Buy article online - an online subscription or single-article purchase is required to access this article.

The two title dinuclear copper(II) complexes, [Cu
2Cl
4(C
17H
20Cl
2N
2)
2], (I), and [Cu
2Cl
4(C
19H
22N
2O
4)
2], (II), have similar coordination environments. In each complex, the asymmetric unit consists of one half-molecule and the two copper centres are bridged by a pair of Cl atoms, resulting in complexes with centrosymmetric structures containing Cu(μ-Cl)
2Cu parallelogram cores; the Cu

Cu separations and Cu—Cl—Cu angles are 3.4285 (8) Å and 83.36 (3)°, respectively, for (I), and 3.565 (2) Å and 84.39 (7)° for (II). Each Cu atom is five-coordinated and the coordination geometry around the Cu atom is best described as a distorted square-pyramid with a τ value of 0.155 (3) for (I) and 0.092 (7) for (II). The apical Cu—Cl bond length is 2.852 (1) Å for (I) and 2.971 (2) Å for (II). The basal Cu—Cl and Cu—N average bonds lengths are 2.2673 (9) and 2.030 (2) Å, respectively, for (I), and 2.280 (2) and 2.038 (6) Å for (II). The molecules of (I) are linked by one C—H

Cl hydrogen bond into a complex [10
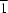
] sheet. The molecules of (II) are linked by one C—H

Cl and one N—H

O hydrogen bond into a complex [100] sheet.
Supporting information
CCDC references: 652293; 654265
For the preparation of complex (I), to a solution containing
N,N-bis(4-chlorobenzyl)propane-1,2-diamine (0.96 g, 3 mmol) and
ethanol (20 ml), a solution of copper chloride (0.48 g, 3 mmol) and ethanol(10 ml) was added dropwise with stirring. The reaction mixture was stirred for 3 h
at 333–343 K and then left to cool at room temperature; the solid obtained
was filtered off, washed successively with chloroform (3 × 3 ml) and
ethanol (3 × 3 ml), and dried at room temperature. Green crystals of
complex (I) suitable for X-ray structure analysis were obtained by slow
evaporation of a dimethylformamide–ethanol (1:10) solution containing the
product over a period of three weeks (yield 0.98 g, 68%; m.p.466–469 K).
Analysis calculated for [Cu2Cl4(C17H20N2Cl2)2]: C 44.61, H 4.40,
N 6.12%; found: C 45.22, H 4.28, N 5.88%. IR (KBr, cm-1): 3450, 3162, 2946,
1599, 1551, 1492, 1440, 1089, 1013, 922, 810, 704, 566.
In same method, green crystals of complex(II) suitable for X-ray structure
analysis also were obtained (yield 1.18 g, 71%; m.p.509– 512 K). Analysis
calculated for [Cu2Cl4(C19H22N2O4)2]: C 47.86%, H 4.65%, N
5.87%; found: C 48.48%, H 4.49%, N 6.02%. IR(KBr, disk,cm-1): 3464, 3250,
2932, 1599, 1553, 1493,1447, 1252, 1034, 922, 812, 767, 704.
All H atoms were located in difference Fourier maps and treated as riding atoms,
with C—H distances of 0.98 Å (methine), 0.93 Å (aryl), 0.96 Å (methyl)
and 0.97 Å (methylene), N—H distances of 0.91 Å, and with
Uiso(H) values of 1.2Ueq(C,N) (aryl, methylene,
imine) and 1.5Ueq(C) (methyl). The value of Rint for complex
(II) is high (Rint = 0.162) because of the poor crystal quality,
resulting in broad diffraction peaks.
For both compounds, data collection: SMART (Siemens, 1996); cell refinement: SAINT (Siemens, 1996); data reduction: SAINT (Siemens, 1996); program(s) used to solve structure: SHELXS97 (Sheldrick, 1997a); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997a); molecular graphics: SHELXTL (Sheldrick, 1997b); software used to prepare material for publication: SHELXTL (Sheldrick, 1997b).
(I) di-µ-chlorido-bis{[
N,
N'-bis(4-chlorobenzyl)propane-1,2-
diamine]chloridocopper(II)}
top
Crystal data top
| [Cu2Cl4(C17H20Cl2N2)2] | F(000) = 932 |
| Mr = 915.38 | Dx = 1.568 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 3324 reflections |
| a = 14.4217 (15) Å | θ = 2.6–27.0° |
| b = 9.5445 (13) Å | µ = 1.68 mm−1 |
| c = 15.6198 (19) Å | T = 298 K |
| β = 115.647 (2)° | Block, green |
| V = 1938.2 (4) Å3 | 0.54 × 0.50 × 0.46 mm |
| Z = 2 | |
Data collection top
Bruker SMART CCD area-detector
diffractometer | 3417 independent reflections |
| Radiation source: fine-focus sealed tube | 2484 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.038 |
| ϕ and ω scans | θmax = 25.0°, θmin = 1.6° |
Absorption correction: multi-scan
(SADABS; Sheldrick, 1996) | h = −15→17 |
| Tmin = 0.464, Tmax = 0.512 | k = −11→6 |
| 8896 measured reflections | l = −17→18 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.031 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.082 | H-atom parameters constrained |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0306P)2 + 0.7906P]
where P = (Fo2 + 2Fc2)/3 |
| 3417 reflections | (Δ/σ)max = 0.001 |
| 218 parameters | Δρmax = 0.39 e Å−3 |
| 0 restraints | Δρmin = −0.28 e Å−3 |
Crystal data top
| [Cu2Cl4(C17H20Cl2N2)2] | V = 1938.2 (4) Å3 |
| Mr = 915.38 | Z = 2 |
| Monoclinic, P21/n | Mo Kα radiation |
| a = 14.4217 (15) Å | µ = 1.68 mm−1 |
| b = 9.5445 (13) Å | T = 298 K |
| c = 15.6198 (19) Å | 0.54 × 0.50 × 0.46 mm |
| β = 115.647 (2)° | |
Data collection top
Bruker SMART CCD area-detector
diffractometer | 3417 independent reflections |
Absorption correction: multi-scan
(SADABS; Sheldrick, 1996) | 2484 reflections with I > 2σ(I) |
| Tmin = 0.464, Tmax = 0.512 | Rint = 0.038 |
| 8896 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.031 | 0 restraints |
| wR(F2) = 0.082 | H-atom parameters constrained |
| S = 1.06 | Δρmax = 0.39 e Å−3 |
| 3417 reflections | Δρmin = −0.28 e Å−3 |
| 218 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Cu1 | 0.56093 (3) | 0.40517 (4) | 0.60255 (3) | 0.03284 (13) | |
| Cl3 | −0.02417 (7) | 0.36440 (11) | 0.64146 (8) | 0.0634 (3) | |
| Cl4 | 0.64329 (11) | 0.57123 (13) | 1.04591 (9) | 0.0884 (4) | |
| Cl2 | 0.71211 (6) | 0.39329 (8) | 0.59008 (6) | 0.0409 (2) | |
| Cl1 | 0.54872 (6) | 0.64141 (8) | 0.59685 (6) | 0.0408 (2) | |
| N1 | 0.41881 (18) | 0.3830 (2) | 0.59727 (18) | 0.0321 (6) | |
| H1 | 0.3741 | 0.4288 | 0.5443 | 0.038* | |
| N2 | 0.58206 (18) | 0.2072 (2) | 0.65562 (18) | 0.0341 (6) | |
| H2 | 0.5980 | 0.1546 | 0.6152 | 0.041* | |
| C1 | 0.3954 (2) | 0.2317 (3) | 0.5811 (2) | 0.0402 (8) | |
| H1A | 0.3831 | 0.2074 | 0.5167 | 0.048* | |
| H1B | 0.3337 | 0.2104 | 0.5887 | 0.048* | |
| C2 | 0.4843 (2) | 0.1465 (3) | 0.6509 (2) | 0.0378 (8) | |
| H2A | 0.4852 | 0.1560 | 0.7137 | 0.045* | |
| C3 | 0.4726 (3) | −0.0085 (3) | 0.6246 (3) | 0.0621 (11) | |
| H3A | 0.4653 | −0.0194 | 0.5609 | 0.093* | |
| H3B | 0.4127 | −0.0451 | 0.6288 | 0.093* | |
| H3C | 0.5324 | −0.0586 | 0.6676 | 0.093* | |
| C4 | 0.4061 (2) | 0.4434 (3) | 0.6791 (2) | 0.0397 (8) | |
| H4A | 0.4227 | 0.5424 | 0.6837 | 0.048* | |
| H4B | 0.4547 | 0.3987 | 0.7369 | 0.048* | |
| C5 | 0.2992 (2) | 0.4263 (3) | 0.6723 (2) | 0.0362 (8) | |
| C6 | 0.2757 (3) | 0.3200 (4) | 0.7195 (2) | 0.0487 (9) | |
| H6 | 0.3277 | 0.2597 | 0.7581 | 0.058* | |
| C7 | 0.1771 (3) | 0.3006 (4) | 0.7109 (3) | 0.0514 (9) | |
| H7 | 0.1624 | 0.2272 | 0.7423 | 0.062* | |
| C8 | 0.1014 (2) | 0.3911 (3) | 0.6555 (2) | 0.0403 (8) | |
| C9 | 0.1223 (2) | 0.4988 (3) | 0.6087 (2) | 0.0415 (8) | |
| H9 | 0.0704 | 0.5605 | 0.5719 | 0.050* | |
| C10 | 0.2204 (2) | 0.5156 (3) | 0.6163 (2) | 0.0389 (8) | |
| H10 | 0.2340 | 0.5878 | 0.5835 | 0.047* | |
| C11 | 0.6707 (2) | 0.1899 (3) | 0.7513 (2) | 0.0419 (8) | |
| H11A | 0.7342 | 0.2094 | 0.7463 | 0.050* | |
| H11B | 0.6732 | 0.0935 | 0.7717 | 0.050* | |
| C12 | 0.6634 (2) | 0.2852 (3) | 0.8247 (2) | 0.0351 (7) | |
| C13 | 0.6297 (2) | 0.2365 (3) | 0.8902 (2) | 0.0428 (8) | |
| H13 | 0.6126 | 0.1423 | 0.8894 | 0.051* | |
| C14 | 0.6210 (3) | 0.3239 (4) | 0.9559 (2) | 0.0485 (9) | |
| H14 | 0.5973 | 0.2898 | 0.9987 | 0.058* | |
| C15 | 0.6478 (3) | 0.4636 (4) | 0.9580 (2) | 0.0485 (9) | |
| C16 | 0.6818 (3) | 0.5153 (4) | 0.8952 (3) | 0.0485 (9) | |
| H16 | 0.6995 | 0.6093 | 0.8970 | 0.058* | |
| C17 | 0.6894 (2) | 0.4259 (3) | 0.8287 (2) | 0.0433 (8) | |
| H17 | 0.7124 | 0.4608 | 0.7858 | 0.052* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cu1 | 0.0315 (2) | 0.0285 (2) | 0.0380 (2) | 0.00195 (16) | 0.01460 (18) | 0.00274 (17) |
| Cl3 | 0.0484 (6) | 0.0729 (7) | 0.0793 (8) | −0.0070 (5) | 0.0374 (6) | −0.0075 (5) |
| Cl4 | 0.1269 (11) | 0.0784 (8) | 0.0701 (8) | −0.0054 (7) | 0.0522 (8) | −0.0269 (6) |
| Cl2 | 0.0319 (4) | 0.0447 (5) | 0.0463 (5) | 0.0029 (3) | 0.0172 (4) | 0.0037 (4) |
| Cl1 | 0.0483 (5) | 0.0277 (4) | 0.0456 (5) | −0.0011 (3) | 0.0196 (4) | −0.0034 (3) |
| N1 | 0.0276 (13) | 0.0317 (15) | 0.0323 (15) | −0.0003 (10) | 0.0087 (12) | 0.0028 (11) |
| N2 | 0.0398 (15) | 0.0279 (14) | 0.0352 (16) | 0.0053 (11) | 0.0168 (13) | 0.0007 (11) |
| C1 | 0.043 (2) | 0.040 (2) | 0.039 (2) | −0.0101 (15) | 0.0188 (17) | −0.0075 (15) |
| C2 | 0.048 (2) | 0.0312 (18) | 0.0366 (19) | −0.0052 (15) | 0.0206 (17) | −0.0005 (14) |
| C3 | 0.086 (3) | 0.035 (2) | 0.072 (3) | −0.0058 (18) | 0.040 (3) | −0.0029 (18) |
| C4 | 0.0383 (19) | 0.041 (2) | 0.038 (2) | 0.0031 (14) | 0.0149 (16) | −0.0040 (15) |
| C5 | 0.0352 (18) | 0.0377 (19) | 0.0348 (19) | 0.0024 (14) | 0.0143 (16) | −0.0036 (15) |
| C6 | 0.048 (2) | 0.053 (2) | 0.045 (2) | 0.0156 (17) | 0.0207 (19) | 0.0130 (18) |
| C7 | 0.061 (2) | 0.050 (2) | 0.056 (2) | −0.0002 (18) | 0.037 (2) | 0.0118 (18) |
| C8 | 0.0390 (19) | 0.045 (2) | 0.040 (2) | −0.0017 (16) | 0.0204 (17) | −0.0097 (16) |
| C9 | 0.039 (2) | 0.038 (2) | 0.042 (2) | 0.0070 (14) | 0.0133 (17) | −0.0002 (15) |
| C10 | 0.043 (2) | 0.0337 (19) | 0.039 (2) | −0.0008 (15) | 0.0166 (17) | 0.0000 (15) |
| C11 | 0.0400 (19) | 0.0366 (19) | 0.045 (2) | 0.0133 (15) | 0.0147 (17) | 0.0104 (16) |
| C12 | 0.0300 (17) | 0.0380 (19) | 0.0298 (18) | 0.0056 (14) | 0.0057 (15) | 0.0031 (14) |
| C13 | 0.049 (2) | 0.0336 (19) | 0.039 (2) | −0.0019 (15) | 0.0127 (17) | 0.0076 (15) |
| C14 | 0.057 (2) | 0.054 (2) | 0.037 (2) | −0.0022 (18) | 0.0220 (19) | 0.0035 (17) |
| C15 | 0.053 (2) | 0.047 (2) | 0.039 (2) | 0.0031 (17) | 0.0142 (18) | −0.0049 (17) |
| C16 | 0.057 (2) | 0.033 (2) | 0.050 (2) | −0.0066 (16) | 0.0184 (19) | −0.0021 (17) |
| C17 | 0.0411 (19) | 0.046 (2) | 0.041 (2) | −0.0028 (15) | 0.0166 (17) | 0.0079 (16) |
Geometric parameters (Å, º) top
| Cu1—N1 | 2.026 (2) | C4—H4B | 0.9700 |
| Cu1—N2 | 2.033 (2) | C5—C6 | 1.380 (4) |
| Cu1—Cl1 | 2.2604 (9) | C5—C10 | 1.386 (4) |
| Cu1—Cl2 | 2.2742 (9) | C6—C7 | 1.382 (4) |
| Cu1—Cl1i | 2.852 (1) | C6—H6 | 0.9300 |
| Cu1—Cu1i | 3.4285 (8) | C7—C8 | 1.367 (5) |
| Cl3—C8 | 1.747 (3) | C7—H7 | 0.9300 |
| Cl4—C15 | 1.739 (4) | C8—C9 | 1.368 (4) |
| Cl1—Cu1i | 2.852 (1) | C9—C10 | 1.377 (4) |
| N1—C1 | 1.480 (4) | C9—H9 | 0.9300 |
| N1—C4 | 1.484 (4) | C10—H10 | 0.9300 |
| N1—H1 | 0.9100 | C11—C12 | 1.503 (4) |
| N2—C2 | 1.496 (4) | C11—H11A | 0.9700 |
| N2—C11 | 1.497 (4) | C11—H11B | 0.9700 |
| N2—H2 | 0.9100 | C12—C13 | 1.388 (4) |
| C1—C2 | 1.511 (4) | C12—C17 | 1.389 (4) |
| C1—H1A | 0.9700 | C13—C14 | 1.371 (4) |
| C1—H1B | 0.9700 | C13—H13 | 0.9300 |
| C2—C3 | 1.525 (4) | C14—C15 | 1.385 (5) |
| C2—H2A | 0.9800 | C14—H14 | 0.9300 |
| C3—H3A | 0.9600 | C15—C16 | 1.365 (5) |
| C3—H3B | 0.9600 | C16—C17 | 1.384 (5) |
| C3—H3C | 0.9600 | C16—H16 | 0.9300 |
| C4—C5 | 1.508 (4) | C17—H17 | 0.9300 |
| C4—H4A | 0.9700 | | |
| | | |
| N1—Cu1—N2 | 83.69 (9) | N1—C4—H4A | 108.8 |
| N1—Cu1—Cl1 | 92.41 (7) | C5—C4—H4A | 108.8 |
| N2—Cu1—Cl1 | 159.84 (8) | N1—C4—H4B | 108.8 |
| N1—Cu1—Cl2 | 169.01 (7) | C5—C4—H4B | 108.8 |
| N2—Cu1—Cl2 | 90.49 (7) | H4A—C4—H4B | 107.7 |
| Cl1—Cu1—Cl2 | 96.18 (3) | C6—C5—C10 | 117.8 (3) |
| N1—Cu1—Cl1i | 82.32 (7) | C6—C5—C4 | 121.4 (3) |
| N2—Cu1—Cl1i | 102.40 (7) | C10—C5—C4 | 120.8 (3) |
| Cl1—Cu1—Cl1i | 96.64 (3) | C5—C6—C7 | 121.8 (3) |
| Cl2—Cu1—Cl1i | 89.87 (3) | C5—C6—H6 | 119.1 |
| N1—Cu1—Cu1i | 85.22 (7) | C7—C6—H6 | 119.1 |
| N2—Cu1—Cu1i | 142.90 (7) | C8—C7—C6 | 118.8 (3) |
| Cl1—Cu1—Cu1i | 55.73 (2) | C8—C7—H7 | 120.6 |
| Cl2—Cu1—Cu1i | 93.96 (3) | C6—C7—H7 | 120.6 |
| Cl1i—Cu1—Cu1i | 40.912 (18) | C7—C8—C9 | 120.9 (3) |
| Cu1—Cl1—Cu1i | 83.36 (3) | C7—C8—Cl3 | 119.2 (3) |
| C1—N1—C4 | 115.0 (2) | C9—C8—Cl3 | 119.9 (3) |
| C1—N1—Cu1 | 105.09 (17) | C8—C9—C10 | 119.8 (3) |
| C4—N1—Cu1 | 115.12 (18) | C8—C9—H9 | 120.1 |
| C1—N1—H1 | 107.1 | C10—C9—H9 | 120.1 |
| C4—N1—H1 | 107.1 | C9—C10—C5 | 120.9 (3) |
| Cu1—N1—H1 | 107.1 | C9—C10—H10 | 119.5 |
| C2—N2—C11 | 112.8 (2) | C5—C10—H10 | 119.5 |
| C2—N2—Cu1 | 111.59 (18) | N2—C11—C12 | 112.5 (2) |
| C11—N2—Cu1 | 114.93 (18) | N2—C11—H11A | 109.1 |
| C2—N2—H2 | 105.5 | C12—C11—H11A | 109.1 |
| C11—N2—H2 | 105.5 | N2—C11—H11B | 109.1 |
| Cu1—N2—H2 | 105.5 | C12—C11—H11B | 109.1 |
| N1—C1—C2 | 110.0 (2) | H11A—C11—H11B | 107.8 |
| N1—C1—H1A | 109.7 | C13—C12—C17 | 117.7 (3) |
| C2—C1—H1A | 109.7 | C13—C12—C11 | 121.5 (3) |
| N1—C1—H1B | 109.7 | C17—C12—C11 | 120.8 (3) |
| C2—C1—H1B | 109.7 | C14—C13—C12 | 121.5 (3) |
| H1A—C1—H1B | 108.2 | C14—C13—H13 | 119.2 |
| N2—C2—C1 | 108.3 (2) | C12—C13—H13 | 119.2 |
| N2—C2—C3 | 112.4 (3) | C13—C14—C15 | 119.2 (3) |
| C1—C2—C3 | 111.5 (3) | C13—C14—H14 | 120.4 |
| N2—C2—H2A | 108.1 | C15—C14—H14 | 120.4 |
| C1—C2—H2A | 108.1 | C16—C15—C14 | 121.0 (3) |
| C3—C2—H2A | 108.1 | C16—C15—Cl4 | 120.1 (3) |
| C2—C3—H3A | 109.5 | C14—C15—Cl4 | 118.8 (3) |
| C2—C3—H3B | 109.5 | C15—C16—C17 | 119.1 (3) |
| H3A—C3—H3B | 109.5 | C15—C16—H16 | 120.5 |
| C2—C3—H3C | 109.5 | C17—C16—H16 | 120.5 |
| H3A—C3—H3C | 109.5 | C16—C17—C12 | 121.5 (3) |
| H3B—C3—H3C | 109.5 | C16—C17—H17 | 119.2 |
| N1—C4—C5 | 113.6 (2) | C12—C17—H17 | 119.2 |
| | | |
| N1—Cu1—Cl1—Cu1i | −82.54 (7) | N1—C1—C2—N2 | −44.7 (3) |
| N2—Cu1—Cl1—Cu1i | −160.8 (2) | N1—C1—C2—C3 | −168.9 (3) |
| Cl2—Cu1—Cl1—Cu1i | 90.59 (3) | C1—N1—C4—C5 | −58.2 (3) |
| Cl1i—Cu1—Cl1—Cu1i | 0.0 | Cu1—N1—C4—C5 | 179.4 (2) |
| N2—Cu1—N1—C1 | −30.59 (19) | N1—C4—C5—C6 | 98.5 (4) |
| Cl1—Cu1—N1—C1 | 169.26 (18) | N1—C4—C5—C10 | −80.0 (4) |
| Cl2—Cu1—N1—C1 | 27.8 (5) | C10—C5—C6—C7 | 0.9 (5) |
| Cl1i—Cu1—N1—C1 | 72.88 (18) | C4—C5—C6—C7 | −177.7 (3) |
| Cu1i—Cu1—N1—C1 | 113.95 (18) | C5—C6—C7—C8 | −1.2 (5) |
| N2—Cu1—N1—C4 | 96.9 (2) | C6—C7—C8—C9 | 0.3 (5) |
| Cl1—Cu1—N1—C4 | −63.2 (2) | C6—C7—C8—Cl3 | 178.2 (3) |
| Cl2—Cu1—N1—C4 | 155.3 (3) | C7—C8—C9—C10 | 0.9 (5) |
| Cl1i—Cu1—N1—C4 | −159.6 (2) | Cl3—C8—C9—C10 | −176.9 (2) |
| Cu1i—Cu1—N1—C4 | −118.5 (2) | C8—C9—C10—C5 | −1.3 (5) |
| N1—Cu1—N2—C2 | 7.23 (19) | C6—C5—C10—C9 | 0.4 (5) |
| Cl1—Cu1—N2—C2 | 87.0 (3) | C4—C5—C10—C9 | 178.9 (3) |
| Cl2—Cu1—N2—C2 | −163.42 (19) | C2—N2—C11—C12 | −72.7 (3) |
| Cl1i—Cu1—N2—C2 | −73.45 (19) | Cu1—N2—C11—C12 | 56.7 (3) |
| Cu1i—Cu1—N2—C2 | −66.2 (2) | N2—C11—C12—C13 | 101.1 (3) |
| N1—Cu1—N2—C11 | −122.7 (2) | N2—C11—C12—C17 | −78.3 (4) |
| Cl1—Cu1—N2—C11 | −43.0 (3) | C17—C12—C13—C14 | 0.8 (5) |
| Cl2—Cu1—N2—C11 | 66.60 (19) | C11—C12—C13—C14 | −178.7 (3) |
| Cl1i—Cu1—N2—C11 | 156.57 (18) | C12—C13—C14—C15 | −1.0 (5) |
| Cu1i—Cu1—N2—C11 | 163.82 (15) | C13—C14—C15—C16 | 0.6 (5) |
| C4—N1—C1—C2 | −78.2 (3) | C13—C14—C15—Cl4 | −176.5 (3) |
| Cu1—N1—C1—C2 | 49.4 (3) | C14—C15—C16—C17 | −0.1 (5) |
| C11—N2—C2—C1 | 148.8 (3) | Cl4—C15—C16—C17 | 176.9 (3) |
| Cu1—N2—C2—C1 | 17.7 (3) | C15—C16—C17—C12 | 0.0 (5) |
| C11—N2—C2—C3 | −87.4 (3) | C13—C12—C17—C16 | −0.3 (5) |
| Cu1—N2—C2—C3 | 141.5 (2) | C11—C12—C17—C16 | 179.2 (3) |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···Cl2i | 0.91 | 2.57 | 3.450 (3) | 164 |
| C11—H11B···Cl2ii | 0.97 | 2.82 | 3.659 (3) | 145 |
| C13—H13···Cl3iii | 0.93 | 2.89 | 3.810 (3) | 170 |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+3/2, y−1/2, −z+3/2; (iii) −x+1/2, y−1/2, −z+3/2. |
(II) di-µ-chlorido-bis{[
N,
N'-bis(3,4-methylenedioxybenzyl)propane-1,2-
diamine]chloridocopper(II)}
top
Crystal data top
| [Cu2Cl4(C19H22N2O4)2] | F(000) = 980 |
| Mr = 953.65 | Dx = 1.487 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 2094 reflections |
| a = 12.2965 (13) Å | θ = 2.5–24.1° |
| b = 11.0554 (12) Å | µ = 1.30 mm−1 |
| c = 15.822 (2) Å | T = 298 K |
| β = 98.140 (2)° | Block, green |
| V = 2129.2 (4) Å3 | 0.45 × 0.37 × 0.31 mm |
| Z = 2 | |
Data collection top
Bruker SMART CCD area-detector
diffractometer | 3730 independent reflections |
| Radiation source: fine-focus sealed tube | 2008 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.162 |
| ϕ and ω scans | θmax = 25.0°, θmin = 1.7° |
Absorption correction: multi-scan
(SADABS; Sheldrick, 1996) | h = −14→6 |
| Tmin = 0.592, Tmax = 0.688 | k = −13→13 |
| 10028 measured reflections | l = −18→18 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.063 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.217 | H-atom parameters constrained |
| S = 1.14 | w = 1/[σ2(Fo2) + (0.059P)2 + 3.0479P]
where P = (Fo2 + 2Fc2)/3 |
| 3730 reflections | (Δ/σ)max < 0.001 |
| 254 parameters | Δρmax = 1.12 e Å−3 |
| 0 restraints | Δρmin = −0.64 e Å−3 |
Crystal data top
| [Cu2Cl4(C19H22N2O4)2] | V = 2129.2 (4) Å3 |
| Mr = 953.65 | Z = 2 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 12.2965 (13) Å | µ = 1.30 mm−1 |
| b = 11.0554 (12) Å | T = 298 K |
| c = 15.822 (2) Å | 0.45 × 0.37 × 0.31 mm |
| β = 98.140 (2)° | |
Data collection top
Bruker SMART CCD area-detector
diffractometer | 3730 independent reflections |
Absorption correction: multi-scan
(SADABS; Sheldrick, 1996) | 2008 reflections with I > 2σ(I) |
| Tmin = 0.592, Tmax = 0.688 | Rint = 0.162 |
| 10028 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.063 | 0 restraints |
| wR(F2) = 0.217 | H-atom parameters constrained |
| S = 1.14 | Δρmax = 1.12 e Å−3 |
| 3730 reflections | Δρmin = −0.64 e Å−3 |
| 254 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Cu1 | 0.54541 (8) | 0.43043 (8) | 0.40956 (6) | 0.0360 (3) | |
| Cl1 | 0.56158 (18) | 0.63216 (17) | 0.43920 (13) | 0.0444 (6) | |
| Cl2 | 0.37692 (16) | 0.44497 (17) | 0.32916 (12) | 0.0384 (5) | |
| O1 | 1.1333 (6) | 0.2222 (7) | 0.5665 (5) | 0.079 (2) | |
| O2 | 1.1506 (6) | 0.2989 (7) | 0.7031 (4) | 0.081 (2) | |
| O3 | 0.6540 (5) | 0.6293 (6) | 0.1355 (4) | 0.067 (2) | |
| O4 | 0.8296 (5) | 0.5707 (6) | 0.1196 (4) | 0.0583 (17) | |
| N1 | 0.6829 (5) | 0.3962 (6) | 0.4935 (4) | 0.0381 (16) | |
| H1 | 0.6713 | 0.4287 | 0.5443 | 0.046* | |
| N2 | 0.5678 (5) | 0.2588 (5) | 0.3672 (4) | 0.0348 (16) | |
| H2 | 0.5091 | 0.2159 | 0.3800 | 0.042* | |
| C1 | 0.6828 (7) | 0.2600 (7) | 0.5049 (5) | 0.042 (2) | |
| H1A | 0.6247 | 0.2363 | 0.5371 | 0.050* | |
| H1B | 0.7524 | 0.2337 | 0.5362 | 0.050* | |
| C2 | 0.6649 (7) | 0.2016 (7) | 0.4170 (5) | 0.039 (2) | |
| H2A | 0.7289 | 0.2201 | 0.3888 | 0.047* | |
| C3 | 0.6573 (9) | 0.0639 (7) | 0.4251 (6) | 0.066 (3) | |
| H3A | 0.5965 | 0.0436 | 0.4543 | 0.099* | |
| H3B | 0.7241 | 0.0336 | 0.4569 | 0.099* | |
| H3C | 0.6467 | 0.0283 | 0.3692 | 0.099* | |
| C4 | 0.7857 (7) | 0.4469 (8) | 0.4740 (6) | 0.049 (2) | |
| H4A | 0.7796 | 0.5344 | 0.4729 | 0.059* | |
| H4B | 0.7969 | 0.4206 | 0.4174 | 0.059* | |
| C5 | 0.8847 (6) | 0.4120 (7) | 0.5363 (5) | 0.041 (2) | |
| C6 | 0.9618 (7) | 0.3294 (8) | 0.5136 (6) | 0.049 (2) | |
| H6 | 0.9549 | 0.2969 | 0.4589 | 0.059* | |
| C7 | 1.0472 (7) | 0.2982 (9) | 0.5739 (6) | 0.052 (2) | |
| C8 | 1.0592 (7) | 0.3451 (9) | 0.6557 (6) | 0.053 (2) | |
| C9 | 0.9841 (7) | 0.4247 (9) | 0.6800 (6) | 0.060 (3) | |
| H9 | 0.9912 | 0.4551 | 0.7353 | 0.072* | |
| C10 | 0.8966 (7) | 0.4585 (8) | 0.6185 (6) | 0.054 (3) | |
| H10 | 0.8451 | 0.5134 | 0.6330 | 0.065* | |
| C11 | 1.1997 (8) | 0.2212 (11) | 0.6477 (7) | 0.075 (3) | |
| H11A | 1.2732 | 0.2491 | 0.6424 | 0.090* | |
| H11B | 1.2048 | 0.1397 | 0.6705 | 0.090* | |
| C12 | 0.5659 (7) | 0.2435 (7) | 0.2716 (5) | 0.042 (2) | |
| H12A | 0.5853 | 0.1608 | 0.2598 | 0.050* | |
| H12B | 0.4920 | 0.2581 | 0.2430 | 0.050* | |
| C13 | 0.6429 (7) | 0.3269 (7) | 0.2362 (5) | 0.0350 (19) | |
| C14 | 0.6058 (7) | 0.4436 (7) | 0.2080 (5) | 0.041 (2) | |
| H14 | 0.5359 | 0.4704 | 0.2149 | 0.049* | |
| C15 | 0.6743 (8) | 0.5138 (7) | 0.1712 (5) | 0.043 (2) | |
| C16 | 0.7790 (7) | 0.4808 (7) | 0.1605 (5) | 0.041 (2) | |
| C17 | 0.8188 (8) | 0.3702 (8) | 0.1886 (5) | 0.050 (2) | |
| H17 | 0.8900 | 0.3463 | 0.1831 | 0.060* | |
| C18 | 0.7476 (7) | 0.2941 (7) | 0.2262 (5) | 0.043 (2) | |
| H18 | 0.7728 | 0.2181 | 0.2451 | 0.051* | |
| C19 | 0.7520 (8) | 0.6666 (8) | 0.1051 (6) | 0.056 (2) | |
| H19A | 0.7810 | 0.7388 | 0.1351 | 0.068* | |
| H19B | 0.7371 | 0.6850 | 0.0447 | 0.068* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cu1 | 0.0484 (7) | 0.0262 (5) | 0.0315 (6) | 0.0033 (5) | −0.0014 (4) | −0.0017 (4) |
| Cl1 | 0.0587 (14) | 0.0258 (10) | 0.0474 (12) | −0.0026 (10) | 0.0033 (10) | −0.0019 (9) |
| Cl2 | 0.0439 (12) | 0.0357 (11) | 0.0339 (11) | 0.0037 (9) | −0.0006 (9) | 0.0004 (9) |
| O1 | 0.068 (5) | 0.100 (6) | 0.069 (5) | 0.028 (4) | 0.003 (4) | −0.019 (4) |
| O2 | 0.063 (5) | 0.102 (6) | 0.068 (5) | 0.026 (4) | −0.020 (4) | −0.024 (4) |
| O3 | 0.072 (5) | 0.044 (4) | 0.089 (5) | 0.010 (4) | 0.026 (4) | 0.028 (4) |
| O4 | 0.064 (4) | 0.051 (4) | 0.064 (4) | −0.004 (4) | 0.022 (3) | 0.006 (3) |
| N1 | 0.053 (4) | 0.034 (4) | 0.027 (3) | 0.015 (3) | 0.003 (3) | −0.001 (3) |
| N2 | 0.049 (4) | 0.028 (3) | 0.025 (3) | 0.003 (3) | −0.004 (3) | −0.001 (3) |
| C1 | 0.061 (6) | 0.034 (4) | 0.029 (4) | 0.004 (4) | 0.000 (4) | 0.011 (4) |
| C2 | 0.049 (5) | 0.031 (4) | 0.034 (5) | 0.008 (4) | −0.004 (4) | 0.004 (4) |
| C3 | 0.096 (8) | 0.029 (5) | 0.067 (7) | 0.004 (5) | −0.011 (6) | 0.002 (4) |
| C4 | 0.050 (6) | 0.046 (5) | 0.055 (6) | −0.006 (5) | 0.016 (5) | 0.005 (4) |
| C5 | 0.037 (5) | 0.033 (5) | 0.051 (5) | −0.003 (4) | 0.000 (4) | −0.004 (4) |
| C6 | 0.047 (5) | 0.059 (6) | 0.041 (5) | −0.008 (5) | 0.008 (4) | −0.004 (5) |
| C7 | 0.039 (5) | 0.054 (6) | 0.062 (6) | 0.001 (5) | 0.010 (5) | −0.009 (5) |
| C8 | 0.037 (5) | 0.059 (6) | 0.062 (6) | −0.003 (5) | 0.000 (5) | −0.010 (5) |
| C9 | 0.051 (6) | 0.066 (7) | 0.059 (6) | 0.004 (5) | −0.004 (5) | −0.027 (5) |
| C10 | 0.041 (5) | 0.045 (5) | 0.075 (7) | 0.005 (4) | 0.002 (5) | −0.019 (5) |
| C11 | 0.055 (7) | 0.084 (8) | 0.083 (8) | 0.022 (6) | 0.004 (6) | 0.010 (7) |
| C12 | 0.063 (6) | 0.036 (5) | 0.023 (4) | 0.009 (4) | −0.005 (4) | −0.006 (4) |
| C13 | 0.044 (5) | 0.037 (4) | 0.022 (4) | 0.005 (4) | 0.000 (4) | 0.003 (3) |
| C14 | 0.043 (5) | 0.040 (5) | 0.041 (5) | 0.008 (4) | 0.012 (4) | 0.004 (4) |
| C15 | 0.058 (6) | 0.034 (4) | 0.040 (5) | 0.008 (4) | 0.014 (4) | 0.006 (4) |
| C16 | 0.047 (5) | 0.039 (5) | 0.037 (5) | 0.001 (4) | 0.006 (4) | −0.002 (4) |
| C17 | 0.053 (6) | 0.054 (6) | 0.044 (5) | 0.007 (5) | 0.011 (5) | −0.011 (5) |
| C18 | 0.053 (6) | 0.036 (5) | 0.036 (5) | 0.006 (4) | −0.001 (4) | 0.000 (4) |
| C19 | 0.067 (7) | 0.045 (5) | 0.060 (6) | −0.004 (5) | 0.015 (5) | 0.006 (5) |
Geometric parameters (Å, º) top
| Cu1—N1 | 2.032 (6) | C4—C5 | 1.504 (11) |
| Cu1—N2 | 2.044 (6) | C4—H4A | 0.9700 |
| Cu1—Cl2 | 2.278 (2) | C4—H4B | 0.9700 |
| Cu1—Cl1 | 2.282 (2) | C5—C10 | 1.387 (12) |
| Cu1—Cl1i | 2.971 (2) | C5—C6 | 1.399 (11) |
| Cu1—Cu1i | 3.565 (2) | C6—C7 | 1.360 (12) |
| Cl1—Cu1i | 2.971 (2) | C6—H6 | 0.9300 |
| O1—C7 | 1.371 (10) | C7—C8 | 1.382 (12) |
| O1—C11 | 1.420 (12) | C8—C9 | 1.369 (12) |
| O2—C8 | 1.360 (10) | C9—C10 | 1.395 (12) |
| O2—C11 | 1.422 (12) | C9—H9 | 0.9300 |
| O3—C15 | 1.404 (10) | C10—H10 | 0.9300 |
| O3—C19 | 1.422 (10) | C11—H11A | 0.9700 |
| O4—C16 | 1.381 (10) | C11—H11B | 0.9700 |
| O4—C19 | 1.423 (10) | C12—C13 | 1.486 (11) |
| N1—C4 | 1.456 (10) | C12—H12A | 0.9700 |
| N1—C1 | 1.517 (10) | C12—H12B | 0.9700 |
| N1—H1 | 0.9100 | C13—C18 | 1.369 (11) |
| N2—C2 | 1.477 (9) | C13—C14 | 1.419 (10) |
| N2—C12 | 1.519 (9) | C14—C15 | 1.338 (11) |
| N2—H2 | 0.9100 | C14—H14 | 0.9300 |
| C1—C2 | 1.521 (10) | C15—C16 | 1.373 (11) |
| C1—H1A | 0.9700 | C16—C17 | 1.368 (11) |
| C1—H1B | 0.9700 | C17—C18 | 1.405 (12) |
| C2—C3 | 1.531 (11) | C17—H17 | 0.9300 |
| C2—H2A | 0.9800 | C18—H18 | 0.9300 |
| C3—H3A | 0.9600 | C19—H19A | 0.9700 |
| C3—H3B | 0.9600 | C19—H19B | 0.9700 |
| C3—H3C | 0.9600 | | |
| | | |
| N1—Cu1—N2 | 84.5 (2) | H4A—C4—H4B | 107.6 |
| N1—Cu1—Cl2 | 170.0 (2) | C10—C5—C6 | 119.8 (8) |
| N2—Cu1—Cl2 | 92.1 (2) | C10—C5—C4 | 119.2 (8) |
| N1—Cu1—Cl1 | 90.4 (2) | C6—C5—C4 | 121.0 (8) |
| N2—Cu1—Cl1 | 164.3 (2) | C7—C6—C5 | 118.0 (8) |
| Cl2—Cu1—Cl1 | 95.26 (8) | C7—C6—H6 | 121.0 |
| N1—Cu1—Cl1i | 81.5 (2) | C5—C6—H6 | 121.0 |
| N2—Cu1—Cl1i | 98.3 (2) | C6—C7—O1 | 128.7 (9) |
| Cl2—Cu1—Cl1i | 89.70 (7) | C6—C7—C8 | 122.1 (9) |
| Cl1—Cu1—Cl1i | 95.61 (7) | O1—C7—C8 | 109.2 (8) |
| N1—Cu1—Cu1i | 83.2 (2) | O2—C8—C9 | 128.6 (9) |
| N2—Cu1—Cu1i | 137.40 (18) | O2—C8—C7 | 110.2 (8) |
| Cl2—Cu1—Cu1i | 93.11 (6) | C9—C8—C7 | 121.2 (9) |
| Cl1—Cu1—Cu1i | 56.04 (6) | C8—C9—C10 | 117.4 (9) |
| Cl1i—Cu1—Cu1i | 39.57 (4) | C8—C9—H9 | 121.3 |
| Cu1—Cl1—Cu1i | 84.39 (7) | C10—C9—H9 | 121.3 |
| C7—O1—C11 | 106.2 (7) | C5—C10—C9 | 121.6 (8) |
| C8—O2—C11 | 106.0 (7) | C5—C10—H10 | 119.2 |
| C15—O3—C19 | 106.8 (7) | C9—C10—H10 | 119.2 |
| C16—O4—C19 | 106.0 (6) | O1—C11—O2 | 108.3 (8) |
| C4—N1—C1 | 115.0 (6) | O1—C11—H11A | 110.0 |
| C4—N1—Cu1 | 117.2 (5) | O2—C11—H11A | 110.0 |
| C1—N1—Cu1 | 104.4 (5) | O1—C11—H11B | 110.0 |
| C4—N1—H1 | 106.5 | O2—C11—H11B | 110.0 |
| C1—N1—H1 | 106.5 | H11A—C11—H11B | 108.4 |
| Cu1—N1—H1 | 106.5 | C13—C12—N2 | 112.7 (6) |
| C2—N2—C12 | 112.4 (6) | C13—C12—H12A | 109.1 |
| C2—N2—Cu1 | 111.0 (4) | N2—C12—H12A | 109.1 |
| C12—N2—Cu1 | 116.5 (4) | C13—C12—H12B | 109.1 |
| C2—N2—H2 | 105.3 | N2—C12—H12B | 109.1 |
| C12—N2—H2 | 105.3 | H12A—C12—H12B | 107.8 |
| Cu1—N2—H2 | 105.3 | C18—C13—C14 | 118.1 (8) |
| N1—C1—C2 | 108.3 (6) | C18—C13—C12 | 122.6 (7) |
| N1—C1—H1A | 110.0 | C14—C13—C12 | 119.3 (7) |
| C2—C1—H1A | 110.0 | C15—C14—C13 | 118.1 (8) |
| N1—C1—H1B | 110.0 | C15—C14—H14 | 121.0 |
| C2—C1—H1B | 110.0 | C13—C14—H14 | 121.0 |
| H1A—C1—H1B | 108.4 | C14—C15—C16 | 124.0 (8) |
| N2—C2—C1 | 108.0 (6) | C14—C15—O3 | 128.0 (8) |
| N2—C2—C3 | 114.6 (7) | C16—C15—O3 | 108.0 (8) |
| C1—C2—C3 | 110.4 (7) | C17—C16—C15 | 119.8 (8) |
| N2—C2—H2A | 107.9 | C17—C16—O4 | 129.2 (8) |
| C1—C2—H2A | 107.9 | C15—C16—O4 | 111.1 (7) |
| C3—C2—H2A | 107.9 | C16—C17—C18 | 117.2 (8) |
| C2—C3—H3A | 109.5 | C16—C17—H17 | 121.4 |
| C2—C3—H3B | 109.5 | C18—C17—H17 | 121.4 |
| H3A—C3—H3B | 109.5 | C13—C18—C17 | 122.9 (8) |
| C2—C3—H3C | 109.5 | C13—C18—H18 | 118.5 |
| H3A—C3—H3C | 109.5 | C17—C18—H18 | 118.5 |
| H3B—C3—H3C | 109.5 | O3—C19—O4 | 108.1 (7) |
| N1—C4—C5 | 114.1 (7) | O3—C19—H19A | 110.1 |
| N1—C4—H4A | 108.7 | O4—C19—H19A | 110.1 |
| C5—C4—H4A | 108.7 | O3—C19—H19B | 110.1 |
| N1—C4—H4B | 108.7 | O4—C19—H19B | 110.1 |
| C5—C4—H4B | 108.7 | H19A—C19—H19B | 108.4 |
| | | |
| N1—Cu1—Cl1—Cu1i | −81.51 (19) | C5—C6—C7—C8 | 0.2 (14) |
| N2—Cu1—Cl1—Cu1i | −152.2 (6) | C11—O1—C7—C6 | 179.7 (10) |
| Cl2—Cu1—Cl1—Cu1i | 90.21 (7) | C11—O1—C7—C8 | 1.0 (11) |
| Cl1i—Cu1—Cl1—Cu1i | 0.0 | C11—O2—C8—C9 | −179.8 (11) |
| N2—Cu1—N1—C4 | 101.1 (6) | C11—O2—C8—C7 | 1.3 (11) |
| Cl2—Cu1—N1—C4 | 171.3 (8) | C6—C7—C8—O2 | 179.8 (9) |
| Cl1—Cu1—N1—C4 | −64.0 (6) | O1—C7—C8—O2 | −1.4 (11) |
| Cl1i—Cu1—N1—C4 | −159.6 (6) | C6—C7—C8—C9 | 0.8 (15) |
| Cu1i—Cu1—N1—C4 | −119.7 (6) | O1—C7—C8—C9 | 179.6 (9) |
| N2—Cu1—N1—C1 | −27.4 (5) | O2—C8—C9—C10 | 179.9 (9) |
| Cl2—Cu1—N1—C1 | 42.8 (13) | C7—C8—C9—C10 | −1.3 (15) |
| Cl1—Cu1—N1—C1 | 167.5 (5) | C6—C5—C10—C9 | −0.1 (14) |
| Cl1i—Cu1—N1—C1 | 71.9 (5) | C4—C5—C10—C9 | 177.0 (8) |
| Cu1i—Cu1—N1—C1 | 111.8 (5) | C8—C9—C10—C5 | 1.0 (14) |
| N1—Cu1—N2—C2 | 0.8 (5) | C7—O1—C11—O2 | −0.2 (11) |
| Cl2—Cu1—N2—C2 | −169.7 (5) | C8—O2—C11—O1 | −0.7 (12) |
| Cl1—Cu1—N2—C2 | 72.3 (8) | C2—N2—C12—C13 | −77.2 (8) |
| Cl1i—Cu1—N2—C2 | −79.7 (5) | Cu1—N2—C12—C13 | 52.6 (8) |
| Cu1i—Cu1—N2—C2 | −72.8 (6) | N2—C12—C13—C18 | 94.1 (9) |
| N1—Cu1—N2—C12 | −129.5 (6) | N2—C12—C13—C14 | −88.7 (8) |
| Cl2—Cu1—N2—C12 | 59.9 (5) | C18—C13—C14—C15 | 1.7 (11) |
| Cl1—Cu1—N2—C12 | −58.1 (9) | C12—C13—C14—C15 | −175.6 (7) |
| Cl1i—Cu1—N2—C12 | 149.9 (5) | C13—C14—C15—C16 | −1.2 (13) |
| Cu1i—Cu1—N2—C12 | 156.8 (4) | C13—C14—C15—O3 | 177.1 (8) |
| C4—N1—C1—C2 | −80.3 (8) | C19—O3—C15—C14 | 179.6 (9) |
| Cu1—N1—C1—C2 | 49.5 (7) | C19—O3—C15—C16 | −1.8 (9) |
| C12—N2—C2—C1 | 158.9 (6) | C14—C15—C16—C17 | −0.2 (13) |
| Cu1—N2—C2—C1 | 26.3 (7) | O3—C15—C16—C17 | −178.9 (7) |
| C12—N2—C2—C3 | −77.7 (9) | C14—C15—C16—O4 | 179.2 (8) |
| Cu1—N2—C2—C3 | 149.8 (6) | O3—C15—C16—O4 | 0.5 (10) |
| N1—C1—C2—N2 | −50.7 (9) | C19—O4—C16—C17 | −179.7 (9) |
| N1—C1—C2—C3 | −176.6 (7) | C19—O4—C16—C15 | 1.0 (9) |
| C1—N1—C4—C5 | −53.5 (9) | C15—C16—C17—C18 | 1.2 (12) |
| Cu1—N1—C4—C5 | −176.7 (5) | O4—C16—C17—C18 | −178.1 (8) |
| N1—C4—C5—C10 | −69.7 (10) | C14—C13—C18—C17 | −0.7 (12) |
| N1—C4—C5—C6 | 107.3 (9) | C12—C13—C18—C17 | 176.5 (7) |
| C10—C5—C6—C7 | −0.5 (13) | C16—C17—C18—C13 | −0.7 (12) |
| C4—C5—C6—C7 | −177.5 (8) | C15—O3—C19—O4 | 2.4 (10) |
| C5—C6—C7—O1 | −178.4 (9) | C16—O4—C19—O3 | −2.1 (9) |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···Cl2i | 0.91 | 2.58 | 3.476 (6) | 169 |
| N2—H2···O3ii | 0.91 | 2.20 | 3.075 (9) | 160 |
| C12—H12A···Cl2ii | 0.97 | 2.84 | 3.774 (8) | 162 |
| C11—H11A···Cl1iii | 0.97 | 2.87 | 3.779 (11) | 156 |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+1, y−1/2, −z+1/2; (iii) −x+2, −y+1, −z+1. |
Experimental details
| | (I) | (II) |
| Crystal data |
| Chemical formula | [Cu2Cl4(C17H20Cl2N2)2] | [Cu2Cl4(C19H22N2O4)2] |
| Mr | 915.38 | 953.65 |
| Crystal system, space group | Monoclinic, P21/n | Monoclinic, P21/c |
| Temperature (K) | 298 | 298 |
| a, b, c (Å) | 14.4217 (15), 9.5445 (13), 15.6198 (19) | 12.2965 (13), 11.0554 (12), 15.822 (2) |
| α, β, γ (°) | 90, 115.647 (2), 90 | 90, 98.140 (2), 90 |
| V (Å3) | 1938.2 (4) | 2129.2 (4) |
| Z | 2 | 2 |
| Radiation type | Mo Kα | Mo Kα |
| µ (mm−1) | 1.68 | 1.30 |
| Crystal size (mm) | 0.54 × 0.50 × 0.46 | 0.45 × 0.37 × 0.31 |
| |
| Data collection |
| Diffractometer | Bruker SMART CCD area-detector
diffractometer | Bruker SMART CCD area-detector
diffractometer |
| Absorption correction | Multi-scan
(SADABS; Sheldrick, 1996) | Multi-scan
(SADABS; Sheldrick, 1996) |
| Tmin, Tmax | 0.464, 0.512 | 0.592, 0.688 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 8896, 3417, 2484 | 10028, 3730, 2008 |
| Rint | 0.038 | 0.162 |
| (sin θ/λ)max (Å−1) | 0.595 | 0.595 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.031, 0.082, 1.06 | 0.063, 0.217, 1.14 |
| No. of reflections | 3417 | 3730 |
| No. of parameters | 218 | 254 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.39, −0.28 | 1.12, −0.64 |
Selected geometric parameters (Å, º) for (I) top| Cu1—N1 | 2.026 (2) | Cu1—Cl2 | 2.2742 (9) |
| Cu1—N2 | 2.033 (2) | Cu1—Cl1i | 2.852 (1) |
| Cu1—Cl1 | 2.2604 (9) | Cu1—Cu1i | 3.4285 (8) |
| | | |
| N1—Cu1—N2 | 83.69 (9) | Cl1—Cu1—Cl2 | 96.18 (3) |
| N1—Cu1—Cl1 | 92.41 (7) | N1—Cu1—Cl1i | 82.32 (7) |
| N2—Cu1—Cl1 | 159.84 (8) | N2—Cu1—Cl1i | 102.40 (7) |
| N1—Cu1—Cl2 | 169.01 (7) | Cl1—Cu1—Cl1i | 96.64 (3) |
| N2—Cu1—Cl2 | 90.49 (7) | Cl2—Cu1—Cl1i | 89.87 (3) |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) for (I) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···Cl2i | 0.91 | 2.57 | 3.450 (3) | 164.4 |
| C11—H11B···Cl2ii | 0.97 | 2.82 | 3.659 (3) | 145.2 |
| C13—H13···Cl3iii | 0.93 | 2.89 | 3.810 (3) | 169.5 |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+3/2, y−1/2, −z+3/2; (iii) −x+1/2, y−1/2, −z+3/2. |
Selected geometric parameters (Å, º) for (II) top| Cu1—N1 | 2.032 (6) | Cu1—Cl1 | 2.282 (2) |
| Cu1—N2 | 2.044 (6) | Cu1—Cl1i | 2.971 (2) |
| Cu1—Cl2 | 2.278 (2) | Cu1—Cu1i | 3.565 (2) |
| | | |
| N1—Cu1—N2 | 84.5 (2) | N1—Cu1—Cl1i | 81.5 (2) |
| N1—Cu1—Cl2 | 170.0 (2) | N2—Cu1—Cl1i | 98.3 (2) |
| N2—Cu1—Cl2 | 92.1 (2) | Cl2—Cu1—Cl1i | 89.70 (7) |
| N1—Cu1—Cl1 | 90.4 (2) | Cl1—Cu1—Cl1i | 95.61 (7) |
| N2—Cu1—Cl1 | 164.3 (2) | Cu1—Cl1—Cu1i | 84.39 (7) |
| Cl2—Cu1—Cl1 | 95.26 (8) | | |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) for (II) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···Cl2i | 0.91 | 2.58 | 3.476 (6) | 169.1 |
| N2—H2···O3ii | 0.91 | 2.20 | 3.075 (9) | 159.9 |
| C12—H12A···Cl2ii | 0.97 | 2.84 | 3.774 (8) | 161.6 |
| C11—H11A···Cl1iii | 0.97 | 2.87 | 3.779 (11) | 155.6 |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+1, y−1/2, −z+1/2; (iii) −x+2, −y+1, −z+1. |

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.
 Cu separations and Cu—Cl—Cu angles are 3.4285 (8) Å and 83.36 (3)°, respectively, for (I), and 3.565 (2) Å and 84.39 (7)° for (II). Each Cu atom is five-coordinated and the coordination geometry around the Cu atom is best described as a distorted square-pyramid with a τ value of 0.155 (3) for (I) and 0.092 (7) for (II). The apical Cu—Cl bond length is 2.852 (1) Å for (I) and 2.971 (2) Å for (II). The basal Cu—Cl and Cu—N average bonds lengths are 2.2673 (9) and 2.030 (2) Å, respectively, for (I), and 2.280 (2) and 2.038 (6) Å for (II). The molecules of (I) are linked by one C—H
Cu separations and Cu—Cl—Cu angles are 3.4285 (8) Å and 83.36 (3)°, respectively, for (I), and 3.565 (2) Å and 84.39 (7)° for (II). Each Cu atom is five-coordinated and the coordination geometry around the Cu atom is best described as a distorted square-pyramid with a τ value of 0.155 (3) for (I) and 0.092 (7) for (II). The apical Cu—Cl bond length is 2.852 (1) Å for (I) and 2.971 (2) Å for (II). The basal Cu—Cl and Cu—N average bonds lengths are 2.2673 (9) and 2.030 (2) Å, respectively, for (I), and 2.280 (2) and 2.038 (6) Å for (II). The molecules of (I) are linked by one C—H Cl hydrogen bond into a complex [10
Cl hydrogen bond into a complex [10 Cl and one N—H
Cl and one N—H O hydrogen bond into a complex [100] sheet.
O hydrogen bond into a complex [100] sheet.
 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2007/12/00/hj3060/hj3060fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2007/12/00/hj3060/hj3060fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2007/12/00/hj3060/hj3060fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/c/issues/2007/12/00/hj3060/hj3060fig4thm.gif)

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry



Dinuclear transition metal complexes have received much attention because of their relevance as models for active sites of biomolecules, such as hemocyanin, tyrosinase and copper oxidases (Pradeep et al., 2005; Sorrell, 1989), and also because of the correlation between the structure and the magnetic behaviour (Schuitema et al., 2002; Rodríguez et al., 2000). Among these dinuclear transition metal complexes, some anions, such as OH-, RO-, RCO2-, Cl- and Br-, are often used as bridging ligands (Christou et al., 2000). For dichloro-bridged dinuclear copper systems, many complexes have been reported in the literature and different magnetic behaviours are observed (Rodríguez et al., 2000; Schuitema et al., 2002; Tuna et al., 1999), and the magneto-structural correlation, which relates the magnetic interaction to ϕ/R, has been established (Marsh et al., 1982). Copper complexes containing diimines also have been extensively studied and reported owing to their potential applications (Alvesa et al., 2004). Previously, our group has reported the structures of some transition metal complexes with diimines (Han et al., 2006; Liu et al., 2007; Xia et al., 2007; Yang et al., 2007). For this reason, we have synthesized two new dichloro-bridged dinuclear copper complexes with diimines, [Cu2Cl2(C17H20N2Cl2)2Cl2], (I), and [Cu2Cl2(C19H22N2O4)2Cl2], (II). In this paper, we report the synthesis and structures of complexes (I) and (II).
Complexes (I) and (II) have similar coordination environments (Fig. 1 and Fig. 2). In each complex, the asymmetric unit consists of one half-molecule; two copper centers are bridged by a pair of chloride atoms, resulting in a complex with a centrosymmetric structure containing a parallelogram Cu(µ-Cl)2Cu core. The Cu···Cu separation of 3.4285 (8) Å for (I) is smaller than those values reported in two dichloro-bridged dinuclear copper(II) complexes, viz. [Cu2 (5-aminomethyl-3-methylpyrazole)2Cl4] [Cu···Cu = 3.479 (4) Å; Schuitema et al., 2002] and {[CuL2Cl2]2[ClO4]2} [where L is 1-(imidazol-4-ylmethyl)-l,5-diazacyclooctane; Cu···Cu = 3.494 (8) Å; Bu et al., 2001], in which magnetic coupling between the copper centers can be observed; but the Cu···Cu separation of 3.565 (2) Å for (II) is longer than those of two dichloro-bridged dinuclear copper(II) complexes mentioned above. The Cu—Cl—Cu angles are 83.36 (3)° for (I) and 84.39 (7)° for (II); these values are smaller than those of the two dichloro-bridged dinuclear copper(II) complexes mentioned above, in which the Cu—Cl—Cu angles range from 85.33 (15) to 88.81 (5)°. Each Cu atom is five-coordinated by two N atoms of one ligand and three Cl atoms, and the coordination polyhedron around the Cu atom may be described as a slightly distorted square pyramid with τ values (Addison et al., 1984) of 0.155 (3) for (I) and 0.092 (7) for (II), where τ is defined as (β - α)/60, and β and α are the largest coordination angles (τ = 0 for a regular square-pyramidal geometry and τ = 1 for trigonal–bipyramidal geometry). The apical position is occupied by one elongated bridging Cl atom, and the apical Cu—Cl bond distances are 2.852 (1) Å for (I), and 2.971 (2) Å for (II); these values are longer than those of the two dichloro-bridged dinuclear copper(II) complexes mentioned above, in which the apical Cu—Cl bond distances range from 2.6571 (14) to 2.829 (4) Å. The basal planes are defined by atoms Cl1, Cl2, N1 and N2; atoms Cu1 are shifted by 0.106 (1) Å for (I) and 0.050 (3) Å for (II) from the basal planes toward the apical Cl1A atom. The basal Cu—Cl and Cu–N average bond lengths are 2.2673 (9) and 2.030 (2) Å in (I), and 2.280 (2) and 2.038 (6) Å in (II) (see in Table 1 and Table 3).
Generally, the dichloro-bridged dinuclear copper complexes with square-pyramidal geometry exhibit three different geometries with regard to the relative arrangement of square pyramids, viz. perpendicular bases (type I), parallel bases (type II) and coplanar bases (type III) (Rodríguez et al., 2000). In the parallel bases case, two square pyramids share a base-to-apex edge, so that a Cl atom situated at the vertex of one base becomes the apical vertex of the other square pyramid. The symmetric arrangements of the Cu(µ-Cl)2Cu cores in complexes (I) and (II) belong to type II; the two intramolecular basal planes are strictly parallel, with an interplanar spacing of 2.731 (6) Å for (I) and 2.82 (3) Å for (II), corresponding to a plane offset of 2.07 (1) Å for (I) and 2.18 (4) Å for (II); the dihedral angles between the parallelogram Cu(µ-Cl)2Cu core and the basal plane are 87.27 (4)° for (I) and 86.85 (8)° for (II).
Some theoretical analysis and real experiments concerning the magneto-structural correlation for dichloro-bridged dinuclear copper(II) complexes can be found in the literature (Schuitema et al., 2002; Rodríguez et al., 2000; Tuna et al., 1999). Marsh et al. (1982) have shown that the exchange coupling interaction depends on the magnitude of the angle at the Cu—Cl—Cu bridge, ϕ, as well as the apical Cu—Cl bond length, R. It was found that for values of the quotient ϕ/R that are lower than 32.6° Å-1 and higher than 34.88° Å-1, the exchange interaction is antiferromagnetic. For values falling between these limits, the exchange interaction was found to be ferromagnetic. In the cases of (I) and (II), the values of the quotient ϕ/R are 29.23 (2) and 28.40 (4)° Å-1, respectively, both quotients are below the border value of 32.6° Å-1, suggesting an antiferromagnetic interaction between the two copper(II) ions.
In the crystal structures of (I) and (II), the molecules are stabilized by a pair of intramolecular N—H···Cl hydrogen bonds (Figs. 1 and 2, and Tables 2 and 4). In (I), the molecules are linked by a single intermolecular C—H···Cl hydrogen bond, atom C11 at (x, y, z) in molecule centred at (1/2, 1/2, 1/2) acts as hydrogen-bond donor, via H11b, to atom Cl2 at (3/2 - x, -1/2 + y, 3/2 - z) in the molecule centred at (1, 0, 1), and propagation is via inversion, the screw axis and translation generating a complex sheet in [101] (Fig. 3 and Table 2).
In (II), the molecules are linked by one intermolecular C—H···Cl and one N—H···O hydrogen bonds, atoms C12 and N2 at (x, y, z) in the molecule centred at (1/2, 1/2, 1/2) act as hydrogen-bond donors, via H12B [12 A in Table 4] and H2, to atoms Cl2 and O3 at (3/2 - x, -1/2 + y, 3/2 - z) [or (-x + 1, y - 1/2, -z + 1/2)?] in the molecule centred at [1, 0, 1], and propagation via a screw axis and translation generates a complex sheet in [100] (Fig. 4 and Table 4).