multi-scan
(SADABS; Bruker, 2009) | h = −14→11 |
| Tmin = 0.890, Tmax = 0.937 | k = −22→22 |
| 69911 measured reflections | l = −32→32 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.042 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.107 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0441P)2 + 1.7339P]
where P = (Fo2 + 2Fc2)/3 |
| 21416 reflections | (Δ/σ)max = 0.002 |
| 900 parameters | Δρmax = 0.66 e Å−3 |
| 0 restraints | Δρmin = −0.39 e Å−3 |
Crystal data top
| C11H8ClNO3 | γ = 94.013 (1)° |
| Mr = 237.63 | V = 2944.12 (6) Å3 |
| Triclinic, P1 | Z = 12 |
| a = 9.4306 (1) Å | Mo Kα radiation |
| b = 14.8892 (2) Å | µ = 0.38 mm−1 |
| c = 21.4949 (2) Å | T = 100 K |
| α = 98.764 (1)° | 0.32 × 0.32 × 0.17 mm |
| β = 97.610 (1)° | |
Data collection top
Bruker SMART APEXII CCD area-detector
diffractometer | 21416 independent reflections |
Absorption correction: multi-scan
(SADABS; Bruker, 2009) | 17626 reflections with I > 2σ(I) |
| Tmin = 0.890, Tmax = 0.937 | Rint = 0.023 |
| 69911 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.042 | 0 restraints |
| wR(F2) = 0.107 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | Δρmax = 0.66 e Å−3 |
| 21416 reflections | Δρmin = −0.39 e Å−3 |
| 900 parameters | |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and
goodness of fit S are based on F2, conventional R-factors R are based
on F, with F set to zero for negative F2. The threshold expression of
F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is
not relevant to the choice of reflections for refinement. R-factors based
on F2 are statistically about twice as large as those based on F, and R-
factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Cl1A | −0.14998 (3) | −0.02223 (2) | 0.341046 (16) | 0.01868 (7) | |
| O1A | 0.3263 (3) | −0.04319 (16) | 0.48527 (17) | 0.0180 (6) | 0.812 (14) |
| O1X | 0.3545 (9) | −0.0222 (8) | 0.4646 (6) | 0.015 (3) | 0.188 (14) |
| O2A | 0.03088 (10) | 0.18351 (6) | 0.50824 (4) | 0.01526 (17) | |
| O3A | 0.36968 (11) | 0.22168 (7) | 0.61306 (5) | 0.0195 (2) | |
| H3OA | 0.379 (2) | 0.2259 (15) | 0.6521 (11) | 0.044 (6)* | |
| N1A | −0.01383 (12) | 0.11003 (7) | 0.45750 (5) | 0.0156 (2) | |
| C1A | 0.09396 (13) | 0.05807 (8) | 0.45789 (6) | 0.0122 (2) | |
| C2A | 0.10778 (13) | −0.02564 (8) | 0.41479 (6) | 0.0125 (2) | |
| C3A | 0.01191 (14) | −0.06649 (9) | 0.36080 (6) | 0.0141 (2) | |
| C4A | 0.04135 (15) | −0.14352 (9) | 0.32123 (6) | 0.0172 (2) | |
| H4A | −0.0251 | −0.1694 | 0.2847 | 0.021* | |
| C5A | 0.16873 (15) | −0.18245 (9) | 0.33552 (7) | 0.0185 (3) | |
| H5A | 0.1900 | −0.2347 | 0.3083 | 0.022* | |
| C6A | 0.26491 (15) | −0.14568 (9) | 0.38919 (7) | 0.0194 (3) | |
| H6A | 0.3514 | −0.1730 | 0.3991 | 0.023* | |
| C7A | 0.23422 (14) | −0.06836 (9) | 0.42877 (7) | 0.0169 (2) | |
| C8A | 0.34386 (15) | 0.04933 (10) | 0.51591 (7) | 0.0205 (3) | |
| H8A1 | 0.4188 | 0.0833 | 0.4982 | 0.025* | 0.81 (3) |
| H8A2 | 0.3767 | 0.0519 | 0.5619 | 0.025* | 0.81 (3) |
| H8X1 | 0.4259 | 0.0961 | 0.5197 | 0.025* | 0.19 (3) |
| H8X2 | 0.3509 | 0.0240 | 0.5561 | 0.025* | 0.19 (3) |
| C9A | 0.20827 (13) | 0.09350 (8) | 0.50748 (6) | 0.0126 (2) | |
| C10A | 0.16384 (13) | 0.17117 (8) | 0.53679 (6) | 0.0129 (2) | |
| C11A | 0.23024 (14) | 0.24317 (9) | 0.59090 (6) | 0.0159 (2) | |
| H11A | 0.2357 | 0.3033 | 0.5767 | 0.019* | |
| H11B | 0.1707 | 0.2463 | 0.6257 | 0.019* | |
| Cl1B | 0.18312 (3) | −0.00439 (2) | 0.007443 (15) | 0.01776 (6) | |
| O1B | 0.65963 (10) | −0.04228 (6) | 0.14643 (5) | 0.01632 (18) | |
| O2B | 0.36463 (10) | 0.18042 (6) | 0.18437 (4) | 0.01489 (17) | |
| O3B | 0.70827 (11) | 0.21754 (7) | 0.28708 (5) | 0.0205 (2) | |
| H3OB | 0.701 (2) | 0.1947 (15) | 0.3188 (11) | 0.045 (7)* | |
| N1B | 0.31605 (12) | 0.10962 (7) | 0.13235 (5) | 0.0146 (2) | |
| C1B | 0.42644 (13) | 0.06067 (8) | 0.12689 (6) | 0.0117 (2) | |
| C2B | 0.43645 (13) | −0.02100 (8) | 0.08103 (6) | 0.0124 (2) | |
| C3B | 0.33814 (14) | −0.05676 (9) | 0.02649 (6) | 0.0140 (2) | |
| C4B | 0.36119 (15) | −0.13382 (9) | −0.01452 (6) | 0.0175 (2) | |
| H4B | 0.2927 | −0.1570 | −0.0510 | 0.021* | |
| C5B | 0.48529 (16) | −0.17698 (9) | −0.00192 (7) | 0.0185 (3) | |
| H5B | 0.5022 | −0.2292 | −0.0303 | 0.022* | |
| C6B | 0.58482 (15) | −0.14450 (9) | 0.05179 (7) | 0.0175 (2) | |
| H6B | 0.6691 | −0.1746 | 0.0604 | 0.021* | |
| C7B | 0.55993 (14) | −0.06715 (8) | 0.09308 (6) | 0.0135 (2) | |
| C8B | 0.68482 (14) | 0.05323 (9) | 0.17478 (6) | 0.0151 (2) | |
| H8B1 | 0.7491 | 0.0856 | 0.1509 | 0.018* | |
| H8B2 | 0.7326 | 0.0589 | 0.2193 | 0.018* | |
| C9B | 0.54673 (13) | 0.09544 (8) | 0.17372 (6) | 0.0126 (2) | |
| C10B | 0.50235 (13) | 0.16982 (8) | 0.20769 (6) | 0.0133 (2) | |
| C11B | 0.57240 (15) | 0.24080 (9) | 0.26190 (6) | 0.0168 (2) | |
| H11C | 0.5837 | 0.3003 | 0.2471 | 0.020* | |
| H11D | 0.5102 | 0.2472 | 0.2957 | 0.020* | |
| Cl1C | 0.09523 (4) | 0.37951 (2) | 0.455536 (18) | 0.02131 (7) | |
| O1C | 0.58055 (10) | 0.52058 (6) | 0.41069 (5) | 0.01714 (18) | |
| O2C | 0.49604 (10) | 0.24970 (6) | 0.47468 (5) | 0.01595 (18) | |
| O3C | 0.71868 (12) | 0.16202 (7) | 0.40291 (5) | 0.0190 (2) | |
| H3OC | 0.787 (2) | 0.1352 (15) | 0.4106 (11) | 0.041 (6)* | |
| N1C | 0.37986 (12) | 0.30420 (7) | 0.47093 (5) | 0.0151 (2) | |
| C1C | 0.42434 (14) | 0.37225 (8) | 0.44320 (6) | 0.0131 (2) | |
| C2C | 0.35011 (14) | 0.45159 (8) | 0.42988 (6) | 0.0130 (2) | |
| C3C | 0.20640 (14) | 0.46471 (9) | 0.43480 (6) | 0.0152 (2) | |
| C4C | 0.14740 (15) | 0.54412 (9) | 0.42267 (7) | 0.0186 (3) | |
| H4C | 0.0494 | 0.5516 | 0.4262 | 0.022* | |
| C5C | 0.23469 (16) | 0.61264 (9) | 0.40524 (7) | 0.0191 (3) | |
| H5C | 0.1951 | 0.6672 | 0.3967 | 0.023* | |
| C6C | 0.37812 (16) | 0.60314 (9) | 0.40009 (6) | 0.0170 (2) | |
| H6C | 0.4363 | 0.6507 | 0.3884 | 0.020* | |
| C7C | 0.43554 (14) | 0.52288 (9) | 0.41238 (6) | 0.0145 (2) | |
| C8C | 0.63505 (15) | 0.43324 (9) | 0.39545 (7) | 0.0185 (3) | |
| H8C1 | 0.7404 | 0.4391 | 0.4086 | 0.022* | |
| H8C2 | 0.6156 | 0.4125 | 0.3489 | 0.022* | |
| C9C | 0.56617 (14) | 0.36461 (9) | 0.42847 (6) | 0.0139 (2) | |
| C10C | 0.60563 (14) | 0.28788 (9) | 0.44945 (6) | 0.0151 (2) | |
| C11C | 0.73922 (15) | 0.24061 (9) | 0.45123 (7) | 0.0183 (2) | |
| H11E | 0.8198 | 0.2824 | 0.4443 | 0.022* | |
| H11F | 0.7635 | 0.2222 | 0.4934 | 0.022* | |
| Cl1D | 0.43836 (4) | 0.39893 (2) | 0.147509 (19) | 0.02281 (7) | |
| O1D | 0.87174 (11) | 0.50326 (7) | 0.04379 (5) | 0.0215 (2) | |
| O2D | 0.83028 (10) | 0.25964 (6) | 0.14473 (5) | 0.01633 (18) | |
| O3D | 1.06485 (12) | 0.18025 (7) | 0.08209 (5) | 0.0193 (2) | |
| H3OD | 1.133 (2) | 0.1533 (15) | 0.0903 (11) | 0.042 (6)* | |
| N1D | 0.70895 (12) | 0.30949 (7) | 0.13738 (5) | 0.0150 (2) | |
| C1D | 0.75425 (13) | 0.38151 (8) | 0.11399 (6) | 0.0128 (2) | |
| C2D | 0.67267 (14) | 0.45686 (8) | 0.09742 (6) | 0.0135 (2) | |
| C3D | 0.53465 (14) | 0.47392 (9) | 0.11069 (6) | 0.0161 (2) | |
| C4D | 0.46861 (16) | 0.54864 (10) | 0.09360 (7) | 0.0205 (3) | |
| H4D | 0.3748 | 0.5588 | 0.1029 | 0.025* | |
| C5D | 0.54207 (17) | 0.60843 (10) | 0.06259 (7) | 0.0223 (3) | |
| H5D | 0.4979 | 0.6601 | 0.0510 | 0.027* | |
| C6D | 0.67863 (17) | 0.59400 (10) | 0.04824 (7) | 0.0211 (3) | |
| H6D | 0.7277 | 0.6355 | 0.0272 | 0.025* | |
| C7D | 0.74285 (15) | 0.51833 (9) | 0.06497 (6) | 0.0169 (2) | |
| C8D | 0.97477 (15) | 0.45798 (10) | 0.08094 (7) | 0.0183 (2) | |
| H8D1 | 1.0261 | 0.5022 | 0.1174 | 0.022* | |
| H8D2 | 1.0464 | 0.4344 | 0.0544 | 0.022* | |
| C9D | 0.90095 (14) | 0.38096 (9) | 0.10479 (6) | 0.0144 (2) | |
| C10D | 0.94288 (14) | 0.30379 (9) | 0.12469 (6) | 0.0155 (2) | |
| C11D | 1.07974 (15) | 0.25993 (10) | 0.12906 (7) | 0.0197 (3) | |
| H11G | 1.1589 | 0.3029 | 0.1222 | 0.024* | |
| H11H | 1.1032 | 0.2434 | 0.1719 | 0.024* | |
| Cl1E | 1.25397 (4) | 0.59857 (2) | 0.228807 (17) | 0.01890 (7) | |
| O1E | 0.74285 (10) | 0.48379 (6) | 0.26095 (5) | 0.01528 (17) | |
| O2E | 0.86544 (10) | 0.73553 (6) | 0.18238 (5) | 0.01656 (18) | |
| O3E | 0.54653 (11) | 0.75356 (7) | 0.25506 (5) | 0.01846 (19) | |
| H3OE | 0.480 (2) | 0.7836 (14) | 0.2481 (10) | 0.030 (5)* | |
| N1E | 0.97307 (13) | 0.67528 (8) | 0.19125 (6) | 0.0162 (2) | |
| C1E | 0.91909 (14) | 0.61778 (8) | 0.22482 (6) | 0.0131 (2) | |
| C2E | 0.98418 (14) | 0.53930 (8) | 0.24620 (6) | 0.0128 (2) | |
| C3E | 1.12800 (14) | 0.52064 (9) | 0.24838 (6) | 0.0145 (2) | |
| C4E | 1.17572 (15) | 0.44091 (9) | 0.26623 (6) | 0.0168 (2) | |
| H4E | 1.2738 | 0.4295 | 0.2672 | 0.020* | |
| C5E | 1.07635 (16) | 0.37797 (9) | 0.28267 (6) | 0.0183 (3) | |
| H5E | 1.1079 | 0.3230 | 0.2946 | 0.022* | |
| C6E | 0.93318 (15) | 0.39328 (9) | 0.28216 (6) | 0.0153 (2) | |
| H6E | 0.8673 | 0.3497 | 0.2937 | 0.018* | |
| C7E | 0.88751 (14) | 0.47437 (8) | 0.26431 (6) | 0.0136 (2) | |
| C8E | 0.69849 (15) | 0.57529 (9) | 0.27309 (6) | 0.0162 (2) | |
| H8E1 | 0.7184 | 0.5992 | 0.3193 | 0.019* | |
| H8E2 | 0.5939 | 0.5742 | 0.2592 | 0.019* | |
| C9E | 0.77880 (14) | 0.63571 (8) | 0.23763 (6) | 0.0133 (2) | |
| C10E | 0.75162 (14) | 0.71038 (9) | 0.21058 (6) | 0.0145 (2) | |
| C11E | 0.63294 (15) | 0.77103 (10) | 0.20794 (7) | 0.0183 (2) | |
| H11I | 0.6726 | 0.8358 | 0.2163 | 0.022* | |
| H11J | 0.5745 | 0.7585 | 0.1653 | 0.022* | |
| Cl1F | 0.50816 (3) | 0.97439 (2) | 0.304531 (16) | 0.01865 (7) | |
| O1F | −0.00859 (10) | 1.00507 (7) | 0.20275 (5) | 0.01909 (19) | |
| O2F | 0.26715 (10) | 0.77997 (6) | 0.14538 (5) | 0.01671 (18) | |
| O3F | −0.06379 (12) | 0.77588 (8) | 0.03786 (5) | 0.0230 (2) | |
| H3OF | −0.058 (2) | 0.7931 (15) | 0.0061 (11) | 0.040 (6)* | |
| N1F | 0.32382 (12) | 0.84505 (8) | 0.19971 (5) | 0.0155 (2) | |
| C1F | 0.23417 (13) | 0.90927 (9) | 0.20075 (6) | 0.0127 (2) | |
| C2F | 0.23864 (13) | 0.99192 (8) | 0.24764 (6) | 0.0127 (2) | |
| C3F | 0.35112 (14) | 1.02804 (9) | 0.29641 (6) | 0.0147 (2) | |
| C4F | 0.34082 (15) | 1.10536 (9) | 0.33993 (6) | 0.0174 (2) | |
| H4F | 0.4186 | 1.1282 | 0.3726 | 0.021* | |
| C5F | 0.21503 (15) | 1.14915 (9) | 0.33524 (7) | 0.0182 (2) | |
| H5F | 0.2069 | 1.2020 | 0.3650 | 0.022* | |
| C6F | 0.10138 (15) | 1.11628 (9) | 0.28742 (7) | 0.0174 (2) | |
| H6F | 0.0163 | 1.1469 | 0.2841 | 0.021* | |
| C7F | 0.11265 (14) | 1.03824 (9) | 0.24435 (6) | 0.0144 (2) | |
| C8F | 0.00595 (14) | 0.95296 (9) | 0.14176 (6) | 0.0168 (2) | |
| H8F1 | 0.0319 | 0.9949 | 0.1127 | 0.020* | |
| H8F2 | −0.0869 | 0.9181 | 0.1227 | 0.020* | |
| C9F | 0.11870 (13) | 0.88901 (9) | 0.14956 (6) | 0.0128 (2) | |
| C10F | 0.14468 (14) | 0.80791 (9) | 0.11709 (6) | 0.0147 (2) | |
| C11F | 0.06913 (15) | 0.74491 (10) | 0.05961 (6) | 0.0187 (3) | |
| H11K | 0.1303 | 0.7412 | 0.0254 | 0.022* | |
| H11L | 0.0526 | 0.6830 | 0.0702 | 0.022* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cl1A | 0.01398 (14) | 0.02265 (15) | 0.01694 (14) | 0.00393 (11) | −0.00300 (11) | −0.00105 (11) |
| O1A | 0.0146 (7) | 0.0152 (7) | 0.0227 (11) | 0.0045 (6) | −0.0027 (7) | 0.0016 (7) |
| O1X | 0.011 (3) | 0.017 (3) | 0.015 (4) | 0.003 (2) | −0.001 (2) | −0.002 (3) |
| O2A | 0.0139 (4) | 0.0142 (4) | 0.0160 (4) | 0.0039 (3) | −0.0012 (3) | −0.0014 (3) |
| O3A | 0.0148 (5) | 0.0270 (5) | 0.0147 (5) | 0.0010 (4) | −0.0016 (4) | 0.0009 (4) |
| N1A | 0.0147 (5) | 0.0128 (5) | 0.0169 (5) | 0.0027 (4) | −0.0018 (4) | −0.0021 (4) |
| C1A | 0.0115 (5) | 0.0122 (5) | 0.0135 (5) | 0.0018 (4) | 0.0020 (4) | 0.0031 (4) |
| C2A | 0.0113 (5) | 0.0121 (5) | 0.0144 (5) | 0.0008 (4) | 0.0020 (4) | 0.0032 (4) |
| C3A | 0.0151 (6) | 0.0145 (5) | 0.0129 (5) | 0.0020 (4) | 0.0024 (4) | 0.0029 (4) |
| C4A | 0.0205 (6) | 0.0155 (6) | 0.0148 (6) | 0.0001 (5) | 0.0025 (5) | 0.0011 (4) |
| C5A | 0.0227 (7) | 0.0128 (5) | 0.0213 (6) | 0.0016 (5) | 0.0087 (5) | 0.0021 (5) |
| C6A | 0.0172 (6) | 0.0142 (6) | 0.0268 (7) | 0.0040 (5) | 0.0046 (5) | 0.0010 (5) |
| C7A | 0.0131 (6) | 0.0147 (5) | 0.0217 (6) | 0.0025 (4) | 0.0002 (5) | 0.0001 (5) |
| C8A | 0.0163 (6) | 0.0206 (6) | 0.0214 (6) | 0.0072 (5) | −0.0036 (5) | −0.0035 (5) |
| C9A | 0.0110 (5) | 0.0139 (5) | 0.0128 (5) | 0.0007 (4) | 0.0002 (4) | 0.0032 (4) |
| C10A | 0.0115 (5) | 0.0142 (5) | 0.0128 (5) | 0.0006 (4) | 0.0003 (4) | 0.0034 (4) |
| C11A | 0.0163 (6) | 0.0157 (5) | 0.0143 (5) | −0.0004 (5) | 0.0013 (4) | −0.0001 (4) |
| Cl1B | 0.01400 (14) | 0.02275 (15) | 0.01492 (13) | 0.00193 (11) | −0.00224 (10) | 0.00164 (11) |
| O1B | 0.0149 (4) | 0.0133 (4) | 0.0194 (5) | 0.0030 (3) | −0.0027 (3) | 0.0020 (3) |
| O2B | 0.0144 (4) | 0.0147 (4) | 0.0142 (4) | 0.0037 (3) | −0.0006 (3) | −0.0007 (3) |
| O3B | 0.0179 (5) | 0.0242 (5) | 0.0177 (5) | −0.0032 (4) | −0.0035 (4) | 0.0061 (4) |
| N1B | 0.0137 (5) | 0.0146 (5) | 0.0139 (5) | 0.0020 (4) | −0.0008 (4) | −0.0007 (4) |
| C1B | 0.0108 (5) | 0.0124 (5) | 0.0120 (5) | 0.0013 (4) | 0.0009 (4) | 0.0032 (4) |
| C2B | 0.0117 (5) | 0.0126 (5) | 0.0130 (5) | 0.0004 (4) | 0.0021 (4) | 0.0029 (4) |
| C3B | 0.0134 (5) | 0.0157 (5) | 0.0128 (5) | 0.0002 (4) | 0.0010 (4) | 0.0035 (4) |
| C4B | 0.0220 (6) | 0.0156 (6) | 0.0136 (6) | −0.0024 (5) | 0.0021 (5) | 0.0007 (4) |
| C5B | 0.0248 (7) | 0.0122 (5) | 0.0189 (6) | 0.0011 (5) | 0.0064 (5) | 0.0010 (5) |
| C6B | 0.0198 (6) | 0.0137 (5) | 0.0207 (6) | 0.0042 (5) | 0.0059 (5) | 0.0041 (5) |
| C7B | 0.0128 (5) | 0.0132 (5) | 0.0149 (5) | 0.0010 (4) | 0.0024 (4) | 0.0031 (4) |
| C8B | 0.0119 (5) | 0.0149 (5) | 0.0177 (6) | 0.0019 (4) | −0.0001 (4) | 0.0019 (4) |
| C9B | 0.0115 (5) | 0.0135 (5) | 0.0130 (5) | 0.0017 (4) | 0.0004 (4) | 0.0035 (4) |
| C10B | 0.0126 (5) | 0.0138 (5) | 0.0133 (5) | 0.0017 (4) | −0.0002 (4) | 0.0031 (4) |
| C11B | 0.0196 (6) | 0.0153 (6) | 0.0140 (5) | −0.0005 (5) | −0.0001 (5) | 0.0009 (4) |
| Cl1C | 0.01531 (14) | 0.02242 (15) | 0.02859 (17) | 0.00173 (12) | 0.00697 (12) | 0.00844 (13) |
| O1C | 0.0165 (4) | 0.0137 (4) | 0.0216 (5) | −0.0001 (3) | 0.0052 (4) | 0.0027 (3) |
| O2C | 0.0168 (4) | 0.0135 (4) | 0.0184 (4) | 0.0038 (3) | 0.0029 (4) | 0.0038 (3) |
| O3C | 0.0207 (5) | 0.0182 (5) | 0.0173 (5) | 0.0094 (4) | −0.0013 (4) | 0.0002 (4) |
| N1C | 0.0149 (5) | 0.0129 (5) | 0.0183 (5) | 0.0028 (4) | 0.0038 (4) | 0.0033 (4) |
| C1C | 0.0141 (5) | 0.0128 (5) | 0.0119 (5) | 0.0008 (4) | 0.0018 (4) | 0.0012 (4) |
| C2C | 0.0150 (6) | 0.0119 (5) | 0.0122 (5) | 0.0016 (4) | 0.0021 (4) | 0.0017 (4) |
| C3C | 0.0151 (6) | 0.0160 (5) | 0.0145 (5) | 0.0013 (4) | 0.0029 (4) | 0.0018 (4) |
| C4C | 0.0190 (6) | 0.0192 (6) | 0.0174 (6) | 0.0058 (5) | 0.0013 (5) | 0.0015 (5) |
| C5C | 0.0264 (7) | 0.0140 (6) | 0.0162 (6) | 0.0057 (5) | −0.0004 (5) | 0.0012 (5) |
| C6C | 0.0240 (7) | 0.0118 (5) | 0.0143 (6) | 0.0021 (5) | −0.0002 (5) | 0.0015 (4) |
| C7C | 0.0173 (6) | 0.0135 (5) | 0.0120 (5) | 0.0001 (4) | 0.0012 (4) | 0.0012 (4) |
| C8C | 0.0191 (6) | 0.0166 (6) | 0.0213 (6) | 0.0010 (5) | 0.0083 (5) | 0.0033 (5) |
| C9C | 0.0137 (5) | 0.0146 (5) | 0.0132 (5) | 0.0008 (4) | 0.0026 (4) | 0.0013 (4) |
| C10C | 0.0153 (6) | 0.0147 (5) | 0.0141 (5) | 0.0019 (4) | 0.0006 (4) | −0.0001 (4) |
| C11C | 0.0157 (6) | 0.0168 (6) | 0.0209 (6) | 0.0044 (5) | 0.0002 (5) | −0.0004 (5) |
| Cl1D | 0.01586 (15) | 0.02262 (16) | 0.03299 (19) | 0.00284 (12) | 0.00784 (13) | 0.00992 (14) |
| O1D | 0.0224 (5) | 0.0256 (5) | 0.0205 (5) | 0.0051 (4) | 0.0084 (4) | 0.0106 (4) |
| O2D | 0.0159 (4) | 0.0137 (4) | 0.0195 (5) | 0.0034 (3) | 0.0016 (4) | 0.0027 (3) |
| O3D | 0.0210 (5) | 0.0186 (5) | 0.0163 (4) | 0.0092 (4) | −0.0023 (4) | −0.0022 (4) |
| N1D | 0.0141 (5) | 0.0136 (5) | 0.0176 (5) | 0.0028 (4) | 0.0025 (4) | 0.0022 (4) |
| C1D | 0.0130 (5) | 0.0131 (5) | 0.0117 (5) | 0.0010 (4) | 0.0016 (4) | 0.0008 (4) |
| C2D | 0.0143 (5) | 0.0126 (5) | 0.0131 (5) | 0.0014 (4) | 0.0006 (4) | 0.0012 (4) |
| C3D | 0.0163 (6) | 0.0153 (5) | 0.0162 (6) | 0.0007 (5) | 0.0013 (5) | 0.0021 (4) |
| C4D | 0.0197 (6) | 0.0192 (6) | 0.0225 (7) | 0.0060 (5) | 0.0017 (5) | 0.0023 (5) |
| C5D | 0.0280 (7) | 0.0172 (6) | 0.0217 (7) | 0.0064 (5) | −0.0008 (6) | 0.0046 (5) |
| C6D | 0.0274 (7) | 0.0176 (6) | 0.0191 (6) | 0.0023 (5) | 0.0025 (5) | 0.0060 (5) |
| C7D | 0.0201 (6) | 0.0166 (6) | 0.0142 (6) | 0.0014 (5) | 0.0027 (5) | 0.0029 (4) |
| C8D | 0.0168 (6) | 0.0209 (6) | 0.0182 (6) | 0.0006 (5) | 0.0051 (5) | 0.0047 (5) |
| C9D | 0.0145 (6) | 0.0161 (5) | 0.0119 (5) | 0.0011 (4) | 0.0020 (4) | 0.0006 (4) |
| C10D | 0.0137 (6) | 0.0158 (5) | 0.0154 (6) | 0.0024 (4) | 0.0005 (4) | −0.0016 (4) |
| C11D | 0.0163 (6) | 0.0192 (6) | 0.0206 (6) | 0.0041 (5) | −0.0022 (5) | −0.0028 (5) |
| Cl1E | 0.01560 (14) | 0.01716 (14) | 0.02477 (16) | 0.00139 (11) | 0.00683 (12) | 0.00269 (12) |
| O1E | 0.0145 (4) | 0.0123 (4) | 0.0194 (4) | 0.0011 (3) | 0.0033 (3) | 0.0030 (3) |
| O2E | 0.0179 (5) | 0.0157 (4) | 0.0183 (4) | 0.0045 (4) | 0.0047 (4) | 0.0069 (3) |
| O3E | 0.0167 (5) | 0.0233 (5) | 0.0176 (5) | 0.0085 (4) | 0.0030 (4) | 0.0068 (4) |
| N1E | 0.0177 (5) | 0.0146 (5) | 0.0179 (5) | 0.0037 (4) | 0.0045 (4) | 0.0048 (4) |
| C1E | 0.0152 (6) | 0.0119 (5) | 0.0120 (5) | 0.0020 (4) | 0.0020 (4) | 0.0006 (4) |
| C2E | 0.0147 (5) | 0.0107 (5) | 0.0126 (5) | 0.0018 (4) | 0.0017 (4) | 0.0009 (4) |
| C3E | 0.0153 (6) | 0.0135 (5) | 0.0140 (5) | 0.0013 (4) | 0.0028 (4) | −0.0002 (4) |
| C4E | 0.0173 (6) | 0.0161 (6) | 0.0163 (6) | 0.0050 (5) | 0.0008 (5) | 0.0006 (5) |
| C5E | 0.0245 (7) | 0.0134 (5) | 0.0166 (6) | 0.0044 (5) | 0.0002 (5) | 0.0017 (5) |
| C6E | 0.0197 (6) | 0.0121 (5) | 0.0133 (5) | 0.0014 (5) | 0.0007 (5) | 0.0012 (4) |
| C7E | 0.0158 (6) | 0.0129 (5) | 0.0114 (5) | 0.0017 (4) | 0.0011 (4) | 0.0007 (4) |
| C8E | 0.0175 (6) | 0.0141 (5) | 0.0185 (6) | 0.0022 (5) | 0.0062 (5) | 0.0035 (5) |
| C9E | 0.0146 (5) | 0.0124 (5) | 0.0129 (5) | 0.0018 (4) | 0.0023 (4) | 0.0015 (4) |
| C10E | 0.0148 (6) | 0.0146 (5) | 0.0138 (5) | 0.0006 (4) | 0.0019 (4) | 0.0020 (4) |
| C11E | 0.0210 (6) | 0.0197 (6) | 0.0167 (6) | 0.0074 (5) | 0.0038 (5) | 0.0066 (5) |
| Cl1F | 0.01254 (13) | 0.02157 (15) | 0.02018 (15) | 0.00260 (11) | −0.00279 (11) | 0.00216 (12) |
| O1F | 0.0127 (4) | 0.0250 (5) | 0.0177 (5) | 0.0059 (4) | −0.0012 (3) | −0.0013 (4) |
| O2F | 0.0173 (4) | 0.0171 (4) | 0.0146 (4) | 0.0048 (4) | −0.0004 (3) | 0.0000 (3) |
| O3F | 0.0182 (5) | 0.0333 (6) | 0.0167 (5) | 0.0006 (4) | −0.0020 (4) | 0.0062 (4) |
| N1F | 0.0149 (5) | 0.0172 (5) | 0.0137 (5) | 0.0048 (4) | −0.0002 (4) | 0.0003 (4) |
| C1F | 0.0095 (5) | 0.0161 (5) | 0.0133 (5) | 0.0014 (4) | 0.0018 (4) | 0.0046 (4) |
| C2F | 0.0105 (5) | 0.0143 (5) | 0.0142 (5) | 0.0012 (4) | 0.0017 (4) | 0.0049 (4) |
| C3F | 0.0125 (5) | 0.0166 (6) | 0.0154 (6) | 0.0011 (4) | 0.0013 (4) | 0.0051 (4) |
| C4F | 0.0188 (6) | 0.0166 (6) | 0.0157 (6) | −0.0019 (5) | 0.0012 (5) | 0.0022 (5) |
| C5F | 0.0231 (7) | 0.0135 (5) | 0.0185 (6) | 0.0015 (5) | 0.0057 (5) | 0.0019 (5) |
| C6F | 0.0182 (6) | 0.0154 (6) | 0.0200 (6) | 0.0048 (5) | 0.0049 (5) | 0.0041 (5) |
| C7F | 0.0123 (5) | 0.0161 (5) | 0.0151 (5) | 0.0027 (4) | 0.0011 (4) | 0.0039 (4) |
| C8F | 0.0136 (6) | 0.0213 (6) | 0.0153 (6) | 0.0054 (5) | −0.0004 (4) | 0.0025 (5) |
| C9F | 0.0098 (5) | 0.0157 (5) | 0.0133 (5) | 0.0010 (4) | 0.0012 (4) | 0.0048 (4) |
| C10F | 0.0138 (5) | 0.0181 (6) | 0.0127 (5) | 0.0017 (4) | 0.0017 (4) | 0.0046 (4) |
| C11F | 0.0198 (6) | 0.0202 (6) | 0.0148 (6) | 0.0001 (5) | 0.0002 (5) | 0.0012 (5) |
Geometric parameters (Å, º) top
| O1A—C8A | 1.422 (2) | C6C—H6C | 0.9500 |
| O1B—C8B | 1.4492 (16) | C8C—C9C | 1.4901 (18) |
| O1C—C8C | 1.4389 (16) | C8C—H8C1 | 0.9900 |
| O1D—C8D | 1.4444 (17) | C8C—H8C2 | 0.9900 |
| O1E—C8E | 1.4490 (16) | C9C—C10C | 1.3487 (18) |
| O1F—C8F | 1.4463 (16) | C10C—C11C | 1.4845 (19) |
| O3A—C11A | 1.4136 (17) | C11C—H11E | 0.9900 |
| O3B—C11B | 1.4108 (17) | C11C—H11F | 0.9900 |
| O3C—C11C | 1.4249 (16) | Cl1D—C3D | 1.7365 (14) |
| O3D—C11D | 1.4214 (16) | O1D—C7D | 1.3743 (17) |
| O3E—C11E | 1.4236 (17) | O2D—C10D | 1.3620 (16) |
| O3F—C11F | 1.4121 (18) | O2D—N1D | 1.4114 (14) |
| Cl1A—C3A | 1.7320 (13) | O3D—H3OD | 0.79 (2) |
| O1A—C7A | 1.381 (2) | N1D—C1D | 1.3199 (16) |
| O1X—C7A | 1.360 (7) | C1D—C9D | 1.4236 (18) |
| O1X—C8A | 1.431 (6) | C1D—C2D | 1.4654 (18) |
| O2A—C10A | 1.3585 (15) | C2D—C3D | 1.4008 (18) |
| O2A—N1A | 1.4193 (14) | C2D—C7D | 1.4123 (18) |
| O3A—H3OA | 0.82 (2) | C3D—C4D | 1.3861 (19) |
| N1A—C1A | 1.3197 (16) | C4D—C5D | 1.391 (2) |
| C1A—C9A | 1.4218 (17) | C4D—H4D | 0.9500 |
| C1A—C2A | 1.4599 (17) | C5D—C6D | 1.387 (2) |
| C2A—C3A | 1.4015 (18) | C5D—H5D | 0.9500 |
| C2A—C7A | 1.4093 (18) | C6D—C7D | 1.3879 (19) |
| C3A—C4A | 1.3878 (18) | C6D—H6D | 0.9500 |
| C4A—C5A | 1.388 (2) | C8D—C9D | 1.4915 (18) |
| C4A—H4A | 0.9500 | C8D—H8D1 | 0.9900 |
| C5A—C6A | 1.384 (2) | C8D—H8D2 | 0.9900 |
| C5A—H5A | 0.9500 | C9D—C10D | 1.3521 (18) |
| C6A—C7A | 1.3950 (19) | C10D—C11D | 1.4851 (19) |
| C6A—H6A | 0.9500 | C11D—H11G | 0.9900 |
| C8A—C9A | 1.4808 (18) | C11D—H11H | 0.9900 |
| C8A—H8A1 | 0.9900 | Cl1E—C3E | 1.7386 (13) |
| C8A—H8A2 | 0.9900 | O1E—C7E | 1.3747 (16) |
| C8A—H8X1 | 0.9900 | O2E—C10E | 1.3583 (15) |
| C8A—H8X2 | 0.9900 | O2E—N1E | 1.4140 (14) |
| C9A—C10A | 1.3512 (17) | O3E—H3OE | 0.81 (2) |
| C10A—C11A | 1.4924 (18) | N1E—C1E | 1.3182 (16) |
| C11A—H11A | 0.9900 | C1E—C9E | 1.4216 (18) |
| C11A—H11B | 0.9900 | C1E—C2E | 1.4638 (17) |
| Cl1B—C3B | 1.7352 (13) | C2E—C3E | 1.3996 (18) |
| O1B—C7B | 1.3685 (16) | C2E—C7E | 1.4152 (17) |
| O2B—C10B | 1.3576 (15) | C3E—C4E | 1.3882 (18) |
| O2B—N1B | 1.4174 (14) | C4E—C5E | 1.3953 (19) |
| O3B—H3OB | 0.82 (2) | C4E—H4E | 0.9500 |
| N1B—C1B | 1.3196 (16) | C5E—C6E | 1.384 (2) |
| C1B—C9B | 1.4229 (17) | C5E—H5E | 0.9500 |
| C1B—C2B | 1.4611 (17) | C6E—C7E | 1.4002 (17) |
| C2B—C3B | 1.4036 (17) | C6E—H6E | 0.9500 |
| C2B—C7B | 1.4076 (17) | C8E—C9E | 1.4935 (17) |
| C3B—C4B | 1.3851 (18) | C8E—H8E1 | 0.9900 |
| C4B—C5B | 1.389 (2) | C8E—H8E2 | 0.9900 |
| C4B—H4B | 0.9500 | C9E—C10E | 1.3554 (17) |
| C5B—C6B | 1.388 (2) | C10E—C11E | 1.4872 (19) |
| C5B—H5B | 0.9500 | C11E—H11I | 0.9900 |
| C6B—C7B | 1.3966 (18) | C11E—H11J | 0.9900 |
| C6B—H6B | 0.9500 | Cl1F—C3F | 1.7324 (13) |
| C8B—C9B | 1.4841 (18) | O1F—C7F | 1.3662 (16) |
| C8B—H8B1 | 0.9900 | O2F—C10F | 1.3547 (16) |
| C8B—H8B2 | 0.9900 | O2F—N1F | 1.4163 (14) |
| C9B—C10B | 1.3542 (17) | O3F—H3OF | 0.77 (2) |
| C10B—C11B | 1.4948 (18) | N1F—C1F | 1.3197 (16) |
| C11B—H11C | 0.9900 | C1F—C9F | 1.4223 (17) |
| C11B—H11D | 0.9900 | C1F—C2F | 1.4613 (18) |
| Cl1C—C3C | 1.7368 (13) | C2F—C3F | 1.4026 (18) |
| O1C—C7C | 1.3755 (16) | C2F—C7F | 1.4141 (18) |
| O2C—C10C | 1.3584 (16) | C3F—C4F | 1.3862 (19) |
| O2C—N1C | 1.4093 (14) | C4F—C5F | 1.393 (2) |
| O3C—H3OC | 0.79 (2) | C4F—H4F | 0.9500 |
| N1C—C1C | 1.3224 (16) | C5F—C6F | 1.388 (2) |
| C1C—C9C | 1.4229 (18) | C5F—H5F | 0.9500 |
| C1C—C2C | 1.4605 (17) | C6F—C7F | 1.3907 (18) |
| C2C—C3C | 1.3982 (18) | C6F—H6F | 0.9500 |
| C2C—C7C | 1.4140 (17) | C8F—C9F | 1.4862 (18) |
| C3C—C4C | 1.3868 (19) | C8F—H8F1 | 0.9900 |
| C4C—C5C | 1.393 (2) | C8F—H8F2 | 0.9900 |
| C4C—H4C | 0.9500 | C9F—C10F | 1.3545 (18) |
| C5C—C6C | 1.387 (2) | C10F—C11F | 1.4949 (19) |
| C5C—H5C | 0.9500 | C11F—H11K | 0.9900 |
| C6C—C7C | 1.3929 (18) | C11F—H11L | 0.9900 |
| | | |
| C7A—O1A—C8A | 119.85 (19) | C9C—C10C—O2C | 109.45 (11) |
| C7A—O1X—C8A | 120.7 (7) | C9C—C10C—C11C | 133.85 (13) |
| C10A—O2A—N1A | 108.74 (9) | O2C—C10C—C11C | 116.69 (11) |
| C11A—O3A—H3OA | 109.0 (16) | O3C—C11C—C10C | 109.42 (11) |
| C1A—N1A—O2A | 104.97 (10) | O3C—C11C—H11E | 109.8 |
| N1A—C1A—C9A | 111.83 (11) | C10C—C11C—H11E | 109.8 |
| N1A—C1A—C2A | 128.59 (11) | O3C—C11C—H11F | 109.8 |
| C9A—C1A—C2A | 119.57 (11) | C10C—C11C—H11F | 109.8 |
| C3A—C2A—C7A | 116.93 (11) | H11E—C11C—H11F | 108.2 |
| C3A—C2A—C1A | 127.48 (11) | C7D—O1D—C8D | 118.66 (10) |
| C7A—C2A—C1A | 115.57 (11) | C10D—O2D—N1D | 109.45 (10) |
| C4A—C3A—C2A | 122.05 (12) | C11D—O3D—H3OD | 106.3 (17) |
| C4A—C3A—Cl1A | 117.21 (10) | C1D—N1D—O2D | 104.47 (10) |
| C2A—C3A—Cl1A | 120.74 (10) | N1D—C1D—C9D | 112.30 (11) |
| C3A—C4A—C5A | 119.45 (13) | N1D—C1D—C2D | 128.00 (12) |
| C3A—C4A—H4A | 120.3 | C9D—C1D—C2D | 119.70 (11) |
| C5A—C4A—H4A | 120.3 | C3D—C2D—C7D | 117.53 (12) |
| C6A—C5A—C4A | 120.44 (13) | C3D—C2D—C1D | 126.93 (12) |
| C6A—C5A—H5A | 119.8 | C7D—C2D—C1D | 115.55 (11) |
| C4A—C5A—H5A | 119.8 | C4D—C3D—C2D | 121.93 (12) |
| C5A—C6A—C7A | 119.68 (13) | C4D—C3D—Cl1D | 117.75 (11) |
| C5A—C6A—H6A | 120.2 | C2D—C3D—Cl1D | 120.29 (10) |
| C7A—C6A—H6A | 120.2 | C3D—C4D—C5D | 118.85 (13) |
| O1X—C7A—O1A | 27.6 (6) | C3D—C4D—H4D | 120.6 |
| O1X—C7A—C6A | 112.2 (3) | C5D—C4D—H4D | 120.6 |
| O1A—C7A—C6A | 115.79 (13) | C6D—C5D—C4D | 121.18 (13) |
| O1X—C7A—C2A | 122.0 (3) | C6D—C5D—H5D | 119.4 |
| O1A—C7A—C2A | 122.43 (13) | C4D—C5D—H5D | 119.4 |
| C6A—C7A—C2A | 121.39 (13) | C5D—C6D—C7D | 119.37 (13) |
| O1A—C8A—O1X | 26.5 (6) | C5D—C6D—H6D | 120.3 |
| O1A—C8A—C9A | 111.68 (12) | C7D—C6D—H6D | 120.3 |
| O1X—C8A—C9A | 113.1 (3) | O1D—C7D—C6D | 116.25 (12) |
| O1A—C8A—H8A1 | 109.3 | O1D—C7D—C2D | 122.44 (12) |
| O1X—C8A—H8A1 | 84.6 | C6D—C7D—C2D | 121.12 (13) |
| C9A—C8A—H8A1 | 109.3 | O1D—C8D—C9D | 110.40 (11) |
| O1A—C8A—H8A2 | 109.3 | O1D—C8D—H8D1 | 109.6 |
| O1X—C8A—H8A2 | 128.3 | C9D—C8D—H8D1 | 109.6 |
| C9A—C8A—H8A2 | 109.3 | O1D—C8D—H8D2 | 109.6 |
| H8A1—C8A—H8A2 | 107.9 | C9D—C8D—H8D2 | 109.6 |
| O1A—C8A—H8X1 | 129.9 | H8D1—C8D—H8D2 | 108.1 |
| O1X—C8A—H8X1 | 109.0 | C10D—C9D—C1D | 104.48 (11) |
| C9A—C8A—H8X1 | 109.0 | C10D—C9D—C8D | 134.54 (12) |
| H8A1—C8A—H8X1 | 27.3 | C1D—C9D—C8D | 120.91 (12) |
| H8A2—C8A—H8X1 | 82.9 | C9D—C10D—O2D | 109.29 (11) |
| O1A—C8A—H8X2 | 85.5 | C9D—C10D—C11D | 134.35 (13) |
| O1X—C8A—H8X2 | 109.0 | O2D—C10D—C11D | 116.36 (12) |
| C9A—C8A—H8X2 | 109.0 | O3D—C11D—C10D | 109.09 (11) |
| H8A1—C8A—H8X2 | 129.4 | O3D—C11D—H11G | 109.9 |
| H8A2—C8A—H8X2 | 26.8 | C10D—C11D—H11G | 109.9 |
| H8X1—C8A—H8X2 | 107.8 | O3D—C11D—H11H | 109.9 |
| C10A—C9A—C1A | 104.75 (11) | C10D—C11D—H11H | 109.9 |
| C10A—C9A—C8A | 133.22 (12) | H11G—C11D—H11H | 108.3 |
| C1A—C9A—C8A | 121.91 (11) | C7E—O1E—C8E | 117.82 (10) |
| C9A—C10A—O2A | 109.71 (11) | C10E—O2E—N1E | 109.45 (9) |
| C9A—C10A—C11A | 133.79 (12) | C11E—O3E—H3OE | 102.8 (14) |
| O2A—C10A—C11A | 116.49 (11) | C1E—N1E—O2E | 104.19 (10) |
| O3A—C11A—C10A | 108.99 (11) | N1E—C1E—C9E | 112.78 (11) |
| O3A—C11A—H11A | 109.9 | N1E—C1E—C2E | 127.53 (12) |
| C10A—C11A—H11A | 109.9 | C9E—C1E—C2E | 119.63 (11) |
| O3A—C11A—H11B | 109.9 | C3E—C2E—C7E | 117.57 (11) |
| C10A—C11A—H11B | 109.9 | C3E—C2E—C1E | 127.35 (11) |
| H11A—C11A—H11B | 108.3 | C7E—C2E—C1E | 115.04 (11) |
| C7B—O1B—C8B | 118.76 (10) | C4E—C3E—C2E | 122.10 (12) |
| C10B—O2B—N1B | 108.86 (9) | C4E—C3E—Cl1E | 117.69 (10) |
| C11B—O3B—H3OB | 109.2 (16) | C2E—C3E—Cl1E | 120.21 (10) |
| C1B—N1B—O2B | 104.87 (10) | C3E—C4E—C5E | 118.47 (13) |
| N1B—C1B—C9B | 112.07 (11) | C3E—C4E—H4E | 120.8 |
| N1B—C1B—C2B | 128.49 (11) | C5E—C4E—H4E | 120.8 |
| C9B—C1B—C2B | 119.44 (11) | C6E—C5E—C4E | 121.97 (12) |
| C3B—C2B—C7B | 117.35 (11) | C6E—C5E—H5E | 119.0 |
| C3B—C2B—C1B | 127.08 (11) | C4E—C5E—H5E | 119.0 |
| C7B—C2B—C1B | 115.56 (11) | C5E—C6E—C7E | 118.62 (12) |
| C4B—C3B—C2B | 121.83 (12) | C5E—C6E—H6E | 120.7 |
| C4B—C3B—Cl1B | 117.41 (10) | C7E—C6E—H6E | 120.7 |
| C2B—C3B—Cl1B | 120.75 (10) | O1E—C7E—C6E | 116.28 (11) |
| C3B—C4B—C5B | 119.46 (12) | O1E—C7E—C2E | 122.29 (11) |
| C3B—C4B—H4B | 120.3 | C6E—C7E—C2E | 121.26 (12) |
| C5B—C4B—H4B | 120.3 | O1E—C8E—C9E | 109.38 (10) |
| C6B—C5B—C4B | 120.66 (12) | O1E—C8E—H8E1 | 109.8 |
| C6B—C5B—H5B | 119.7 | C9E—C8E—H8E1 | 109.8 |
| C4B—C5B—H5B | 119.7 | O1E—C8E—H8E2 | 109.8 |
| C5B—C6B—C7B | 119.38 (13) | C9E—C8E—H8E2 | 109.8 |
| C5B—C6B—H6B | 120.3 | H8E1—C8E—H8E2 | 108.2 |
| C7B—C6B—H6B | 120.3 | C10E—C9E—C1E | 104.06 (11) |
| O1B—C7B—C6B | 115.66 (11) | C10E—C9E—C8E | 135.66 (12) |
| O1B—C7B—C2B | 122.99 (11) | C1E—C9E—C8E | 120.28 (11) |
| C6B—C7B—C2B | 121.30 (12) | C9E—C10E—O2E | 109.51 (11) |
| O1B—C8B—C9B | 110.18 (10) | C9E—C10E—C11E | 135.14 (12) |
| O1B—C8B—H8B1 | 109.6 | O2E—C10E—C11E | 115.27 (11) |
| C9B—C8B—H8B1 | 109.6 | O3E—C11E—C10E | 107.97 (11) |
| O1B—C8B—H8B2 | 109.6 | O3E—C11E—H11I | 110.1 |
| C9B—C8B—H8B2 | 109.6 | C10E—C11E—H11I | 110.1 |
| H8B1—C8B—H8B2 | 108.1 | O3E—C11E—H11J | 110.1 |
| C10B—C9B—C1B | 104.40 (11) | C10E—C11E—H11J | 110.1 |
| C10B—C9B—C8B | 134.25 (12) | H11I—C11E—H11J | 108.4 |
| C1B—C9B—C8B | 121.24 (11) | C7F—O1F—C8F | 118.90 (10) |
| C9B—C10B—O2B | 109.79 (11) | C10F—O2F—N1F | 108.98 (10) |
| C9B—C10B—C11B | 134.18 (12) | C11F—O3F—H3OF | 108.3 (17) |
| O2B—C10B—C11B | 116.00 (11) | C1F—N1F—O2F | 104.82 (10) |
| O3B—C11B—C10B | 111.18 (11) | N1F—C1F—C9F | 112.03 (11) |
| O3B—C11B—H11C | 109.4 | N1F—C1F—C2F | 128.30 (11) |
| C10B—C11B—H11C | 109.4 | C9F—C1F—C2F | 119.63 (11) |
| O3B—C11B—H11D | 109.4 | C3F—C2F—C7F | 116.97 (12) |
| C10B—C11B—H11D | 109.4 | C3F—C2F—C1F | 127.36 (11) |
| H11C—C11B—H11D | 108.0 | C7F—C2F—C1F | 115.63 (11) |
| C7C—O1C—C8C | 118.28 (10) | C4F—C3F—C2F | 122.22 (12) |
| C10C—O2C—N1C | 109.41 (9) | C4F—C3F—Cl1F | 117.88 (10) |
| C11C—O3C—H3OC | 104.2 (17) | C2F—C3F—Cl1F | 119.89 (10) |
| C1C—N1C—O2C | 104.56 (10) | C3F—C4F—C5F | 119.20 (13) |
| N1C—C1C—C9C | 111.99 (11) | C3F—C4F—H4F | 120.4 |
| N1C—C1C—C2C | 128.45 (12) | C5F—C4F—H4F | 120.4 |
| C9C—C1C—C2C | 119.48 (11) | C6F—C5F—C4F | 120.58 (13) |
| C3C—C2C—C7C | 117.71 (11) | C6F—C5F—H5F | 119.7 |
| C3C—C2C—C1C | 126.64 (11) | C4F—C5F—H5F | 119.7 |
| C7C—C2C—C1C | 115.61 (11) | C5F—C6F—C7F | 119.65 (13) |
| C4C—C3C—C2C | 121.98 (12) | C5F—C6F—H6F | 120.2 |
| C4C—C3C—Cl1C | 117.96 (10) | C7F—C6F—H6F | 120.2 |
| C2C—C3C—Cl1C | 120.05 (10) | O1F—C7F—C6F | 116.08 (12) |
| C3C—C4C—C5C | 118.67 (13) | O1F—C7F—C2F | 122.30 (12) |
| C3C—C4C—H4C | 120.7 | C6F—C7F—C2F | 121.37 (12) |
| C5C—C4C—H4C | 120.7 | O1F—C8F—C9F | 110.21 (10) |
| C6C—C5C—C4C | 121.55 (13) | O1F—C8F—H8F1 | 109.6 |
| C6C—C5C—H5C | 119.2 | C9F—C8F—H8F1 | 109.6 |
| C4C—C5C—H5C | 119.2 | O1F—C8F—H8F2 | 109.6 |
| C5C—C6C—C7C | 119.01 (13) | C9F—C8F—H8F2 | 109.6 |
| C5C—C6C—H6C | 120.5 | H8F1—C8F—H8F2 | 108.1 |
| C7C—C6C—H6C | 120.5 | C10F—C9F—C1F | 104.40 (11) |
| O1C—C7C—C6C | 115.90 (11) | C10F—C9F—C8F | 134.93 (12) |
| O1C—C7C—C2C | 122.86 (11) | C1F—C9F—C8F | 120.66 (11) |
| C6C—C7C—C2C | 121.08 (12) | C9F—C10F—O2F | 109.76 (11) |
| O1C—C8C—C9C | 110.57 (11) | C9F—C10F—C11F | 134.67 (12) |
| O1C—C8C—H8C1 | 109.5 | O2F—C10F—C11F | 115.56 (11) |
| C9C—C8C—H8C1 | 109.5 | O3F—C11F—C10F | 110.80 (12) |
| O1C—C8C—H8C2 | 109.5 | O3F—C11F—H11K | 109.5 |
| C9C—C8C—H8C2 | 109.5 | C10F—C11F—H11K | 109.5 |
| H8C1—C8C—H8C2 | 108.1 | O3F—C11F—H11L | 109.5 |
| C10C—C9C—C1C | 104.59 (11) | C10F—C11F—H11L | 109.5 |
| C10C—C9C—C8C | 134.88 (12) | H11K—C11F—H11L | 108.1 |
| C1C—C9C—C8C | 120.52 (11) | | |
| | | |
| C10A—O2A—N1A—C1A | −0.38 (13) | C1C—C9C—C10C—C11C | −178.04 (14) |
| O2A—N1A—C1A—C9A | 0.71 (14) | C8C—C9C—C10C—C11C | 3.4 (3) |
| O2A—N1A—C1A—C2A | −178.21 (12) | N1C—O2C—C10C—C9C | −0.79 (14) |
| N1A—C1A—C2A—C3A | 4.1 (2) | N1C—O2C—C10C—C11C | 178.23 (11) |
| C9A—C1A—C2A—C3A | −174.78 (12) | C9C—C10C—C11C—O3C | −104.66 (17) |
| N1A—C1A—C2A—C7A | −177.58 (13) | O2C—C10C—C11C—O3C | 76.62 (15) |
| C9A—C1A—C2A—C7A | 3.57 (17) | C10D—O2D—N1D—C1D | −0.64 (13) |
| C7A—C2A—C3A—C4A | −2.34 (18) | O2D—N1D—C1D—C9D | 0.68 (14) |
| C1A—C2A—C3A—C4A | 175.99 (12) | O2D—N1D—C1D—C2D | −179.61 (12) |
| C7A—C2A—C3A—Cl1A | 177.42 (10) | N1D—C1D—C2D—C3D | 8.9 (2) |
| C1A—C2A—C3A—Cl1A | −4.25 (18) | C9D—C1D—C2D—C3D | −171.38 (12) |
| C2A—C3A—C4A—C5A | 0.72 (19) | N1D—C1D—C2D—C7D | −171.45 (13) |
| Cl1A—C3A—C4A—C5A | −179.05 (10) | C9D—C1D—C2D—C7D | 8.24 (17) |
| C3A—C4A—C5A—C6A | 0.9 (2) | C7D—C2D—C3D—C4D | −0.8 (2) |
| C4A—C5A—C6A—C7A | −0.7 (2) | C1D—C2D—C3D—C4D | 178.85 (13) |
| C8A—O1X—C7A—O1A | −65.5 (12) | C7D—C2D—C3D—Cl1D | 177.34 (10) |
| C8A—O1X—C7A—C6A | −169.0 (10) | C1D—C2D—C3D—Cl1D | −3.04 (19) |
| C8A—O1X—C7A—C2A | 34.2 (18) | C2D—C3D—C4D—C5D | −0.2 (2) |
| C8A—O1A—C7A—O1X | 65.3 (6) | Cl1D—C3D—C4D—C5D | −178.37 (11) |
| C8A—O1A—C7A—C6A | 154.3 (3) | C3D—C4D—C5D—C6D | 0.5 (2) |
| C8A—O1A—C7A—C2A | −32.8 (5) | C4D—C5D—C6D—C7D | 0.2 (2) |
| C5A—C6A—C7A—O1X | −158.0 (9) | C8D—O1D—C7D—C6D | 150.90 (13) |
| C5A—C6A—C7A—O1A | 172.0 (2) | C8D—O1D—C7D—C2D | −34.17 (18) |
| C5A—C6A—C7A—C2A | −1.0 (2) | C5D—C6D—C7D—O1D | 173.75 (13) |
| C3A—C2A—C7A—O1X | 157.2 (9) | C5D—C6D—C7D—C2D | −1.3 (2) |
| C1A—C2A—C7A—O1X | −21.3 (9) | C3D—C2D—C7D—O1D | −173.18 (12) |
| C3A—C2A—C7A—O1A | −170.1 (3) | C1D—C2D—C7D—O1D | 7.16 (19) |
| C1A—C2A—C7A—O1A | 11.4 (3) | C3D—C2D—C7D—C6D | 1.51 (19) |
| C3A—C2A—C7A—C6A | 2.47 (19) | C1D—C2D—C7D—C6D | −178.15 (12) |
| C1A—C2A—C7A—C6A | −176.06 (12) | C7D—O1D—C8D—C9D | 41.39 (16) |
| C7A—O1A—C8A—O1X | −63.7 (6) | N1D—C1D—C9D—C10D | −0.48 (15) |
| C7A—O1A—C8A—C9A | 34.9 (4) | C2D—C1D—C9D—C10D | 179.78 (11) |
| C7A—O1X—C8A—O1A | 66.6 (13) | N1D—C1D—C9D—C8D | −177.76 (12) |
| C7A—O1X—C8A—C9A | −26.0 (17) | C2D—C1D—C9D—C8D | 2.50 (18) |
| N1A—C1A—C9A—C10A | −0.78 (14) | O1D—C8D—C9D—C10D | 157.63 (14) |
| C2A—C1A—C9A—C10A | 178.25 (11) | O1D—C8D—C9D—C1D | −26.06 (17) |
| N1A—C1A—C9A—C8A | −177.31 (12) | C1D—C9D—C10D—O2D | 0.05 (14) |
| C2A—C1A—C9A—C8A | 1.72 (18) | C8D—C9D—C10D—O2D | 176.78 (14) |
| O1A—C8A—C9A—C10A | 164.6 (2) | C1D—C9D—C10D—C11D | −179.74 (14) |
| O1X—C8A—C9A—C10A | −166.8 (9) | C8D—C9D—C10D—C11D | −3.0 (3) |
| O1A—C8A—C9A—C1A | −20.0 (3) | N1D—O2D—C10D—C9D | 0.36 (14) |
| O1X—C8A—C9A—C1A | 8.6 (9) | N1D—O2D—C10D—C11D | −179.81 (11) |
| C1A—C9A—C10A—O2A | 0.51 (14) | C9D—C10D—C11D—O3D | −108.84 (17) |
| C8A—C9A—C10A—O2A | 176.46 (14) | O2D—C10D—C11D—O3D | 71.38 (15) |
| C1A—C9A—C10A—C11A | −178.22 (13) | C10E—O2E—N1E—C1E | −0.65 (13) |
| C8A—C9A—C10A—C11A | −2.3 (3) | O2E—N1E—C1E—C9E | 1.28 (14) |
| N1A—O2A—C10A—C9A | −0.10 (14) | O2E—N1E—C1E—C2E | 178.31 (12) |
| N1A—O2A—C10A—C11A | 178.87 (10) | N1E—C1E—C2E—C3E | 15.8 (2) |
| C9A—C10A—C11A—O3A | 1.1 (2) | C9E—C1E—C2E—C3E | −167.35 (12) |
| O2A—C10A—C11A—O3A | −177.53 (10) | N1E—C1E—C2E—C7E | −161.79 (13) |
| C10B—O2B—N1B—C1B | −0.28 (13) | C9E—C1E—C2E—C7E | 15.06 (17) |
| O2B—N1B—C1B—C9B | 0.60 (13) | C7E—C2E—C3E—C4E | 1.33 (19) |
| O2B—N1B—C1B—C2B | 179.88 (11) | C1E—C2E—C3E—C4E | −176.21 (12) |
| N1B—C1B—C2B—C3B | 10.9 (2) | C7E—C2E—C3E—Cl1E | −178.33 (9) |
| C9B—C1B—C2B—C3B | −169.83 (12) | C1E—C2E—C3E—Cl1E | 4.13 (18) |
| N1B—C1B—C2B—C7B | −170.13 (12) | C2E—C3E—C4E—C5E | −0.2 (2) |
| C9B—C1B—C2B—C7B | 9.09 (16) | Cl1E—C3E—C4E—C5E | 179.42 (10) |
| C7B—C2B—C3B—C4B | −0.60 (18) | C3E—C4E—C5E—C6E | −0.6 (2) |
| C1B—C2B—C3B—C4B | 178.31 (12) | C4E—C5E—C6E—C7E | 0.3 (2) |
| C7B—C2B—C3B—Cl1B | −179.73 (9) | C8E—O1E—C7E—C6E | 151.01 (11) |
| C1B—C2B—C3B—Cl1B | −0.82 (18) | C8E—O1E—C7E—C2E | −33.66 (17) |
| C2B—C3B—C4B—C5B | −0.46 (19) | C5E—C6E—C7E—O1E | 176.26 (11) |
| Cl1B—C3B—C4B—C5B | 178.69 (10) | C5E—C6E—C7E—C2E | 0.88 (19) |
| C3B—C4B—C5B—C6B | 1.1 (2) | C3E—C2E—C7E—O1E | −176.75 (11) |
| C4B—C5B—C6B—C7B | −0.6 (2) | C1E—C2E—C7E—O1E | 1.09 (17) |
| C8B—O1B—C7B—C6B | 150.88 (11) | C3E—C2E—C7E—C6E | −1.65 (18) |
| C8B—O1B—C7B—C2B | −31.71 (17) | C1E—C2E—C7E—C6E | 176.19 (11) |
| C5B—C6B—C7B—O1B | 176.94 (11) | C7E—O1E—C8E—C9E | 45.73 (15) |
| C5B—C6B—C7B—C2B | −0.52 (19) | N1E—C1E—C9E—C10E | −1.44 (15) |
| C3B—C2B—C7B—O1B | −176.18 (11) | C2E—C1E—C9E—C10E | −178.73 (11) |
| C1B—C2B—C7B—O1B | 4.79 (17) | N1E—C1E—C9E—C8E | 177.74 (12) |
| C3B—C2B—C7B—C6B | 1.10 (18) | C2E—C1E—C9E—C8E | 0.44 (18) |
| C1B—C2B—C7B—C6B | −177.94 (11) | O1E—C8E—C9E—C10E | 149.36 (15) |
| C7B—O1B—C8B—C9B | 40.50 (15) | O1E—C8E—C9E—C1E | −29.49 (16) |
| N1B—C1B—C9B—C10B | −0.71 (14) | C1E—C9E—C10E—O2E | 0.95 (14) |
| C2B—C1B—C9B—C10B | 179.95 (11) | C8E—C9E—C10E—O2E | −178.03 (14) |
| N1B—C1B—C9B—C8B | −177.38 (11) | C1E—C9E—C10E—C11E | −175.36 (15) |
| C2B—C1B—C9B—C8B | 3.27 (17) | C8E—C9E—C10E—C11E | 5.7 (3) |
| O1B—C8B—C9B—C10B | 157.70 (14) | N1E—O2E—C10E—C9E | −0.24 (14) |
| O1B—C8B—C9B—C1B | −26.79 (16) | N1E—O2E—C10E—C11E | 176.89 (11) |
| C1B—C9B—C10B—O2B | 0.50 (14) | C9E—C10E—C11E—O3E | 14.2 (2) |
| C8B—C9B—C10B—O2B | 176.53 (13) | O2E—C10E—C11E—O3E | −161.96 (11) |
| C1B—C9B—C10B—C11B | −177.46 (14) | C10F—O2F—N1F—C1F | −0.97 (13) |
| C8B—C9B—C10B—C11B | −1.4 (3) | O2F—N1F—C1F—C9F | 0.71 (14) |
| N1B—O2B—C10B—C9B | −0.16 (14) | O2F—N1F—C1F—C2F | 178.25 (12) |
| N1B—O2B—C10B—C11B | 178.21 (10) | N1F—C1F—C2F—C3F | 10.6 (2) |
| C9B—C10B—C11B—O3B | −9.0 (2) | C9F—C1F—C2F—C3F | −171.99 (12) |
| O2B—C10B—C11B—O3B | 173.12 (11) | N1F—C1F—C2F—C7F | −166.92 (12) |
| C10C—O2C—N1C—C1C | 0.47 (13) | C9F—C1F—C2F—C7F | 10.45 (17) |
| O2C—N1C—C1C—C9C | −0.01 (14) | C7F—C2F—C3F—C4F | 0.24 (18) |
| O2C—N1C—C1C—C2C | −176.90 (12) | C1F—C2F—C3F—C4F | −177.29 (12) |
| N1C—C1C—C2C—C3C | −10.9 (2) | C7F—C2F—C3F—Cl1F | 179.01 (9) |
| C9C—C1C—C2C—C3C | 172.43 (12) | C1F—C2F—C3F—Cl1F | 1.48 (18) |
| N1C—C1C—C2C—C7C | 166.83 (13) | C2F—C3F—C4F—C5F | −0.20 (19) |
| C9C—C1C—C2C—C7C | −9.87 (17) | Cl1F—C3F—C4F—C5F | −179.00 (10) |
| C7C—C2C—C3C—C4C | 0.49 (19) | C3F—C4F—C5F—C6F | −0.3 (2) |
| C1C—C2C—C3C—C4C | 178.14 (13) | C4F—C5F—C6F—C7F | 0.8 (2) |
| C7C—C2C—C3C—Cl1C | 179.53 (10) | C8F—O1F—C7F—C6F | 153.31 (12) |
| C1C—C2C—C3C—Cl1C | −2.81 (19) | C8F—O1F—C7F—C2F | −32.25 (18) |
| C2C—C3C—C4C—C5C | −0.2 (2) | C5F—C6F—C7F—O1F | 173.71 (12) |
| Cl1C—C3C—C4C—C5C | −179.24 (10) | C5F—C6F—C7F—C2F | −0.8 (2) |
| C3C—C4C—C5C—C6C | −0.3 (2) | C3F—C2F—C7F—O1F | −173.89 (12) |
| C4C—C5C—C6C—C7C | 0.3 (2) | C1F—C2F—C7F—O1F | 3.93 (18) |
| C8C—O1C—C7C—C6C | −153.54 (12) | C3F—C2F—C7F—C6F | 0.26 (18) |
| C8C—O1C—C7C—C2C | 30.87 (18) | C1F—C2F—C7F—C6F | 178.09 (12) |
| C5C—C6C—C7C—O1C | −175.67 (12) | C7F—O1F—C8F—C9F | 42.00 (16) |
| C5C—C6C—C7C—C2C | 0.00 (19) | N1F—C1F—C9F—C10F | −0.20 (14) |
| C3C—C2C—C7C—O1C | 174.97 (12) | C2F—C1F—C9F—C10F | −177.98 (11) |
| C1C—C2C—C7C—O1C | −2.94 (18) | N1F—C1F—C9F—C8F | −179.63 (11) |
| C3C—C2C—C7C—C6C | −0.40 (19) | C2F—C1F—C9F—C8F | 2.60 (17) |
| C1C—C2C—C7C—C6C | −178.31 (12) | O1F—C8F—C9F—C10F | 153.38 (14) |
| C7C—O1C—C8C—C9C | −41.90 (16) | O1F—C8F—C9F—C1F | −27.41 (16) |
| N1C—C1C—C9C—C10C | −0.46 (15) | C1F—C9F—C10F—O2F | −0.43 (14) |
| C2C—C1C—C9C—C10C | 176.75 (11) | C8F—C9F—C10F—O2F | 178.87 (13) |
| N1C—C1C—C9C—C8C | 178.39 (12) | C1F—C9F—C10F—C11F | 178.69 (14) |
| C2C—C1C—C9C—C8C | −4.40 (18) | C8F—C9F—C10F—C11F | −2.0 (3) |
| O1C—C8C—C9C—C10C | −152.37 (15) | N1F—O2F—C10F—C9F | 0.88 (14) |
| O1C—C8C—C9C—C1C | 29.21 (17) | N1F—O2F—C10F—C11F | −178.42 (11) |
| C1C—C9C—C10C—O2C | 0.74 (14) | C9F—C10F—C11F—O3F | −5.0 (2) |
| C8C—C9C—C10C—O2C | −177.86 (14) | O2F—C10F—C11F—O3F | 174.06 (11) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3A—H3OA···O3Ei | 0.82 (2) | 1.99 (2) | 2.8006 (14) | 165 (2) |
| O3B—H3OB···O3C | 0.82 (2) | 1.93 (2) | 2.7321 (15) | 167 (2) |
| O3F—H3OF···O3Dii | 0.77 (2) | 1.99 (2) | 2.7535 (15) | 170 (2) |
| O3C—H3OC···Cl1Aiii | 0.79 (2) | 2.73 (2) | 3.2601 (11) | 126 (2) |
| O3D—H3OD···Cl1Biii | 0.79 (2) | 2.83 (2) | 3.3070 (11) | 120.7 (19) |
| O3E—H3OE···Cl1F | 0.81 (2) | 2.89 (2) | 3.3580 (11) | 118.8 (16) |
| O3C—H3OC···N1Aiii | 0.79 (2) | 2.10 (2) | 2.8471 (15) | 158 (2) |
| O3D—H3OD···N1Biii | 0.79 (2) | 2.04 (2) | 2.8110 (15) | 165 (2) |
| O3E—H3OE···N1F | 0.81 (2) | 2.04 (2) | 2.8132 (15) | 160 (2) |
| C4A—H4A···O2Eiv | 0.95 | 2.48 | 3.3964 (17) | 162 |
| C4B—H4B···O2Dv | 0.95 | 2.44 | 3.3457 (16) | 160 |
| C4F—H4F···O2Cvi | 0.95 | 2.61 | 3.4190 (17) | 144 |
| C8A—H8A1···O1Avii | 0.99 | 2.51 | 3.122 (3) | 120 |
| C8D—H8D2···O1Dviii | 0.99 | 2.61 | 3.3164 (16) | 128 |
| C8F—H8F2···O1Bix | 0.99 | 2.60 | 3.2861 (16) | 127 |
| C6C—H6C···O3Ai | 0.95 | 2.55 | 3.4748 (17) | 164 |
| C6E—H6E···O3B | 0.95 | 2.36 | 3.2748 (17) | 161 |
| C11E—H11I···O1Bvi | 0.99 | 2.53 | 3.2632 (16) | 131 |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+1, −y+1, −z; (iii) x+1, y, z; (iv) x−1, y−1, z; (v) −x+1, −y, −z; (vi) x, y+1, z; (vii) −x+1, −y, −z+1; (viii) −x+2, −y+1, −z; (ix) x−1, y+1, z. |
Experimental details
| | (I) | (II) |
| Crystal data |
| Chemical formula | C11H8FNO3 | C11H8ClNO3 |
| Mr | 221.18 | 237.63 |
| Crystal system, space group | Orthorhombic, Pbca | Triclinic, P1 |
| Temperature (K) | 100 | 100 |
| a, b, c (Å) | 14.1516 (2), 7.0064 (1), 18.3134 (2) | 9.4306 (1), 14.8892 (2), 21.4949 (2) |
| α, β, γ (°) | 90, 90, 90 | 98.764 (1), 97.610 (1), 94.013 (1) |
| V (Å3) | 1815.81 (4) | 2944.12 (6) |
| Z | 8 | 12 |
| Radiation type | Mo Kα | Mo Kα |
| µ (mm−1) | 0.13 | 0.38 |
| Crystal size (mm) | 0.35 × 0.10 × 0.07 | 0.32 × 0.32 × 0.17 |
| |
| Data collection |
| Diffractometer | Bruker SMART APEXII CCD area-detector
diffractometer | Bruker SMART APEXII CCD area-detector
diffractometer |
| Absorption correction | Multi-scan
(SADABS; Bruker, 2009) | Multi-scan
(SADABS; Bruker, 2009) |
| Tmin, Tmax | 0.955, 0.991 | 0.890, 0.937 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 18961, 3017, 2279 | 69911, 21416, 17626 |
| Rint | 0.040 | 0.023 |
| (sin θ/λ)max (Å−1) | 0.735 | 0.760 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.044, 0.123, 1.05 | 0.042, 0.107, 1.04 |
| No. of reflections | 3017 | 21416 |
| No. of parameters | 149 | 900 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.54, −0.24 | 0.66, −0.39 |
Selected geometric parameters (Å, º) for (I) top| O1—C8 | 1.4526 (14) | O3—C11 | 1.4107 (15) |
| | | |
| C7—O1—C8—C9 | 44.47 (14) | O2—C10—C11—O3 | 175.24 (11) |
Hydrogen-bond geometry (Å, º) for (I) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3···N1i | 0.81 (2) | 2.07 (2) | 2.8557 (14) | 163 (2) |
| O3—H3···F1i | 0.81 (2) | 2.73 (2) | 3.2644 (12) | 125.3 (18) |
| C8—H8A···F1ii | 0.99 | 2.60 | 3.5095 (16) | 153.2 |
| C4—H4A···F1iii | 0.95 | 2.58 | 3.4674 (14) | 155.4 |
| Symmetry codes: (i) x−1/2, y, −z+3/2; (ii) −x+1/2, y−1/2, z; (iii) −x+1, −y, −z+1. |
Selected geometric parameters (Å, º) for (II) top| O1A—C8A | 1.422 (2) | O3A—C11A | 1.4136 (17) |
| O1B—C8B | 1.4492 (16) | O3B—C11B | 1.4108 (17) |
| O1C—C8C | 1.4389 (16) | O3C—C11C | 1.4249 (16) |
| O1D—C8D | 1.4444 (17) | O3D—C11D | 1.4214 (16) |
| O1E—C8E | 1.4490 (16) | O3E—C11E | 1.4236 (17) |
| O1F—C8F | 1.4463 (16) | O3F—C11F | 1.4121 (18) |
| | | |
| C7A—O1A—C8A—C9A | 34.9 (4) | C7D—O1D—C8D—C9D | 41.39 (16) |
| O2A—C10A—C11A—O3A | −177.53 (10) | O2D—C10D—C11D—O3D | 71.38 (15) |
| C7B—O1B—C8B—C9B | 40.50 (15) | C7E—O1E—C8E—C9E | 45.73 (15) |
| O2B—C10B—C11B—O3B | 173.12 (11) | O2E—C10E—C11E—O3E | −161.96 (11) |
| C7C—O1C—C8C—C9C | −41.90 (16) | C7F—O1F—C8F—C9F | 42.00 (16) |
| O2C—C10C—C11C—O3C | 76.62 (15) | O2F—C10F—C11F—O3F | 174.06 (11) |
Hydrogen-bond geometry (Å, º) for (II) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3A—H3OA···O3Ei | 0.82 (2) | 1.99 (2) | 2.8006 (14) | 165 (2) |
| O3B—H3OB···O3C | 0.82 (2) | 1.93 (2) | 2.7321 (15) | 167 (2) |
| O3F—H3OF···O3Dii | 0.77 (2) | 1.99 (2) | 2.7535 (15) | 170 (2) |
| O3C—H3OC···Cl1Aiii | 0.79 (2) | 2.73 (2) | 3.2601 (11) | 126 (2) |
| O3D—H3OD···Cl1Biii | 0.79 (2) | 2.83 (2) | 3.3070 (11) | 120.7 (19) |
| O3E—H3OE···Cl1F | 0.81 (2) | 2.89 (2) | 3.3580 (11) | 118.8 (16) |
| O3C—H3OC···N1Aiii | 0.79 (2) | 2.10 (2) | 2.8471 (15) | 158 (2) |
| O3D—H3OD···N1Biii | 0.79 (2) | 2.04 (2) | 2.8110 (15) | 165 (2) |
| O3E—H3OE···N1F | 0.81 (2) | 2.04 (2) | 2.8132 (15) | 160 (2) |
| C4A—H4A···O2Eiv | 0.95 | 2.48 | 3.3964 (17) | 162.1 |
| C4B—H4B···O2Dv | 0.95 | 2.44 | 3.3457 (16) | 160.1 |
| C4F—H4F···O2Cvi | 0.95 | 2.61 | 3.4190 (17) | 143.5 |
| C8A—H8A1···O1Avii | 0.99 | 2.51 | 3.122 (3) | 119.7 |
| C8D—H8D2···O1Dviii | 0.99 | 2.61 | 3.3164 (16) | 127.9 |
| C8F—H8F2···O1Bix | 0.99 | 2.60 | 3.2861 (16) | 126.6 |
| C6C—H6C···O3Ai | 0.95 | 2.55 | 3.4748 (17) | 164.3 |
| C6E—H6E···O3B | 0.95 | 2.36 | 3.2748 (17) | 160.6 |
| C11E—H11I···O1Bvi | 0.99 | 2.53 | 3.2632 (16) | 131.1 |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+1, −y+1, −z; (iii) x+1, y, z; (iv) x−1, y−1, z; (v) −x+1, −y, −z; (vi) x, y+1, z; (vii) −x+1, −y, −z+1; (viii) −x+2, −y+1, −z; (ix) x−1, y+1, z. |
Selected geometric parameters (Å, °) for (II) top| Molecule | O1—C8 | O3—C11 | O2—C10—C11—O3 | C7—O1—C8—C9 |
| A | 1.422 (2) | 1.4136 (17) | -177.53 (10) | 34.9 (4) |
| B | 1.4492 (16) | 1.4108 (17) | 173.12 (11) | 40.50 (15) |
| C | 1.4389 (16) | 1.4249 (16) | 76.62 (15) | -41.90 (16) |
| D | 1.4444 (17) | 1.4214 (16) | 71.38 (15) | 41.39 (16) |
| E | 1.4490 (16) | 1.4236 (17) | -161.96 (11) | 45.73 (15) |
| F | 1.4463 (16) | 1.4121 (18) | 174.06 (11) | 42.00 (16) |
 F and the absence of O—H
F and the absence of O—H O and C—H
O and C—H O interactions in (I), and the absence of C—H
O interactions in (I), and the absence of C—H Cl and the presence of O—H
Cl and the presence of O—H O and C—H
O and C—H O interactions in (II). However, the geometry of the synthons formed by the O—H
O interactions in (II). However, the geometry of the synthons formed by the O—H N and O—H
N and O—H X (X = F or Cl) interactions observed in the constitution of the supramolecular networks of both (I) and (II) remains similar. Also, C—H
X (X = F or Cl) interactions observed in the constitution of the supramolecular networks of both (I) and (II) remains similar. Also, C—H O interactions are not preferred in the presence of F in (I), while they are much preferred in the presence of Cl in (II).
O interactions are not preferred in the presence of F in (I), while they are much preferred in the presence of Cl in (II).
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2012/08/00/ku3066/ku3066fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2012/08/00/ku3066/ku3066fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2012/08/00/ku3066/ku3066fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/c/issues/2012/08/00/ku3066/ku3066fig4thm.gif)
![[Figure 5]](http://journals.iucr.org/c/issues/2012/08/00/ku3066/ku3066fig5thm.gif)
![[Figure 6]](http://journals.iucr.org/c/issues/2012/08/00/ku3066/ku3066fig6thm.gif)
![[Figure 7]](http://journals.iucr.org/c/issues/2012/08/00/ku3066/ku3066fig7thm.gif)
![[Figure 8]](http://journals.iucr.org/c/issues/2012/08/00/ku3066/ku3066fig8thm.gif)




Isoxazole derivatives have interesting biological and medicinal applications (Lin et al.,1996, Hu et al., 2004). Leflunomide [systematic name: N-(4'-trifluoromethylphenyl)-5-methylisoxazole-4-carboxamide] is an isoxazole drug used for the treatment of rheumatoid arthritis (Rozman et al., 2002). The study of fluorine-, bromine- and chlorine-substituted leflunomide metabolite analogues has shown that the presence of an F atom at the 2-position of the phenyl ring disrupts the intermolecular hydrogen bonding that is observed for the other derivatives, due to differences in the crystal packing for these molecules (Venkatachalam et al., 2005). Since interactions involving halogens are known to contribute to characteristic supramolecular synthons (Pedireddi et al. 1992; Desiraju et al. 1993), the present description of the crystal strucures of the two title isoxazole derivatives, (I) and (II), may contribute to the development of crystal-engineering strategies. Some of the recent reports of structures that are relevant to the present study are: 3-(3-chlorophenyl)-1-methyl-3,3a,4,9b-tetrahydro-1H-chromeno[4,3-c]isoxazole-3a-carbonitrile (Swaminathan et al., 2011a), its 1-methyl-3-p-tolyl-substituted analogue (Gangadharan et al., 2011), and 1-methyl-3-(2-methylphenyl)-3,3a,4,9b-tetrahydro-1H-chromeno[4,3-c][1,2] oxazole-3a-carbonitrile (Swaminathan et al., 2011b).
The simple replacement of F by Cl lowers the point-group symmetry of the crystal from D2h in (I) to Ci = S2 in (II). The unit cell of (II) presents a complex picture, with six molecules in its asymmetric unit (A–F), each differing from the others in conformation and related by pseudo-translations and pseudo-inversion centres. There are pseudo-inversion centres at [0.34 (2), 0.50 (1), 0.16 (1)] between molecules B and F, at [0.83 (3), 0.50 (2), 0.17 (1)] between molecules D and E, and at [0.67 (3), 0.50 (2), 0.37 (1)] between molecules C and E. The pseudo-translations are [-0.33 (3), -0.01 (2), 0.33 (2)], relating molecules C and D, and [-0.33 (1), -0.01 (1), 0.33 (1)], relating molecules A and B. The molecular structures of (I) and (II) are shown in Figs. 1 and 2, and selected bond lengths, bond angles and conformation angles are listed in Tables 1 and 3, respectively.
The geometric parameters of the 4H-chromeno[4,3-c]isoxazole rings of (I) and (II) are comparable with those reported for similar structures retrieved from the Cambridge Structural Database (Version 5.32: Allen, 2002). However, in (I) the O1—C8 [1.4526 (14) Å; Raihan et al., 2010; Liaskopoulos et al., 2007] and O3—C11 [1.4107 (15) Å] bond lengths differ significantly from the respective mean values of 1.425 and 1.400 Å. The increase in O1–C8 may be attributed to the inductive negative effect of the halogen atom on chroman atom O1. The increase in O3—C11 is possibly due to the participation of atom O3 in a relatively strong O—H···N hydrogen bond. In (II), these values are in the range 1.4210 (2)–1.4492 (16) Å for O1—C8, and 1.4109 (17)–1.4248 (17) Å for O3—C11.
In both (I) and (II), the chroman ring (C1/C2/C7/O1/C8/C9) adopts a screw-boat conformation (Cremer & Pople, 1975), with atoms C8 and O1 deviating from the plane defined by the rest of the atoms. Of the six molecules in the asymmetric unit of (II), the puckering of the chroman ring of molecule A switches between nearly envelope and screw-boat configurations, owing to the disordered atom O1, which was modelled using two sets of atomic sites with refined occupancies of 0.814 (14) and 0.189 (14). The puckering of six-membered rings is measured using three parameters, Q, which provides a measure of the magnitude or amplitude of the puckering, θ, which is a descriptor of the type of conformation, viz. chair (C), half-chair (H), envelope (E), twist or twist-boat (T), boat (B) or screw-boat (S), and ϕ, which is an estimate of puckering distortion. While θ can take values from 0 to 90°, 0° represents an ideal chair and 90° an ideal twist conformation; ϕ ranges from 0 to 360° (Cremer & Pople, 1975). In (II), the puckering of molecules A–F is described by θ values of 57.6 (4), 63.0 (2), 63.5 (2), 61.8 (2), 67.8 (2) and 64.2 (2)°, respectively. It is seen that molecule E, with θ = 67.8 (2)°, conforms closely to the ideal value of 67.5° for a screw-boat conformation.
The hydroxyl group and isoxazole ring exhibit an antiperiplanar conformation with respect to atoms O3/C11/C10/O2, defined by a torsion angle of 175.24 (11)° in (I). However, in (II), atom O3 of the hydroxyl group has synclinal, synclinal and (-)-antiperiplanar conformations with the isoxazole ring in molecules C, D and E, respectively, with values of 76.61 (15), 71.39 (15) and -161.96 (11)°, respectively (Table 3). These differences in conformation may be explained in terms of the mode of participation of atom O3 in the hydrogen-bonding network. Atoms O3 of molecules C, D and E participate as donors in O—H···N and O—H···Cl hydrogen bonds, and as acceptors in O—H···O hydrogen bonds and stronger C—H···O interactions (Table 4). In molecules A, B and F, O—H···N and O—H···Cl bonds involving atom O3 are absent. A similar synclinal conformation of -70.23° for the hydroxyl O atom has been reported for a closely related isoxazoline derivative (Denmark et al., 1999). Fig. 3 shows the superimposition of molecules of (I) and the six molecules of (II), in which the differences in the conformations of the invididual molecules can readily be seen.
In (I), the intermolecular interaction pattern is characterized by conventional O—H···N and O—H···F bonded supramolecular motifs. In addition, C—H···F bonds between centrosymmetrically related molecules and π–π (arene) interactions combine to generate a two-dimensional hydrogen-bonding network. The overall picture of the intermolecular interactions may be visualized as due to two simple graph-set motifs (Bernstein et al., 1995), R21(6) (Fig. 4) and R22(8) (Fig.5). Firstly, the R21(6) motif is generated through O3—H3···N1i [symmetry code: (i) x - 1/2, y, -z + 3/2] and O3—H3···F1i hydrogen bonds, which link the molecules into one-dimensional chains running parallel to the a axis (Fig. 4). These one-dimensional chains have aromatic π–π stacking interactions between the centroids Cg and Cg(1/2 - x, 1/2 + y, z) of the fluoro-substituted phenyl ring; the centroid-to-centroid distance is 3.592 (7) Å, the interplanar spacing is ca 3.51 Å and the ring offset is ca 0.76 Å. Secondly, C8—H8A···F1(-x + 1/2, y - 1/2, z) and C4—H4A···F1(-x + 1, -y, -z + 1) interactions between centrosymetric pairs form an R22(8) motif. These rings are linked through C(7) chains characterized by C—H···F interactions, leading to a two-dimensional network. The C—H···F interactions seem to play a proactive role by participating in intermolecular interaction patterns in the absence of conventional hydrogen bonds (Choudhury et al., 2004).
In (II), the geometry of the intermolecular interactions appears complicated due to the presence of six molecules in the asymmetric unit. The interactions are characterized by a combination of conventional O—H···N, O—H···O and O—H···Cl bonds. In addition, nonconventional C—H···O, C—H···π and π–π (arene) interactions lead to a three-dimensional hydrogen-bonding network. The complex picture of the intermolecular interaction pattern (Fig. 6) may be simplified in terms of groups of molecules and their roles as donors or acceptors in interactions other than C—H···O. While C—H···O type interactions are present involving all six molecules in the asymmetric unit in (II), such commonality is not seen in the cases of the O—H···O, O—H···N and O—H···Cl interactions. In the strong O—H···O bonds, atoms O3 of molecules A, B and F participate as donors and those of molecules C, D and E as acceptors. Similarly, in the strong O—H···N bonds atoms O3 of molecules C, D and E are donors and atoms N1 of molecules A, B and F are acceptors. Atoms O3 of molecules C, D and E are engaged in O—H ··· Cl interactions, with the Cl atoms of molecules A, B and F as acceptors. The fundamental characteristic motifs are an R22(6) motif generated through the C—H···O hydrogen bonds C8A—H5A···O1Avii [symmetry code: (vii) -x + 1, -y, -z + 1] (Fig. 7) and C8D—H4D···O1Dviii [symmetry code: (viii) -x + 2, -y + 1, -z] between the chroman rings of centrosymmetric pairs of molecules, an R21(6) motif involving O—H···N and O—H···Cl hydrogen bonds in both [Both what?] (Fig. 7 ), an R44(18) motif generated through C4B—H1B···O2Dv [symmetry code: (v) -x + 1, -y, -z] and O3D—H8D···Cl1Biii [symmetry code: (iii) x + 1, y, z] hydrogen bonds, and an R44(32) motif involving C6C—H3C···O3Ai [symmetry code: (i) -x + 1, -y + 1, -z + 1] and O3C—H8C···N1Aiii [symmetry code: (iii) x + 1, y, z] hydrogen bonds.
The packing of the six molecules in the asymmetric unit of (II) (Fig. 6) shows a ladder-like arrangement in which groups of molecules ABF and CDE alternate with FAAFBBF and ECCEDDE, respectively, as repeat units. The FAAFBBF and ECCEDDE sequences due to molecules ABF (molecular group I) and CDE (molecular group II) are further illustrated by the fact that, in group I, the O atom of the hydroxyl group is coplanar with the isoxazole ring with an estimated standard deviation of about 0.2 Å. Such coplanarity is not observed in molecular group II. Furthermore, (I) and molecules B and F of (II) have similar conformations, with slight differences in the O3—C11—C10—O2 angles of the respective molecules.
In (I), F···F interactions are absent, which agrees with an earlier observation that the C—F group prefers to form C—H···F interactions rather than F···F contacts and C—H···F interactions prevail over C—H···O interactions (Thalladi et al., 1998). The F1···O1(-x + 1/2, y - 1/2, z) [3.1344 (13) Å] interaction gives rise to the interaction of the lone pair of atom O1 with the electron-deficient chroman system, and this type of interaction has also been observed for 6-chloro-2-oxo-2H-chromene-3-carboxylate derivatives (Santos-Contreras et al., 2007).
In the crystal packing of (II), three different π–π interactions are observed, with Cg···Cg distances, interplanar spacings and ring offsets of, respectively: (i) 3.5145 (8) Å, ca 3.40 Å and ca 0.87 Å between the centroids of the chloro-substituted phenyl rings of molecules A and F; (ii) 3.5784 (8) Å, ca 3.21 Å and ca 1.58 Å between the isoxazole ring and the chloro-substituted phenyl ring of symmetry-related molecules C; and (iii) 3.5597 (8) Å, ca 3.29 Å and ca 1.35 Å between the isoxazole ring of molecule D and the chloro-substituted ring of molecule E. A C—H···π interaction is also observed between atom H1A [H4A?] and the isoxazole ring of molecule E, C4A—H4A···Cgiv = 2.71 Å and 146° [symmetry code: (iv) x - 1, y - 1, z].
C—H···Cl interactions are absent in (II), probably due to the presence of Cl···Cl contacts. Both Cl···Cl interactions observed in (II) are of type I trans geometry and hence generate strong and weak hydrogen bonds other than C—H···Cl (Hathwar et al., 2010; Nayak et al., 2011). In (II), a C3F—Cl1F···Cl1A—C3A interaction is observed between molecules A and F, with Cl1F···Cl1A = 3.2113 (5) Å, C3F—Cl1F···Cl1A = 152.03 (5)° and Cl1F···Cl1A—C3A = 156.51 (5)° [symmetry code: (?) x - 1, y - 1, z] (Fig. 8). There is also a C3B—Cl1B···Cl1B—C3B interaction between two molecules B, with Cl1B···Cl1B = 3.4396 (5) Å, C3B—Cl1B···Cl1B = 152.22 (5)° and Cl1B···Cl1B—C3B = 152.22 (5)° [Identical angle?] [symmetry code: (?) -x, -y, -z] (Fig. 8). It may be inferred that the geometry of both Cl···Cl interactions of type I trans geometry generates both strong and weak hydrogen bonds and appears to add stability to the crystal packing. [Please supply missing symop indicators and add to relevant atom labels]
From the above, it may be concluded that the structures of (I) and (II) together demonstrate the subtle nature of the mechanism behind intermolecular interactions and how a simple exchange of substituents may drastically alter the molecular interaction patterns and effect changes in the symmetry of crystal structures. Thus, the structures remain a good example of why crystal structure prediction of small molecules remains a problem as difficult as that of protein folding and, in fact, much more difficult than had been expected.