Buy article online - an online subscription or single-article purchase is required to access this article.

The crystal structures of isostructural 2,6-naphthalenedicarboxylate tetrahydrate salts of nickel(II) and cobalt(II) have been determined using Monte Carlo simulated annealing techniques and laboratory X-ray powder diffraction data. These compounds crystallize in the triclinic space group
P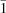
, with
Z = 2;
a = 10.0851 (4),
b = 10.9429 (5),
c = 6.2639 (3) Å, α = 98.989 (2), β = 87.428 (3), γ = 108.015 (2)°,
V = 649.32 (5) Å
3 for [Ni(C
12H
6O
4)(H
2O)
4], and
a = 10.1855 (6),
b = 10.8921 (6),
c = 6.2908 (5) Å, α = 98.519 (4), β = 87.563 (4), γ = 108.304 (3)°,
V = 655.28 (8) Å
3 for [Co(C
12H
6O
4)(H
2O)
4]. The water-molecule H atoms were located by quantum chemical geometry optimization using
CASTEP. The structure consists of alternating hydrocarbon and metal/oxygen layers parallel to the
ac plane. Each naphthalenedicarboxylate anion bridges two metal cations; each carboxyl group is monodentate. The resulting structure contains infinite chains parallel to [111]. The octahedral coordination sphere of the metal cations contains
trans carboxylates and four equatorial water molecules. The carboxyl groups are rotated by 15–20° out of the naphthalene plane. The metal/oxygen layers are characterized by an extensive hydrogen-bonding network. The orientations of the carboxyl groups are determined by the formation of short (O

O = 2.53 Å) hydrogen bonds between the carbonyl O atoms and the
cis water molecules. Molecular mechanics energy minimizations suggest that coordination and hydrogen-bonding interactions are most important in determining the crystal packing.
Supporting information
CCDC references: 214227; 214228
For both compounds, program(s) used to refine structure: GSAS.
Crystal data top
| Triclinic, P1 | α = 98.987 (2)° |
| a = 10.0848 (4) Å | β = 87.427 (3)° |
| b = 10.9428 (5) Å | γ = 108.0158 (18)° |
| c = 6.2639 (3) Å | KG1, KG2 radiation |
Data collection top
| 2θmin = 13.043°, 2θmax = 89.983°, 2θstep = 0.02° | |
Refinement top
| Rp = 0.049 | 3848 data points |
| Rwp = 0.065 | 52 parameters |
| Rexp = 0.022 | 30 restraints |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.3539 (10) | 1.1253 (7) | 0.7132 (10) | 0.031 (2)* | |
| C2 | 0.4652 (9) | 1.1749 (7) | 0.5782 (11) | 0.031 (2)* | |
| C3 | 0.4631 (9) | 1.1189 (8) | 0.3616 (11) | 0.031 (2)* | |
| C4 | 0.3497 (10) | 1.0133 (8) | 0.2805 (9) | 0.031 (2)* | |
| C5 | 0.1241 (10) | 0.8574 (7) | 0.3347 (10) | 0.031 (2)* | |
| C6 | 0.0128 (10) | 0.8077 (7) | 0.4697 (13) | 0.031 (2)* | |
| C7 | 0.0148 (10) | 0.8638 (7) | 0.6862 (12) | 0.031 (2)* | |
| C8 | 0.1282 (10) | 0.9694 (7) | 0.7673 (10) | 0.031 (2)* | |
| C9 | 0.2400 (9) | 1.0195 (7) | 0.6326 (9) | 0.031 (2)* | |
| C10 | 0.2379 (9) | 0.9632 (7) | 0.4153 (9) | 0.031 (2)* | |
| H1 | 0.3568 (11) | 1.1698 (8) | 0.8812 (11) | 0.05* | |
| H3 | 0.5496 (10) | 1.1574 (9) | 0.2565 (13) | 0.05* | |
| H4 | 0.3495 (11) | 0.9707 (9) | 0.1119 (9) | 0.05* | |
| H5 | 0.1211 (11) | 0.8129 (9) | 0.1667 (11) | 0.05* | |
| H7 | −0.0716 (10) | 0.8253 (8) | 0.7914 (14) | 0.05* | |
| H8 | 0.1284 (11) | 1.0120 (9) | 0.9360 (11) | 0.05* | |
| C11 | 0.5877 (10) | 1.2891 (8) | 0.6662 (14) | 0.022 (2)* | |
| C12 | −0.1097 (10) | 0.6936 (8) | 0.3817 (16) | 0.022 (2)* | |
| O1 | 0.5949 (12) | 1.3121 (12) | 0.868 (2) | 0.022 (2)* | |
| O2 | 0.6826 (15) | 1.3411 (12) | 0.5260 (19) | 0.022 (2)* | |
| O3 | −0.2203 (17) | 0.6798 (13) | 0.4952 (18) | 0.022 (2)* | |
| O4 | −0.0989 (15) | 0.6339 (12) | 0.214 (3) | 0.022 (2)* | |
| Ni | 0.7479 (6) | 1.4790 (5) | 1.0272 (9) | 0.0357 (17)* | |
| O5 | 0.8846 (15) | 1.4778 (14) | 0.760 (2) | 0.028 (3)* | |
| O6 | 0.6051 (15) | 1.4822 (12) | 1.278 (2) | 0.028 (3)* | |
| O7 | 0.8289 (13) | 1.3466 (11) | 1.145 (2) | 0.028 (3)* | |
| O8 | 0.6873 (16) | 1.6231 (11) | 0.898 (2) | 0.028 (3)* | |
| H5a | 0.86528 | 1.38675 | 0.74713 | 0.03* | |
| H5b | 0.97172 | 1.50115 | 0.85866 | 0.03* | |
| H6a | 0.53731 | 1.39076 | 1.26716 | 0.03* | |
| H6b | 0.55396 | 1.54789 | 1.2619 | 0.03* | |
| H7a | 0.84631 | 1.38881 | 1.30045 | 0.03* | |
| H7b | 0.74702 | 1.27221 | 1.15524 | 0.03* | |
| H8a | 0.67419 | 1.57228 | 0.75349 | 0.03* | |
| H8b | 0.58858 | 1.59919 | 0.9664 | 0.03* | |
Crystal data top
| Triclinic, P1 | α = 98.522 (4)° |
| a = 10.1856 (6) Å | β = 87.573 (4)° |
| b = 10.8931 (6) Å | γ = 108.300 (3)° |
| c = 6.2902 (5) Å | KG1, KG2 radiation |
Data collection top
| 2θmin = 13.043°, 2θmax = 89.983°, 2θstep = 0.02° | |
Refinement top
| Rp = 0.030 | 3848 data points |
| Rwp = 0.040 | 52 parameters |
| Rexp = 0.026 | 30 restraints |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.3455 (19) | 1.1260 (15) | 0.708 (2) | 0.041 (4)* | |
| C2 | 0.4611 (18) | 1.1748 (15) | 0.581 (2) | 0.041 (4)* | |
| C3 | 0.4669 (19) | 1.1159 (15) | 0.370 (2) | 0.041 (4)* | |
| C4 | 0.357 (2) | 1.0084 (15) | 0.2846 (18) | 0.041 (4)* | |
| C5 | 0.131 (2) | 0.8515 (14) | 0.326 (2) | 0.041 (4)* | |
| C6 | 0.015 (2) | 0.8027 (13) | 0.452 (2) | 0.041 (4)* | |
| C7 | 0.0092 (19) | 0.8615 (14) | 0.664 (3) | 0.041 (4)* | |
| C8 | 0.1191 (19) | 0.9690 (15) | 0.749 (2) | 0.041 (4)* | |
| C9 | 0.2352 (19) | 1.0183 (14) | 0.6229 (19) | 0.041 (4)* | |
| C10 | 0.241 (2) | 0.9592 (13) | 0.4105 (18) | 0.041 (4)* | |
| H1 | 0.342 (2) | 1.1727 (18) | 0.872 (2) | 0.05* | |
| H3 | 0.557 (2) | 1.1538 (16) | 0.272 (2) | 0.05* | |
| H4 | 0.363 (2) | 0.9637 (17) | 0.1198 (19) | 0.05* | |
| H5 | 0.134 (2) | 0.8048 (16) | 0.162 (2) | 0.05* | |
| H7 | −0.081 (2) | 0.8237 (15) | 0.762 (3) | 0.05* | |
| H8 | 0.113 (2) | 1.0138 (18) | 0.914 (2) | 0.05* | |
| C11 | 0.5798 (19) | 1.2910 (17) | 0.674 (3) | 0.003 (4)* | |
| C12 | −0.104 (2) | 0.6865 (14) | 0.360 (3) | 0.003 (4)* | |
| O1 | 0.587 (2) | 1.315 (3) | 0.864 (4) | 0.003 (4)* | |
| O2 | 0.679 (2) | 1.333 (2) | 0.535 (4) | 0.003 (4)* | |
| O3 | −0.227 (3) | 0.687 (2) | 0.491 (4) | 0.003 (4)* | |
| O4 | −0.101 (3) | 0.635 (3) | 0.207 (4) | 0.003 (4)* | |
| Co | 0.7467 (12) | 1.4819 (11) | 1.0291 (19) | 0.052 (4)* | |
| O5 | 0.883 (3) | 1.466 (3) | 0.746 (4) | 0.023 (5)* | |
| O6 | 0.602 (3) | 1.490 (2) | 1.272 (4) | 0.023 (5)* | |
| O7 | 0.827 (2) | 1.348 (2) | 1.137 (4) | 0.023 (5)* | |
| O8 | 0.694 (3) | 1.623 (2) | 0.902 (4) | 0.023 (5)* | |
| H5a | 0.86416 | 1.38101 | 0.75995 | 0.03* | |
| H5b | 0.96329 | 1.50775 | 0.86141 | 0.03* | |
| H6a | 0.54693 | 1.39876 | 1.25994 | 0.03* | |
| H6b | 0.66144 | 1.49091 | 1.39108 | 0.03* | |
| H7a | 0.84693 | 1.37086 | 1.28164 | 0.03* | |
| H7b | 0.75374 | 1.26519 | 1.11385 | 0.03* | |
| H8a | 0.66284 | 1.56483 | 0.7441 | 0.03* | |
| H8b | 0.59358 | 1.59916 | 0.96558 | 0.03* | |

 Subscribe to Journal of Applied Crystallography
Subscribe to Journal of Applied Crystallography
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.
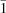 , with Z = 2; a = 10.0851 (4), b = 10.9429 (5), c = 6.2639 (3) Å, α = 98.989 (2), β = 87.428 (3), γ = 108.015 (2)°, V = 649.32 (5) Å3 for [Ni(C12H6O4)(H2O)4], and a = 10.1855 (6), b = 10.8921 (6), c = 6.2908 (5) Å, α = 98.519 (4), β = 87.563 (4), γ = 108.304 (3)°, V = 655.28 (8) Å3 for [Co(C12H6O4)(H2O)4]. The water-molecule H atoms were located by quantum chemical geometry optimization using CASTEP. The structure consists of alternating hydrocarbon and metal/oxygen layers parallel to the ac plane. Each naphthalenedicarboxylate anion bridges two metal cations; each carboxyl group is monodentate. The resulting structure contains infinite chains parallel to [111]. The octahedral coordination sphere of the metal cations contains trans carboxylates and four equatorial water molecules. The carboxyl groups are rotated by 15–20° out of the naphthalene plane. The metal/oxygen layers are characterized by an extensive hydrogen-bonding network. The orientations of the carboxyl groups are determined by the formation of short (O
, with Z = 2; a = 10.0851 (4), b = 10.9429 (5), c = 6.2639 (3) Å, α = 98.989 (2), β = 87.428 (3), γ = 108.015 (2)°, V = 649.32 (5) Å3 for [Ni(C12H6O4)(H2O)4], and a = 10.1855 (6), b = 10.8921 (6), c = 6.2908 (5) Å, α = 98.519 (4), β = 87.563 (4), γ = 108.304 (3)°, V = 655.28 (8) Å3 for [Co(C12H6O4)(H2O)4]. The water-molecule H atoms were located by quantum chemical geometry optimization using CASTEP. The structure consists of alternating hydrocarbon and metal/oxygen layers parallel to the ac plane. Each naphthalenedicarboxylate anion bridges two metal cations; each carboxyl group is monodentate. The resulting structure contains infinite chains parallel to [111]. The octahedral coordination sphere of the metal cations contains trans carboxylates and four equatorial water molecules. The carboxyl groups are rotated by 15–20° out of the naphthalene plane. The metal/oxygen layers are characterized by an extensive hydrogen-bonding network. The orientations of the carboxyl groups are determined by the formation of short (O O = 2.53 Å) hydrogen bonds between the carbonyl O atoms and the cis water molecules. Molecular mechanics energy minimizations suggest that coordination and hydrogen-bonding interactions are most important in determining the crystal packing.
O = 2.53 Å) hydrogen bonds between the carbonyl O atoms and the cis water molecules. Molecular mechanics energy minimizations suggest that coordination and hydrogen-bonding interactions are most important in determining the crystal packing. O = 2.53 Å) hydrogen bonds between the carbonyl O atoms and the cis water molecules. Molecular mechanics energy minimizations suggest that coordination and hydrogen-bonding interactions are most important in determining the crystal packing.
O = 2.53 Å) hydrogen bonds between the carbonyl O atoms and the cis water molecules. Molecular mechanics energy minimizations suggest that coordination and hydrogen-bonding interactions are most important in determining the crystal packing.
 journal menu
journal menu









 Subscribe to Journal of Applied Crystallography
Subscribe to Journal of Applied Crystallography


