Buy article online - an online subscription or single-article purchase is required to access this article.
The title compound, alternatively sodium pyridin-2-olate trihydrate, Na
+·C
5H
3N
2O
3−·3H
2O, crystallizes in the
P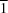
space group. It is made up of edge-shared chains of NaO
6 octahedra with five water molecules and one 5-nitro-2-pyridonate anion. Four of these water molecules are bicoordinating, involved in connecting the adjacent octahedra, and the fifth is coordinated to only one octahedron. The crystal structure is stabilized by a network of strong O—H

O and O—H

N interactions. The organic moieties occupy the space between the chains with an antiparallel alignment.
Supporting information
CCDC reference: 150761
Data collection: KappaCCD (Enraf-Nonius, 1998); cell refinement: KappaCCD; data reduction: TEXSAN (Molecular Structure Corporation, 1997); program(s) used to solve structure: SIR92 (Altomare et al., 1993); program(s) used to refine structure: TEXSAN; molecular graphics: ORTEP (Johnson, 1976) and MOLVIEW (Cense, 1990); software used to prepare material for publication: TEXSAN.
Crystal data top
| Na(C5H3N2O3)·3H2O | Z = 2 |
| Mr = 216.13 | F(000) = 224.00 |
| Triclinic, P1 | Dx = 1.562 Mg m−3 |
| a = 7.0044 (5) Å | Ag Kα radiation, λ = 0.5608 Å |
| b = 11.182 (1) Å | Cell parameters from 2258 reflections |
| c = 6.3971 (8) Å | θ = 2,81–22,61° |
| α = 102.978 (8)° | µ = 0.10 mm−1 |
| β = 104.697 (4)° | T = 296 K |
| γ = 74.017 (8)° | Needle, yellow |
| V = 459.40 (9) Å3 | 0.34 × 0.14 × 0.11 mm |
Data collection top
KappaCCD
diffractometer | 1573 reflections with I > 3.00σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.000 |
| Graphite monochromator | θmax = 22.6°, θmin = 2.8° |
| φ scans | h = −9→8 |
| 2258 measured reflections | k = −15→14 |
| 2258 independent reflections | l = 0→8 |
Refinement top
| Refinement on F | 0 restraints |
| Least-squares matrix: full | 0 constraints |
| R[F2 > 2σ(F2)] = 0.051 | H-atom parameters not refined |
| wR(F2) = 0.057 | Weighting scheme based on measured s.u.'s w = 1/[σ2(Fo)] |
| S = 1.91 | (Δ/σ)max < 0.001 |
| 1573 reflections | Δρmax = 0.31 e Å−3 |
| 127 parameters | Δρmin = −0.28 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Na1 | 0.60107 (8) | 0.44741 (5) | 0.75827 (8) | 0.0414 (2) | |
| O1 | 0.9347 (2) | −0.30740 (9) | 0.6454 (2) | 0.0450 (3) | |
| O2 | 0.6122 (2) | 0.13815 (11) | 0.1563 (2) | 0.0690 (4) | |
| O3 | 0.6263 (2) | 0.24290 (10) | 0.4802 (2) | 0.0698 (4) | |
| O4 | 0.9128 (2) | 0.3444 (2) | 0.9266 (2) | 0.0906 (5) | |
| O5 | 0.5640 (2) | 0.63327 (9) | 1.0299 (2) | 0.0470 (3) | |
| O6 | 0.27311 (13) | 0.50049 (9) | 0.5196 (2) | 0.0406 (3) | |
| N1 | 0.7957 (2) | −0.19659 (11) | 0.3649 (2) | 0.0501 (4) | |
| N2 | 0.6485 (2) | 0.14286 (11) | 0.3564 (2) | 0.0391 (4) | |
| C1 | 0.8650 (2) | −0.20009 (12) | 0.5839 (2) | 0.0328 (4) | |
| C2 | 0.8558 (2) | −0.08602 (13) | 0.7355 (2) | 0.0411 (4) | |
| C3 | 0.7846 (2) | 0.02772 (13) | 0.6660 (2) | 0.0406 (4) | |
| C4 | 0.7209 (2) | 0.02791 (12) | 0.4430 (2) | 0.0310 (4) | |
| C5 | 0.7277 (2) | −0.08527 (13) | 0.3005 (2) | 0.0464 (4) | |
| H1 | 0.9064 | −0.0890 | 0.9084 | 0.062* | |
| H2 | 0.7776 | 0.1143 | 0.7810 | 0.061* | |
| H3 | 0.6745 | −0.0824 | 0.1273 | 0.070* | |
| H4 | 1.0199 | 0.3138 | 0.8470 | 0.137* | |
| H5 | 0.9576 | 0.3296 | 1.0336 | 0.137* | |
| H6 | 0.6662 | 0.6673 | 1.1405 | 0.071* | |
| H7 | 0.5131 | 0.6862 | 0.9925 | 0.071* | |
| H8 | 0.1888 | 0.5589 | 0.5766 | 0.061* | |
| H9 | 0.2200 | 0.4385 | 0.4619 | 0.061* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Na1 | 0.0456 (4) | 0.0411 (4) | 0.0359 (4) | −0.0055 (3) | 0.0073 (2) | 0.0100 (3) |
| O1 | 0.0548 (6) | 0.0331 (6) | 0.0442 (6) | −0.0059 (5) | 0.0039 (5) | 0.0133 (5) |
| O2 | 0.1104 (10) | 0.0446 (7) | 0.0462 (8) | −0.0076 (7) | 0.0053 (6) | 0.0208 (6) |
| O3 | 0.1015 (10) | 0.0264 (6) | 0.0678 (9) | −0.0067 (6) | 0.0088 (7) | 0.0003 (6) |
| O4 | 0.0630 (8) | 0.1352 (13) | 0.0460 (8) | 0.0288 (8) | 0.0081 (6) | 0.0276 (8) |
| O5 | 0.0608 (7) | 0.0361 (6) | 0.0439 (6) | −0.0153 (5) | 0.0000 (5) | 0.0131 (5) |
| O6 | 0.0380 (5) | 0.0329 (5) | 0.0501 (6) | −0.0059 (4) | 0.0103 (4) | 0.0066 (4) |
| N1 | 0.0832 (9) | 0.0269 (7) | 0.0315 (7) | −0.0061 (6) | 0.0049 (6) | 0.0031 (5) |
| N2 | 0.0413 (7) | 0.0301 (7) | 0.0448 (8) | −0.0076 (5) | 0.0062 (5) | 0.0079 (6) |
| C1 | 0.0327 (7) | 0.0330 (8) | 0.0332 (8) | −0.0067 (5) | 0.0061 (5) | 0.0083 (6) |
| C2 | 0.0513 (8) | 0.0393 (9) | 0.0291 (8) | −0.0105 (7) | 0.0019 (6) | 0.0055 (6) |
| C3 | 0.0505 (8) | 0.0334 (8) | 0.0335 (8) | −0.0104 (6) | 0.0058 (6) | −0.0012 (6) |
| C4 | 0.0327 (7) | 0.0264 (7) | 0.0338 (8) | −0.0058 (5) | 0.0071 (5) | 0.0053 (6) |
| C5 | 0.0724 (10) | 0.0318 (8) | 0.0294 (8) | −0.0059 (7) | 0.0080 (7) | 0.0043 (6) |
Geometric parameters (Å, º) top
| Na1—O3 | 2.556 (2) | O6—H8 | 0.84 |
| Na1—O4 | 2.288 (2) | O6—H9 | 0.84 |
| Na1—O5 | 2.391 (2) | N1—C1 | 1.367 (2) |
| Na1—O5i | 2.430 (2) | N1—C5 | 1.323 (3) |
| Na1—O6 | 2.416 (1) | N2—C4 | 1.426 (2) |
| Na1—O6ii | 2.412 (1) | C1—C2 | 1.415 (3) |
| O1—C1 | 1.275 (2) | C2—C3 | 1.362 (3) |
| O2—N2 | 1.232 (2) | C2—H1 | 1.08 |
| O3—N2 | 1.213 (2) | C3—C4 | 1.383 (3) |
| O4—H4 | 0.96 | C3—H2 | 1.07 |
| O4—H5 | 0.71 | C4—C5 | 1.380 (3) |
| O5—H6 | 0.97 | C5—H3 | 1.08 |
| O5—H7 | 0.66 | | |
| | | |
| O1···H9iii | 1.9598 | O2···H3vi | 2.4589 |
| O1···H5iv | 2.0465 | O2···C5vi | 3.320 (3) |
| O1···H8v | 2.0571 | O4···N1vii | 3.011 (2) |
| O1···O4iv | 2.755 (2) | O5···N1viii | 2.992 (2) |
| O1···O6iii | 2.797 (2) | N1···H6ix | 2.0774 |
| O1···O6v | 2.875 (2) | N1···H4vii | 2.0931 |
| | | |
| O3—Na1—O4 | 83.10 (7) | Na1—O6—Na1 | 95.45 (5) |
| O3—Na1—O5 | 176.08 (6) | Na1—O6—H8 | 114.5 |
| O3—Na1—O5i | 91.06 (6) | Na1—O6—H9 | 114.2 |
| O3—Na1—O6 | 81.50 (6) | Na1—O6ii—H8ii | 115.3 |
| O3—Na1—O6ii | 78.25 (6) | Na1—O6ii—H9ii | 107.7 |
| O4—Na1—O5 | 97.38 (7) | H8—O6—H9 | 109.1 |
| O4—Na1—O5i | 90.69 (6) | C1—N1—C5 | 118.4 (2) |
| O4—Na1—O6 | 164.41 (7) | O2—N2—O3 | 121.3 (2) |
| O4—Na1—O6ii | 94.65 (6) | O2—N2—C4 | 118.9 (2) |
| O5—Na1—O5 | 85.05 (5) | O3—N2—C4 | 119.7 (2) |
| O5—Na1—O6 | 97.82 (5) | O1—C1—N1 | 118.4 (2) |
| O5—Na1—O6ii | 105.56 (5) | O1—C1—C2 | 121.9 (2) |
| O5i—Na1—O6 | 87.22 (5) | N1—C1—C2 | 119.7 (2) |
| O5i—Na1—O6ii | 167.35 (5) | C1—C2—C3 | 120.9 (2) |
| O6—Na1—O6 | 84.55 (5) | C1—C2—H1 | 119.6 |
| Na1—O3—N2 | 176.2 (1) | C3—C2—H1 | 119.4 |
| Na1—O4—H4 | 119.2 | C2—C3—C4 | 117.8 (2) |
| Na1—O4—H5 | 135.7 | C2—C3—H2 | 120.9 |
| H4—O4—H5 | 105.1 | C4—C3—H2 | 121.2 |
| Na1—O5—Na1 | 94.95 (5) | N2—C4—C3 | 121.4 (2) |
| Na1—O5—H6 | 129.6 | N2—C4—C5 | 119.1 (2) |
| Na1—O5—H7 | 115.6 | C3—C4—C5 | 119.5 (2) |
| Na1—O5i—H6i | 102.9 | N1—C5—C4 | 123.5 (2) |
| Na1—O5i—H7i | 116.7 | N1—C5—H3 | 118.5 |
| H6—O5—H7 | 97.7 | C4—C5—H3 | 118.0 |
| Symmetry codes: (i) −x+1, −y+1, −z+2; (ii) −x+1, −y+1, −z+1; (iii) −x+1, −y, −z+1; (iv) −x+2, −y, −z+2; (v) x+1, y−1, z; (vi) −x+1, −y, −z; (vii) −x+2, −y, −z+1; (viii) x, y+1, z+1; (ix) x, y−1, z−1. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O4—H5···O1iv | 0.71 | 2.05 | 2.755 (2) | 173 |
| O6—H8···O1x | 0.84 | 2.06 | 2.875 (2) | 166 |
| O6—H9···O1iii | 0.84 | 1.96 | 2.797 (2) | 171 |
| O5—H7···O2ii | 0.66 | 2.24 | 2.891 (2) | 170 |
| C5—H3···O2vi | 1.08 | 2.46 | 3.320 (3) | 136 |
| O4—H4···N1vii | 0.96 | 2.09 | 3.011 (2) | 160 |
| O5—H6···N1viii | 0.97 | 2.08 | 2.992 (2) | 157 |
| Symmetry codes: (ii) −x+1, −y+1, −z+1; (iii) −x+1, −y, −z+1; (iv) −x+2, −y, −z+2; (vi) −x+1, −y, −z; (vii) −x+2, −y, −z+1; (viii) x, y+1, z+1; (x) x−1, y+1, z. |

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.
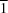 space group. It is made up of edge-shared chains of NaO6 octahedra with five water molecules and one 5-nitro-2-pyridonate anion. Four of these water molecules are bicoordinating, involved in connecting the adjacent octahedra, and the fifth is coordinated to only one octahedron. The crystal structure is stabilized by a network of strong O—H
space group. It is made up of edge-shared chains of NaO6 octahedra with five water molecules and one 5-nitro-2-pyridonate anion. Four of these water molecules are bicoordinating, involved in connecting the adjacent octahedra, and the fifth is coordinated to only one octahedron. The crystal structure is stabilized by a network of strong O—H O and O—H
O and O—H N interactions. The organic moieties occupy the space between the chains with an antiparallel alignment.
N interactions. The organic moieties occupy the space between the chains with an antiparallel alignment. O and O—H
O and O—H N interactions. The organic moieties occupy the space between the chains with an antiparallel alignment.
N interactions. The organic moieties occupy the space between the chains with an antiparallel alignment.
 journal menu
journal menu









 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry


