Buy article online - an online subscription or single-article purchase is required to access this article.
The occurrence of a first-order reversible phase transition in glycine-glutaric acid co-crystals at 220-230 K has been confirmed by three different techniques - single-crystal X-ray diffraction, polarized Raman spectroscopy and differential scanning calorimetry. The most interesting feature of this phase transition is that every second glutaric acid molecule changes its conformation, and this fact results in the space-group symmetry change from
P2
1/
c to
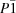
. The topology of the hydrogen-bonded motifs remains almost the same and hydrogen bonds do not switch to other atoms, although the hydrogen bond lengths do change and some of the bonds become inequivalent.
Supporting information
CCDC references: 889409; 889410; 889411; 889412; 889413; 889414; 889415; 889416; 889417
For all compounds, data collection: STOE X-AREA (Stoe & Cie, 2006); cell refinement: STOE X-AREA (Stoe & Cie, 2006); data reduction: STOE X-RED (Stoe & Cie, 2006); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008) and X-STEP32 (Stoe & Cie, 2000); molecular graphics: Mercury (Macrae et al., 2006); software used to prepare material for publication: Mercury (Macrae et al., 2006), PLATON (Spek, 2009), enCIFer (Allen et al., 2004).
(300) Propane-1,3-dicarboxylic acid – Aminoethanoic acid (1/1)
top
Crystal data top
| C5H8O4·C2H5NO2 | F(000) = 440 |
| Mr = 207.18 | Dx = 1.434 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3307 reflections |
| a = 4.9110 (7) Å | θ = 2.0–29.2° |
| b = 20.922 (3) Å | µ = 0.13 mm−1 |
| c = 10.2744 (15) Å | T = 300 K |
| β = 114.586 (10)° | Prism, colourless |
| V = 959.9 (2) Å3 | 0.32 × 0.17 × 0.10 mm |
| Z = 4 | |
Data collection top
STOE IPDS 2
diffractometer | 1421 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.046 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 29.2°, θmin = 2.0° |
| rotation method scans | h = −5→6 |
| 6828 measured reflections | k = −28→24 |
| 2578 independent reflections | l = −14→14 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.048 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.096 | H-atom parameters constrained |
| S = 0.89 | w = 1/[σ2(Fo2) + (0.0419P)2]
where P = (Fo2 + 2Fc2)/3 |
| 2578 reflections | (Δ/σ)max < 0.001 |
| 130 parameters | Δρmax = 0.19 e Å−3 |
| 0 restraints | Δρmin = −0.17 e Å−3 |
Crystal data top
| C5H8O4·C2H5NO2 | V = 959.9 (2) Å3 |
| Mr = 207.18 | Z = 4 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 4.9110 (7) Å | µ = 0.13 mm−1 |
| b = 20.922 (3) Å | T = 300 K |
| c = 10.2744 (15) Å | 0.32 × 0.17 × 0.10 mm |
| β = 114.586 (10)° | |
Data collection top
STOE IPDS 2
diffractometer | 1421 reflections with I > 2σ(I) |
| 6828 measured reflections | Rint = 0.046 |
| 2578 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.048 | 0 restraints |
| wR(F2) = 0.096 | H-atom parameters constrained |
| S = 0.89 | Δρmax = 0.19 e Å−3 |
| 2578 reflections | Δρmin = −0.17 e Å−3 |
| 130 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.1303 (3) | 0.44553 (6) | 0.33353 (12) | 0.0431 (3) | |
| O5 | 0.6180 (3) | 0.64113 (6) | 0.22044 (13) | 0.0492 (4) | |
| O3 | 0.2017 (3) | 0.91549 (6) | −0.03314 (12) | 0.0412 (3) | |
| N1 | 0.5566 (4) | 0.52554 (7) | 0.33624 (14) | 0.0360 (4) | |
| H1A | 0.6599 | 0.5565 | 0.3179 | 0.054* | |
| H1B | 0.6819 | 0.5000 | 0.4039 | 0.054* | |
| H1C | 0.4268 | 0.5426 | 0.3661 | 0.054* | |
| C2 | 0.3932 (4) | 0.48823 (8) | 0.20491 (17) | 0.0334 (4) | |
| H2A | 0.5357 | 0.4649 | 0.1802 | 0.040* | |
| H2B | 0.2843 | 0.5172 | 0.1268 | 0.040* | |
| O6 | 0.8167 (4) | 0.59463 (6) | 0.09106 (14) | 0.0501 (4) | |
| H6 | 0.8418 | 0.6018 | 0.0184 | 0.075* | |
| O2 | 0.0510 (3) | 0.40246 (7) | 0.12345 (14) | 0.0543 (4) | |
| C1 | 0.1753 (4) | 0.44149 (8) | 0.22259 (17) | 0.0332 (4) | |
| C7 | 0.6740 (4) | 0.64281 (8) | 0.11621 (17) | 0.0347 (4) | |
| O4 | 0.1063 (4) | 0.84677 (6) | 0.10598 (16) | 0.0565 (4) | |
| H4 | 0.0356 | 0.8788 | 0.1260 | 0.085* | |
| C5 | 0.4645 (5) | 0.75376 (9) | 0.06721 (18) | 0.0423 (5) | |
| H5A | 0.3195 | 0.7383 | 0.1012 | 0.051* | |
| H5B | 0.6295 | 0.7725 | 0.1481 | 0.051* | |
| C6 | 0.5810 (5) | 0.69752 (8) | 0.01277 (18) | 0.0379 (4) | |
| H6B | 0.4261 | 0.6831 | −0.0774 | 0.045* | |
| H6A | 0.7512 | 0.7112 | −0.0050 | 0.045* | |
| C3 | 0.2064 (4) | 0.86107 (8) | 0.01003 (17) | 0.0345 (4) | |
| C4 | 0.3204 (5) | 0.80483 (9) | −0.04245 (19) | 0.0443 (5) | |
| H4A | 0.4651 | 0.8199 | −0.0771 | 0.053* | |
| H4B | 0.1546 | 0.7861 | −0.1230 | 0.053* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0537 (9) | 0.0417 (7) | 0.0454 (7) | −0.0153 (7) | 0.0319 (6) | −0.0063 (6) |
| O5 | 0.0761 (11) | 0.0384 (7) | 0.0480 (7) | 0.0049 (7) | 0.0405 (7) | 0.0067 (6) |
| O3 | 0.0536 (9) | 0.0293 (6) | 0.0473 (7) | 0.0004 (6) | 0.0275 (6) | 0.0027 (5) |
| N1 | 0.0422 (10) | 0.0321 (8) | 0.0396 (7) | −0.0084 (7) | 0.0228 (7) | 0.0000 (6) |
| C2 | 0.0348 (11) | 0.0350 (9) | 0.0344 (8) | −0.0021 (8) | 0.0186 (8) | −0.0002 (7) |
| O6 | 0.0726 (11) | 0.0360 (7) | 0.0510 (8) | 0.0163 (7) | 0.0349 (8) | 0.0061 (6) |
| O2 | 0.0612 (10) | 0.0556 (9) | 0.0575 (8) | −0.0243 (8) | 0.0358 (7) | −0.0237 (7) |
| C1 | 0.0331 (11) | 0.0306 (9) | 0.0389 (9) | 0.0014 (8) | 0.0180 (8) | −0.0015 (7) |
| C7 | 0.0408 (12) | 0.0277 (9) | 0.0377 (8) | −0.0027 (8) | 0.0184 (8) | −0.0020 (7) |
| O4 | 0.0853 (12) | 0.0378 (8) | 0.0758 (9) | 0.0195 (8) | 0.0627 (9) | 0.0160 (7) |
| C5 | 0.0587 (14) | 0.0335 (10) | 0.0419 (9) | 0.0103 (9) | 0.0280 (9) | 0.0057 (7) |
| C6 | 0.0483 (13) | 0.0325 (9) | 0.0398 (9) | 0.0048 (9) | 0.0252 (9) | 0.0037 (7) |
| C3 | 0.0374 (11) | 0.0328 (9) | 0.0367 (9) | 0.0041 (8) | 0.0189 (8) | 0.0041 (7) |
| C4 | 0.0581 (14) | 0.0389 (11) | 0.0461 (10) | 0.0129 (10) | 0.0318 (10) | 0.0087 (8) |
Geometric parameters (Å, º) top
| O1—C1 | 1.2508 (19) | C7—C6 | 1.498 (2) |
| O5—C7 | 1.212 (2) | O4—C3 | 1.306 (2) |
| O3—C3 | 1.219 (2) | O4—H4 | 0.8200 |
| N1—C2 | 1.474 (2) | C5—C4 | 1.500 (2) |
| N1—H1A | 0.8900 | C5—C6 | 1.514 (2) |
| N1—H1B | 0.8900 | C5—H5A | 0.9700 |
| N1—H1C | 0.8900 | C5—H5B | 0.9700 |
| C2—C1 | 1.515 (2) | C6—H6B | 0.9700 |
| C2—H2A | 0.9700 | C6—H6A | 0.9700 |
| C2—H2B | 0.9700 | C3—C4 | 1.497 (2) |
| O6—C7 | 1.313 (2) | C4—H4A | 0.9700 |
| O6—H6 | 0.8200 | C4—H4B | 0.9700 |
| O2—C1 | 1.249 (2) | | |
| | | |
| C2—N1—H1A | 109.5 | C4—C5—H5A | 108.7 |
| C2—N1—H1B | 109.5 | C6—C5—H5A | 108.7 |
| H1A—N1—H1B | 109.5 | C4—C5—H5B | 108.7 |
| C2—N1—H1C | 109.5 | C6—C5—H5B | 108.7 |
| H1A—N1—H1C | 109.5 | H5A—C5—H5B | 107.6 |
| H1B—N1—H1C | 109.5 | C7—C6—C5 | 112.48 (13) |
| N1—C2—C1 | 112.00 (13) | C7—C6—H6B | 109.1 |
| N1—C2—H2A | 109.2 | C5—C6—H6B | 109.1 |
| C1—C2—H2A | 109.2 | C7—C6—H6A | 109.1 |
| N1—C2—H2B | 109.2 | C5—C6—H6A | 109.1 |
| C1—C2—H2B | 109.2 | H6B—C6—H6A | 107.8 |
| H2A—C2—H2B | 107.9 | O3—C3—O4 | 122.19 (16) |
| C7—O6—H6 | 109.5 | O3—C3—C4 | 123.82 (15) |
| O2—C1—O1 | 125.15 (17) | O4—C3—C4 | 113.98 (15) |
| O2—C1—C2 | 117.00 (15) | C3—C4—C5 | 114.82 (14) |
| O1—C1—C2 | 117.83 (15) | C3—C4—H4A | 108.6 |
| O5—C7—O6 | 119.17 (15) | C5—C4—H4A | 108.6 |
| O5—C7—C6 | 122.33 (16) | C3—C4—H4B | 108.6 |
| O6—C7—C6 | 118.49 (14) | C5—C4—H4B | 108.6 |
| C3—O4—H4 | 109.5 | H4A—C4—H4B | 107.5 |
| C4—C5—C6 | 114.14 (14) | | |
| | | |
| N1—C2—C1—O2 | −171.95 (16) | C4—C5—C6—C7 | −170.01 (18) |
| N1—C2—C1—O1 | 9.1 (2) | O3—C3—C4—C5 | 147.7 (2) |
| O5—C7—C6—C5 | 10.4 (3) | O4—C3—C4—C5 | −33.3 (3) |
| O6—C7—C6—C5 | −170.82 (17) | C6—C5—C4—C3 | −179.48 (18) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1B···O3i | 0.89 | 2.14 | 2.9671 (19) | 154 |
| N1—H1C···O3ii | 0.89 | 2.00 | 2.8824 (19) | 169 |
| N1—H1A···O5 | 0.89 | 2.00 | 2.7665 (18) | 143 |
| O4—H4···O1iii | 0.82 | 1.75 | 2.5706 (18) | 178 |
| O6—H6···O2iv | 0.82 | 1.75 | 2.5444 (18) | 164 |
| Symmetry codes: (i) −x+1, y−1/2, −z+1/2; (ii) x, −y+3/2, z+1/2; (iii) −x, y+1/2, −z+1/2; (iv) −x+1, −y+1, −z. |
(275) Propane-1,3-dicarboxylic acid – Aminoethanoic acid (1/1)
top
Crystal data top
| C5H8O4·C2H5NO2 | F(000) = 440 |
| Mr = 207.18 | Dx = 1.438 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3205 reflections |
| a = 4.9022 (7) Å | θ = 2.0–29.2° |
| b = 20.901 (3) Å | µ = 0.13 mm−1 |
| c = 10.2730 (15) Å | T = 275 K |
| β = 114.60 (1)° | Prism, colourless |
| V = 957.0 (2) Å3 | 0.32 × 0.17 × 0.10 mm |
| Z = 4 | |
Data collection top
STOE IPDS 2
diffractometer | 1461 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.047 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 29.2°, θmin = 2.0° |
| rotation method scans | h = −5→6 |
| 6792 measured reflections | k = −28→24 |
| 2570 independent reflections | l = −14→14 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.043 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.085 | H-atom parameters constrained |
| S = 0.87 | w = 1/[σ2(Fo2) + (0.0364P)2]
where P = (Fo2 + 2Fc2)/3 |
| 2570 reflections | (Δ/σ)max = 0.001 |
| 130 parameters | Δρmax = 0.16 e Å−3 |
| 0 restraints | Δρmin = −0.18 e Å−3 |
Crystal data top
| C5H8O4·C2H5NO2 | V = 957.0 (2) Å3 |
| Mr = 207.18 | Z = 4 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 4.9022 (7) Å | µ = 0.13 mm−1 |
| b = 20.901 (3) Å | T = 275 K |
| c = 10.2730 (15) Å | 0.32 × 0.17 × 0.10 mm |
| β = 114.60 (1)° | |
Data collection top
STOE IPDS 2
diffractometer | 1461 reflections with I > 2σ(I) |
| 6792 measured reflections | Rint = 0.047 |
| 2570 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.043 | 0 restraints |
| wR(F2) = 0.085 | H-atom parameters constrained |
| S = 0.87 | Δρmax = 0.16 e Å−3 |
| 2570 reflections | Δρmin = −0.18 e Å−3 |
| 130 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.1302 (3) | 0.44534 (6) | 0.33384 (11) | 0.0391 (3) | |
| O5 | 0.6205 (3) | 0.64085 (6) | 0.22111 (12) | 0.0434 (3) | |
| O3 | 0.2021 (3) | 0.91559 (5) | −0.03340 (11) | 0.0368 (3) | |
| N1 | 0.5575 (3) | 0.52520 (6) | 0.33659 (13) | 0.0325 (3) | |
| H1A | 0.6613 | 0.5562 | 0.3184 | 0.049* | |
| H1B | 0.6827 | 0.4996 | 0.4043 | 0.049* | |
| H1C | 0.4274 | 0.5424 | 0.3664 | 0.049* | |
| C2 | 0.3937 (4) | 0.48788 (8) | 0.20508 (15) | 0.0302 (4) | |
| H2A | 0.5364 | 0.4643 | 0.1807 | 0.036* | |
| H2B | 0.2857 | 0.5170 | 0.1269 | 0.036* | |
| O6 | 0.8202 (3) | 0.59420 (6) | 0.09161 (12) | 0.0452 (3) | |
| H6 | 0.8488 | 0.6018 | 0.0200 | 0.068* | |
| O2 | 0.0470 (3) | 0.40245 (6) | 0.12262 (13) | 0.0489 (4) | |
| C1 | 0.1736 (4) | 0.44130 (8) | 0.22219 (16) | 0.0304 (4) | |
| C7 | 0.6768 (4) | 0.64243 (8) | 0.11682 (16) | 0.0310 (4) | |
| O4 | 0.1064 (4) | 0.84659 (6) | 0.10554 (15) | 0.0518 (4) | |
| H4 | 0.0380 | 0.8788 | 0.1264 | 0.078* | |
| C5 | 0.4674 (4) | 0.75361 (8) | 0.06781 (16) | 0.0377 (4) | |
| H5A | 0.3234 | 0.7382 | 0.1026 | 0.045* | |
| H5B | 0.6335 | 0.7725 | 0.1482 | 0.045* | |
| C6 | 0.5829 (4) | 0.69733 (7) | 0.01302 (17) | 0.0341 (4) | |
| H6B | 0.4271 | 0.6830 | −0.0770 | 0.041* | |
| H6A | 0.7530 | 0.7110 | −0.0051 | 0.041* | |
| C3 | 0.2068 (4) | 0.86087 (8) | 0.00940 (16) | 0.0321 (4) | |
| C4 | 0.3207 (5) | 0.80467 (8) | −0.04307 (17) | 0.0398 (5) | |
| H4A | 0.4646 | 0.8197 | −0.0783 | 0.048* | |
| H4B | 0.1543 | 0.7857 | −0.1231 | 0.048* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0477 (8) | 0.0394 (7) | 0.0401 (6) | −0.0147 (6) | 0.0281 (6) | −0.0068 (5) |
| O5 | 0.0665 (10) | 0.0342 (7) | 0.0416 (6) | 0.0049 (7) | 0.0346 (7) | 0.0065 (5) |
| O3 | 0.0474 (8) | 0.0267 (6) | 0.0414 (6) | 0.0001 (6) | 0.0234 (6) | 0.0021 (5) |
| N1 | 0.0377 (9) | 0.0291 (7) | 0.0369 (7) | −0.0079 (7) | 0.0218 (6) | 0.0001 (6) |
| C2 | 0.0314 (10) | 0.0312 (8) | 0.0310 (8) | −0.0021 (8) | 0.0158 (7) | −0.0006 (6) |
| O6 | 0.0660 (10) | 0.0323 (7) | 0.0462 (7) | 0.0155 (7) | 0.0321 (7) | 0.0058 (5) |
| O2 | 0.0540 (9) | 0.0498 (8) | 0.0525 (7) | −0.0223 (7) | 0.0318 (7) | −0.0231 (6) |
| C1 | 0.0294 (10) | 0.0269 (8) | 0.0379 (8) | 0.0016 (8) | 0.0168 (7) | −0.0005 (7) |
| C7 | 0.0349 (10) | 0.0251 (8) | 0.0340 (8) | −0.0020 (8) | 0.0153 (7) | −0.0016 (6) |
| O4 | 0.0787 (11) | 0.0346 (7) | 0.0694 (8) | 0.0180 (7) | 0.0579 (8) | 0.0149 (6) |
| C5 | 0.0517 (12) | 0.0303 (9) | 0.0382 (8) | 0.0092 (9) | 0.0256 (8) | 0.0048 (7) |
| C6 | 0.0438 (12) | 0.0285 (8) | 0.0368 (8) | 0.0053 (8) | 0.0234 (8) | 0.0035 (7) |
| C3 | 0.0347 (11) | 0.0317 (9) | 0.0326 (8) | 0.0033 (8) | 0.0166 (8) | 0.0037 (7) |
| C4 | 0.0525 (13) | 0.0345 (10) | 0.0410 (9) | 0.0109 (9) | 0.0279 (9) | 0.0073 (7) |
Geometric parameters (Å, º) top
| O1—C1 | 1.2534 (18) | C7—C6 | 1.502 (2) |
| O5—C7 | 1.2132 (18) | O4—C3 | 1.3082 (19) |
| O3—C3 | 1.2221 (19) | O4—H4 | 0.8200 |
| N1—C2 | 1.4749 (19) | C5—C4 | 1.507 (2) |
| N1—H1A | 0.8900 | C5—C6 | 1.512 (2) |
| N1—H1B | 0.8900 | C5—H5A | 0.9700 |
| N1—H1C | 0.8900 | C5—H5B | 0.9700 |
| C2—C1 | 1.517 (2) | C6—H6B | 0.9700 |
| C2—H2A | 0.9700 | C6—H6A | 0.9700 |
| C2—H2B | 0.9700 | C3—C4 | 1.494 (2) |
| O6—C7 | 1.3149 (19) | C4—H4A | 0.9700 |
| O6—H6 | 0.8200 | C4—H4B | 0.9700 |
| O2—C1 | 1.2499 (19) | | |
| | | |
| C2—N1—H1A | 109.5 | C4—C5—H5A | 108.8 |
| C2—N1—H1B | 109.5 | C6—C5—H5A | 108.8 |
| H1A—N1—H1B | 109.5 | C4—C5—H5B | 108.8 |
| C2—N1—H1C | 109.5 | C6—C5—H5B | 108.8 |
| H1A—N1—H1C | 109.5 | H5A—C5—H5B | 107.7 |
| H1B—N1—H1C | 109.5 | C7—C6—C5 | 112.43 (12) |
| N1—C2—C1 | 112.07 (12) | C7—C6—H6B | 109.1 |
| N1—C2—H2A | 109.2 | C5—C6—H6B | 109.1 |
| C1—C2—H2A | 109.2 | C7—C6—H6A | 109.1 |
| N1—C2—H2B | 109.2 | C5—C6—H6A | 109.1 |
| C1—C2—H2B | 109.2 | H6B—C6—H6A | 107.9 |
| H2A—C2—H2B | 107.9 | O3—C3—O4 | 121.90 (15) |
| C7—O6—H6 | 109.5 | O3—C3—C4 | 124.04 (14) |
| O2—C1—O1 | 125.25 (16) | O4—C3—C4 | 114.06 (14) |
| O2—C1—C2 | 117.19 (14) | C3—C4—C5 | 114.62 (13) |
| O1—C1—C2 | 117.56 (14) | C3—C4—H4A | 108.6 |
| O5—C7—O6 | 119.35 (14) | C5—C4—H4A | 108.6 |
| O5—C7—C6 | 122.17 (15) | C3—C4—H4B | 108.6 |
| O6—C7—C6 | 118.47 (13) | C5—C4—H4B | 108.6 |
| C3—O4—H4 | 109.5 | H4A—C4—H4B | 107.6 |
| C4—C5—C6 | 113.85 (13) | | |
| | | |
| N1—C2—C1—O2 | −172.37 (15) | C4—C5—C6—C7 | −169.69 (17) |
| N1—C2—C1—O1 | 8.5 (2) | O3—C3—C4—C5 | 147.16 (18) |
| O5—C7—C6—C5 | 10.6 (3) | O4—C3—C4—C5 | −33.7 (2) |
| O6—C7—C6—C5 | −170.55 (16) | C6—C5—C4—C3 | −179.64 (16) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1B···O3i | 0.89 | 2.13 | 2.9564 (18) | 154 |
| N1—H1C···O3ii | 0.89 | 2.00 | 2.8789 (18) | 169 |
| N1—H1A···O5 | 0.89 | 2.00 | 2.7658 (17) | 143 |
| O4—H4···O1iii | 0.82 | 1.75 | 2.5680 (17) | 176 |
| O6—H6···O2iv | 0.82 | 1.74 | 2.5425 (17) | 165 |
| Symmetry codes: (i) −x+1, y−1/2, −z+1/2; (ii) x, −y+3/2, z+1/2; (iii) −x, y+1/2, −z+1/2; (iv) −x+1, −y+1, −z. |
(250) Propane-1,3-dicarboxylic acid – Aminoethanoic acid (1/1)
top
Crystal data top
| C5H8O4·C2H5NO2 | F(000) = 440 |
| Mr = 207.18 | Dx = 1.442 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3499 reflections |
| a = 4.8960 (6) Å | θ = 2.0–29.5° |
| b = 20.878 (2) Å | µ = 0.13 mm−1 |
| c = 10.2701 (13) Å | T = 250 K |
| β = 114.634 (9)° | Prism, colourless |
| V = 954.3 (2) Å3 | 0.32 × 0.17 × 0.10 mm |
| Z = 4 | |
Data collection top
STOE IPDS 2
diffractometer | 1535 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.045 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 29.2°, θmin = 2.0° |
| rotation method scans | h = −5→6 |
| 6775 measured reflections | k = −28→24 |
| 2562 independent reflections | l = −14→14 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.044 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.086 | H-atom parameters constrained |
| S = 0.88 | w = 1/[σ2(Fo2) + (0.0377P)2]
where P = (Fo2 + 2Fc2)/3 |
| 2562 reflections | (Δ/σ)max < 0.001 |
| 130 parameters | Δρmax = 0.16 e Å−3 |
| 0 restraints | Δρmin = −0.20 e Å−3 |
Crystal data top
| C5H8O4·C2H5NO2 | V = 954.3 (2) Å3 |
| Mr = 207.18 | Z = 4 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 4.8960 (6) Å | µ = 0.13 mm−1 |
| b = 20.878 (2) Å | T = 250 K |
| c = 10.2701 (13) Å | 0.32 × 0.17 × 0.10 mm |
| β = 114.634 (9)° | |
Data collection top
STOE IPDS 2
diffractometer | 1535 reflections with I > 2σ(I) |
| 6775 measured reflections | Rint = 0.045 |
| 2562 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.044 | 0 restraints |
| wR(F2) = 0.086 | H-atom parameters constrained |
| S = 0.88 | Δρmax = 0.16 e Å−3 |
| 2562 reflections | Δρmin = −0.20 e Å−3 |
| 130 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.1300 (3) | 0.44533 (6) | 0.33405 (11) | 0.0352 (3) | |
| O5 | 0.6231 (3) | 0.64055 (5) | 0.22156 (11) | 0.0386 (3) | |
| O3 | 0.2029 (3) | 0.91557 (5) | −0.03367 (11) | 0.0332 (3) | |
| N1 | 0.5587 (3) | 0.52504 (6) | 0.33685 (13) | 0.0294 (3) | |
| H1A | 0.6627 | 0.5566 | 0.3182 | 0.044* | |
| H1B | 0.6866 | 0.4992 | 0.4053 | 0.044* | |
| H1C | 0.4270 | 0.5422 | 0.3673 | 0.044* | |
| C2 | 0.3943 (4) | 0.48752 (7) | 0.20504 (15) | 0.0277 (3) | |
| H2A | 0.5386 | 0.4635 | 0.1808 | 0.033* | |
| H2B | 0.2860 | 0.5169 | 0.1258 | 0.033* | |
| O6 | 0.8236 (3) | 0.59387 (6) | 0.09217 (12) | 0.0411 (3) | |
| H6 | 0.8544 | 0.6018 | 0.0202 | 0.062* | |
| O2 | 0.0439 (3) | 0.40249 (6) | 0.12210 (13) | 0.0446 (3) | |
| C1 | 0.1725 (4) | 0.44122 (8) | 0.22194 (15) | 0.0275 (3) | |
| C7 | 0.6793 (4) | 0.64212 (7) | 0.11717 (15) | 0.0280 (3) | |
| O4 | 0.1064 (3) | 0.84645 (6) | 0.10527 (14) | 0.0464 (4) | |
| H4 | 0.0338 | 0.8789 | 0.1252 | 0.070* | |
| C5 | 0.4699 (4) | 0.75347 (8) | 0.06795 (16) | 0.0331 (4) | |
| H5A | 0.3249 | 0.7379 | 0.1036 | 0.040* | |
| H5B | 0.6381 | 0.7728 | 0.1488 | 0.040* | |
| C6 | 0.5856 (4) | 0.69709 (7) | 0.01315 (16) | 0.0304 (4) | |
| H6B | 0.4280 | 0.6826 | −0.0779 | 0.036* | |
| H6A | 0.7579 | 0.7109 | −0.0050 | 0.036* | |
| C3 | 0.2068 (4) | 0.86084 (8) | 0.00894 (15) | 0.0284 (4) | |
| C4 | 0.3212 (4) | 0.80446 (8) | −0.04366 (17) | 0.0348 (4) | |
| H4A | 0.4662 | 0.8197 | −0.0797 | 0.042* | |
| H4B | 0.1528 | 0.7851 | −0.1243 | 0.042* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0427 (8) | 0.0349 (6) | 0.0366 (6) | −0.0127 (6) | 0.0251 (5) | −0.0051 (5) |
| O5 | 0.0582 (9) | 0.0310 (6) | 0.0371 (6) | 0.0045 (6) | 0.0304 (6) | 0.0058 (5) |
| O3 | 0.0418 (8) | 0.0242 (6) | 0.0384 (6) | −0.0003 (6) | 0.0216 (5) | 0.0020 (5) |
| N1 | 0.0346 (8) | 0.0264 (7) | 0.0326 (6) | −0.0066 (6) | 0.0193 (6) | 0.0002 (5) |
| C2 | 0.0288 (9) | 0.0284 (8) | 0.0288 (7) | −0.0002 (7) | 0.0151 (7) | 0.0000 (6) |
| O6 | 0.0596 (9) | 0.0301 (6) | 0.0415 (6) | 0.0143 (6) | 0.0291 (6) | 0.0057 (5) |
| O2 | 0.0492 (9) | 0.0463 (7) | 0.0468 (7) | −0.0206 (7) | 0.0285 (6) | −0.0213 (6) |
| C1 | 0.0272 (9) | 0.0254 (8) | 0.0326 (7) | 0.0016 (7) | 0.0152 (7) | 0.0007 (6) |
| C7 | 0.0327 (10) | 0.0223 (7) | 0.0298 (7) | −0.0022 (7) | 0.0139 (7) | −0.0016 (6) |
| O4 | 0.0712 (10) | 0.0300 (7) | 0.0632 (8) | 0.0166 (7) | 0.0529 (7) | 0.0134 (6) |
| C5 | 0.0459 (11) | 0.0253 (8) | 0.0336 (8) | 0.0076 (8) | 0.0219 (7) | 0.0044 (6) |
| C6 | 0.0391 (11) | 0.0256 (8) | 0.0321 (8) | 0.0036 (7) | 0.0205 (7) | 0.0029 (6) |
| C3 | 0.0301 (10) | 0.0276 (8) | 0.0300 (7) | 0.0020 (7) | 0.0150 (7) | 0.0032 (6) |
| C4 | 0.0460 (12) | 0.0296 (9) | 0.0352 (8) | 0.0106 (8) | 0.0234 (8) | 0.0069 (6) |
Geometric parameters (Å, º) top
| O1—C1 | 1.2553 (17) | C7—C6 | 1.503 (2) |
| O5—C7 | 1.2133 (18) | O4—C3 | 1.3098 (19) |
| O3—C3 | 1.2210 (18) | O4—H4 | 0.8300 |
| N1—C2 | 1.4783 (19) | C5—C4 | 1.510 (2) |
| N1—H1A | 0.9000 | C5—C6 | 1.513 (2) |
| N1—H1B | 0.9000 | C5—H5A | 0.9800 |
| N1—H1C | 0.9000 | C5—H5B | 0.9800 |
| C2—C1 | 1.517 (2) | C6—H6B | 0.9800 |
| C2—H2A | 0.9800 | C6—H6A | 0.9800 |
| C2—H2B | 0.9800 | C3—C4 | 1.498 (2) |
| O6—C7 | 1.3157 (19) | C4—H4A | 0.9800 |
| O6—H6 | 0.8300 | C4—H4B | 0.9800 |
| O2—C1 | 1.2503 (19) | | |
| | | |
| C2—N1—H1A | 109.5 | C4—C5—H5A | 108.8 |
| C2—N1—H1B | 109.5 | C6—C5—H5A | 108.8 |
| H1A—N1—H1B | 109.5 | C4—C5—H5B | 108.8 |
| C2—N1—H1C | 109.5 | C6—C5—H5B | 108.8 |
| H1A—N1—H1C | 109.5 | H5A—C5—H5B | 107.7 |
| H1B—N1—H1C | 109.5 | C7—C6—C5 | 112.35 (12) |
| N1—C2—C1 | 112.06 (12) | C7—C6—H6B | 109.1 |
| N1—C2—H2A | 109.2 | C5—C6—H6B | 109.1 |
| C1—C2—H2A | 109.2 | C7—C6—H6A | 109.1 |
| N1—C2—H2B | 109.2 | C5—C6—H6A | 109.1 |
| C1—C2—H2B | 109.2 | H6B—C6—H6A | 107.9 |
| H2A—C2—H2B | 107.9 | O3—C3—O4 | 122.06 (15) |
| C7—O6—H6 | 109.5 | O3—C3—C4 | 123.95 (14) |
| O2—C1—O1 | 125.25 (15) | O4—C3—C4 | 113.99 (13) |
| O2—C1—C2 | 117.26 (13) | C3—C4—C5 | 114.49 (12) |
| O1—C1—C2 | 117.48 (13) | C3—C4—H4A | 108.6 |
| O5—C7—O6 | 119.32 (14) | C5—C4—H4A | 108.6 |
| O5—C7—C6 | 122.28 (14) | C3—C4—H4B | 108.6 |
| O6—C7—C6 | 118.39 (13) | C5—C4—H4B | 108.6 |
| C3—O4—H4 | 109.5 | H4A—C4—H4B | 107.6 |
| C4—C5—C6 | 113.75 (12) | | |
| | | |
| N1—C2—C1—O2 | −172.80 (15) | C4—C5—C6—C7 | −169.33 (16) |
| N1—C2—C1—O1 | 8.0 (2) | O3—C3—C4—C5 | 146.67 (17) |
| O5—C7—C6—C5 | 10.5 (2) | O4—C3—C4—C5 | −33.9 (2) |
| O6—C7—C6—C5 | −170.40 (15) | C6—C5—C4—C3 | −179.68 (16) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1B···O3i | 0.90 | 2.12 | 2.9504 (17) | 153 |
| N1—H1C···O3ii | 0.90 | 1.99 | 2.8760 (18) | 170 |
| N1—H1A···O5 | 0.90 | 1.98 | 2.7621 (17) | 144 |
| O4—H4···O1iii | 0.83 | 1.74 | 2.5676 (16) | 178 |
| O6—H6···O2iv | 0.83 | 1.73 | 2.5414 (16) | 165 |
| Symmetry codes: (i) −x+1, y−1/2, −z+1/2; (ii) x, −y+3/2, z+1/2; (iii) −x, y+1/2, −z+1/2; (iv) −x+1, −y+1, −z. |
(225) Propane-1,3-dicarboxylic acid – Aminoethanoic acid (1/1)
top
Crystal data top
| C5H8O4·C2H5NO2 | F(000) = 440 |
| Mr = 207.18 | Dx = 1.446 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3782 reflections |
| a = 4.8888 (6) Å | θ = 2.0–29.5° |
| b = 20.859 (2) Å | µ = 0.13 mm−1 |
| c = 10.2664 (13) Å | T = 225 K |
| β = 114.648 (9)° | Prism, colourless |
| V = 951.6 (2) Å3 | 0.32 × 0.17 × 0.10 mm |
| Z = 4 | |
Data collection top
STOE IPDS 2
diffractometer | 1594 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.043 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 29.2°, θmin = 2.0° |
| rotation method scans | h = −5→6 |
| 6775 measured reflections | k = −28→24 |
| 2558 independent reflections | l = −14→14 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.042 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.081 | H-atom parameters constrained |
| S = 0.88 | w = 1/[σ2(Fo2) + (0.037P)2]
where P = (Fo2 + 2Fc2)/3 |
| 2558 reflections | (Δ/σ)max < 0.001 |
| 130 parameters | Δρmax = 0.21 e Å−3 |
| 0 restraints | Δρmin = −0.18 e Å−3 |
Crystal data top
| C5H8O4·C2H5NO2 | V = 951.6 (2) Å3 |
| Mr = 207.18 | Z = 4 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 4.8888 (6) Å | µ = 0.13 mm−1 |
| b = 20.859 (2) Å | T = 225 K |
| c = 10.2664 (13) Å | 0.32 × 0.17 × 0.10 mm |
| β = 114.648 (9)° | |
Data collection top
STOE IPDS 2
diffractometer | 1594 reflections with I > 2σ(I) |
| 6775 measured reflections | Rint = 0.043 |
| 2558 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.042 | 0 restraints |
| wR(F2) = 0.081 | H-atom parameters constrained |
| S = 0.88 | Δρmax = 0.21 e Å−3 |
| 2558 reflections | Δρmin = −0.18 e Å−3 |
| 130 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.1297 (3) | 0.44519 (5) | 0.33441 (10) | 0.0314 (3) | |
| O5 | 0.6253 (3) | 0.64046 (5) | 0.22218 (10) | 0.0346 (3) | |
| O3 | 0.2033 (3) | 0.91561 (5) | −0.03396 (10) | 0.0296 (3) | |
| N1 | 0.5594 (3) | 0.52477 (6) | 0.33723 (12) | 0.0263 (3) | |
| H1A | 0.6652 | 0.5561 | 0.3190 | 0.039* | |
| H1B | 0.6859 | 0.4987 | 0.4058 | 0.039* | |
| H1C | 0.4275 | 0.5422 | 0.3673 | 0.039* | |
| C2 | 0.3944 (3) | 0.48745 (7) | 0.20527 (14) | 0.0251 (3) | |
| H2A | 0.5387 | 0.4633 | 0.1809 | 0.030* | |
| H2B | 0.2864 | 0.5170 | 0.1261 | 0.030* | |
| O6 | 0.8268 (3) | 0.59350 (5) | 0.09291 (11) | 0.0366 (3) | |
| H6 | 0.8563 | 0.6012 | 0.0205 | 0.055* | |
| O2 | 0.0402 (3) | 0.40272 (6) | 0.12133 (12) | 0.0403 (3) | |
| C1 | 0.1715 (3) | 0.44119 (7) | 0.22204 (14) | 0.0247 (3) | |
| C7 | 0.6820 (4) | 0.64190 (7) | 0.11764 (14) | 0.0249 (3) | |
| O4 | 0.1061 (3) | 0.84624 (5) | 0.10478 (13) | 0.0423 (3) | |
| H4 | 0.0407 | 0.8790 | 0.1277 | 0.063* | |
| C5 | 0.4719 (4) | 0.75334 (7) | 0.06844 (14) | 0.0300 (4) | |
| H5A | 0.6403 | 0.7727 | 0.1493 | 0.036* | |
| H5B | 0.3267 | 0.7377 | 0.1041 | 0.036* | |
| C6 | 0.5880 (4) | 0.69689 (7) | 0.01334 (15) | 0.0276 (3) | |
| H6B | 0.4301 | 0.6823 | −0.0778 | 0.033* | |
| H6A | 0.7604 | 0.7107 | −0.0049 | 0.033* | |
| C3 | 0.2072 (3) | 0.86082 (7) | 0.00868 (14) | 0.0259 (3) | |
| C4 | 0.3223 (4) | 0.80435 (7) | −0.04410 (15) | 0.0320 (4) | |
| H4A | 0.4675 | 0.8196 | −0.0801 | 0.038* | |
| H4B | 0.1537 | 0.7849 | −0.1247 | 0.038* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0382 (7) | 0.0317 (6) | 0.0323 (5) | −0.0113 (5) | 0.0225 (5) | −0.0052 (4) |
| O5 | 0.0512 (8) | 0.0277 (6) | 0.0338 (5) | 0.0037 (5) | 0.0266 (5) | 0.0051 (4) |
| O3 | 0.0376 (7) | 0.0217 (5) | 0.0332 (5) | 0.0002 (5) | 0.0183 (5) | 0.0016 (4) |
| N1 | 0.0308 (7) | 0.0234 (6) | 0.0295 (6) | −0.0054 (6) | 0.0173 (5) | 0.0006 (5) |
| C2 | 0.0259 (8) | 0.0269 (7) | 0.0255 (6) | −0.0005 (7) | 0.0136 (6) | 0.0002 (6) |
| O6 | 0.0515 (8) | 0.0273 (6) | 0.0376 (6) | 0.0121 (6) | 0.0251 (6) | 0.0044 (5) |
| O2 | 0.0431 (8) | 0.0427 (7) | 0.0421 (6) | −0.0173 (6) | 0.0249 (6) | −0.0182 (5) |
| C1 | 0.0237 (8) | 0.0231 (7) | 0.0296 (7) | 0.0015 (6) | 0.0133 (6) | −0.0012 (6) |
| C7 | 0.0282 (9) | 0.0207 (7) | 0.0268 (6) | −0.0022 (6) | 0.0124 (6) | −0.0011 (5) |
| O4 | 0.0647 (9) | 0.0280 (6) | 0.0565 (7) | 0.0156 (6) | 0.0475 (7) | 0.0125 (5) |
| C5 | 0.0402 (10) | 0.0249 (8) | 0.0294 (7) | 0.0073 (7) | 0.0191 (7) | 0.0038 (6) |
| C6 | 0.0347 (9) | 0.0234 (7) | 0.0294 (7) | 0.0037 (7) | 0.0181 (6) | 0.0027 (6) |
| C3 | 0.0263 (8) | 0.0262 (7) | 0.0265 (7) | 0.0017 (7) | 0.0123 (6) | 0.0023 (6) |
| C4 | 0.0434 (11) | 0.0272 (8) | 0.0322 (7) | 0.0089 (7) | 0.0225 (7) | 0.0062 (6) |
Geometric parameters (Å, º) top
| O1—C1 | 1.2554 (16) | C7—C6 | 1.5042 (19) |
| O5—C7 | 1.2159 (17) | O4—C3 | 1.3103 (17) |
| O3—C3 | 1.2212 (17) | O4—H4 | 0.8300 |
| N1—C2 | 1.4766 (18) | C5—C6 | 1.515 (2) |
| N1—H1A | 0.9000 | C5—C4 | 1.516 (2) |
| N1—H1B | 0.9000 | C5—H5A | 0.9800 |
| N1—H1C | 0.9000 | C5—H5B | 0.9800 |
| C2—C1 | 1.518 (2) | C6—H6B | 0.9800 |
| C2—H2A | 0.9800 | C6—H6A | 0.9800 |
| C2—H2B | 0.9800 | C3—C4 | 1.501 (2) |
| O6—C7 | 1.3170 (17) | C4—H4A | 0.9800 |
| O6—H6 | 0.8300 | C4—H4B | 0.9800 |
| O2—C1 | 1.2536 (17) | | |
| | | |
| C2—N1—H1A | 109.5 | C6—C5—H5A | 108.9 |
| C2—N1—H1B | 109.5 | C4—C5—H5A | 108.9 |
| H1A—N1—H1B | 109.5 | C6—C5—H5B | 108.9 |
| C2—N1—H1C | 109.5 | C4—C5—H5B | 108.9 |
| H1A—N1—H1C | 109.5 | H5A—C5—H5B | 107.7 |
| H1B—N1—H1C | 109.5 | C7—C6—C5 | 112.19 (11) |
| N1—C2—C1 | 111.98 (11) | C7—C6—H6B | 109.2 |
| N1—C2—H2A | 109.2 | C5—C6—H6B | 109.2 |
| C1—C2—H2A | 109.2 | C7—C6—H6A | 109.2 |
| N1—C2—H2B | 109.2 | C5—C6—H6A | 109.2 |
| C1—C2—H2B | 109.2 | H6B—C6—H6A | 107.9 |
| H2A—C2—H2B | 107.9 | O3—C3—O4 | 122.19 (14) |
| C7—O6—H6 | 109.5 | O3—C3—C4 | 123.87 (13) |
| O2—C1—O1 | 125.24 (14) | O4—C3—C4 | 113.93 (12) |
| O2—C1—C2 | 117.27 (12) | C3—C4—C5 | 114.22 (11) |
| O1—C1—C2 | 117.49 (12) | C3—C4—H4A | 108.7 |
| O5—C7—O6 | 119.33 (13) | C5—C4—H4A | 108.7 |
| O5—C7—C6 | 122.19 (13) | C3—C4—H4B | 108.7 |
| O6—C7—C6 | 118.47 (12) | C5—C4—H4B | 108.7 |
| C3—O4—H4 | 109.5 | H4A—C4—H4B | 107.6 |
| C6—C5—C4 | 113.44 (11) | | |
| | | |
| N1—C2—C1—O2 | −173.27 (13) | C4—C5—C6—C7 | −169.30 (14) |
| N1—C2—C1—O1 | 7.84 (19) | O3—C3—C4—C5 | 146.60 (16) |
| O5—C7—C6—C5 | 10.6 (2) | O4—C3—C4—C5 | −34.2 (2) |
| O6—C7—C6—C5 | −170.43 (14) | C6—C5—C4—C3 | −179.71 (14) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1B···O3i | 0.90 | 2.11 | 2.9420 (16) | 154 |
| N1—H1C···O3ii | 0.90 | 1.98 | 2.8716 (16) | 170 |
| N1—H1A···O5 | 0.90 | 1.99 | 2.7641 (15) | 143 |
| O4—H4···O1iii | 0.83 | 1.74 | 2.5650 (15) | 175 |
| O6—H6···O2iv | 0.83 | 1.73 | 2.5417 (15) | 165 |
| Symmetry codes: (i) −x+1, y−1/2, −z+1/2; (ii) x, −y+3/2, z+1/2; (iii) −x, y+1/2, −z+1/2; (iv) −x+1, −y+1, −z. |
(200) Propane-1,3-dicarboxylic acid – Aminoethanoic acid (1/1)
top
Crystal data top
| C5H8O4·C2H5NO2 | Z = 4 |
| Mr = 207.18 | F(000) = 440 |
| Triclinic, P1 | Dx = 1.486 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 4.9155 (7) Å | Cell parameters from 6919 reflections |
| b = 20.215 (3) Å | θ = 2.0–29.6° |
| c = 10.1862 (16) Å | µ = 0.13 mm−1 |
| α = 85.578 (14)° | T = 200 K |
| β = 113.133 (11)° | Prism, colourless |
| γ = 88.301 (13)° | 0.40 × 0.23 × 0.10 mm |
| V = 926.1 (2) Å3 | |
Data collection top
STOE IPDS 2
diffractometer | 2491 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.055 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 29.2°, θmin = 2.0° |
| rotation method scans | h = −6→5 |
| 9663 measured reflections | k = −27→27 |
| 4312 independent reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.049 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.081 | H-atom parameters constrained |
| S = 0.89 | w = 1/[σ2(Fo2) + (0.0266P)2]
where P = (Fo2 + 2Fc2)/3 |
| 4312 reflections | (Δ/σ)max = 0.001 |
| 259 parameters | Δρmax = 0.19 e Å−3 |
| 0 restraints | Δρmin = −0.21 e Å−3 |
Crystal data top
| C5H8O4·C2H5NO2 | γ = 88.301 (13)° |
| Mr = 207.18 | V = 926.1 (2) Å3 |
| Triclinic, P1 | Z = 4 |
| a = 4.9155 (7) Å | Mo Kα radiation |
| b = 20.215 (3) Å | µ = 0.13 mm−1 |
| c = 10.1862 (16) Å | T = 200 K |
| α = 85.578 (14)° | 0.40 × 0.23 × 0.10 mm |
| β = 113.133 (11)° | |
Data collection top
STOE IPDS 2
diffractometer | 2491 reflections with I > 2σ(I) |
| 9663 measured reflections | Rint = 0.055 |
| 4312 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.049 | 0 restraints |
| wR(F2) = 0.081 | H-atom parameters constrained |
| S = 0.89 | Δρmax = 0.19 e Å−3 |
| 4312 reflections | Δρmin = −0.21 e Å−3 |
| 259 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O23 | 0.1871 (4) | 0.58896 (6) | 0.45166 (16) | 0.0260 (4) | |
| O25 | 0.6524 (4) | 0.84815 (6) | 0.75064 (16) | 0.0284 (4) | |
| O26 | 0.8511 (4) | 0.90410 (6) | 0.62247 (16) | 0.0289 (4) | |
| H26 | 0.8788 | 0.9011 | 0.5468 | 0.043* | |
| O21 | 0.1282 (3) | 1.05514 (6) | 0.82424 (16) | 0.0261 (3) | |
| O22 | 0.0124 (4) | 1.09123 (7) | 0.59539 (16) | 0.0297 (4) | |
| O13 | 0.7719 (3) | 0.08769 (6) | 1.01431 (16) | 0.0250 (3) | |
| O24 | 0.0718 (4) | 0.65453 (6) | 0.59001 (18) | 0.0316 (4) | |
| H24 | 0.0020 | 0.6197 | 0.6105 | 0.047* | |
| O16 | 0.1845 (4) | 0.40381 (7) | 0.90921 (17) | 0.0328 (4) | |
| H16 | 0.1809 | 0.4042 | 0.9909 | 0.049* | |
| O14 | 0.7927 (4) | 0.15354 (6) | 0.83429 (16) | 0.0297 (4) | |
| H14 | 0.8949 | 0.1206 | 0.8291 | 0.045* | |
| O15 | 0.4073 (4) | 0.34554 (6) | 0.80336 (18) | 0.0374 (4) | |
| N21 | 0.5779 (4) | 0.97095 (7) | 0.84561 (18) | 0.0225 (4) | |
| H21A | 0.6957 | 0.9380 | 0.8351 | 0.034* | |
| H21B | 0.4520 | 0.9529 | 0.8822 | 0.034* | |
| H21C | 0.6926 | 0.9992 | 0.9064 | 0.034* | |
| C22 | 0.4041 (5) | 1.00798 (9) | 0.7040 (2) | 0.0200 (4) | |
| H22A | 0.3119 | 0.9759 | 0.6331 | 0.024* | |
| H22B | 0.5394 | 1.0335 | 0.6720 | 0.024* | |
| C13 | 0.7020 (5) | 0.14025 (8) | 0.9381 (2) | 0.0197 (4) | |
| C25 | 0.4718 (5) | 0.74512 (9) | 0.5767 (2) | 0.0219 (4) | |
| H25A | 0.3256 | 0.7615 | 0.6133 | 0.026* | |
| H25B | 0.6275 | 0.7168 | 0.6557 | 0.026* | |
| C23 | 0.1863 (5) | 0.64326 (9) | 0.4972 (2) | 0.0204 (4) | |
| C21 | 0.1633 (5) | 1.05543 (8) | 0.7089 (2) | 0.0190 (4) | |
| C26 | 0.6120 (5) | 0.80423 (8) | 0.5320 (2) | 0.0209 (4) | |
| H26A | 0.7865 | 0.7880 | 0.5151 | 0.025* | |
| H26B | 0.4674 | 0.8273 | 0.4408 | 0.025* | |
| C24 | 0.3174 (5) | 0.70327 (9) | 0.4526 (2) | 0.0239 (5) | |
| H24A | 0.1576 | 0.7313 | 0.3749 | 0.029* | |
| H24B | 0.4623 | 0.6882 | 0.4139 | 0.029* | |
| C16 | 0.5015 (5) | 0.30815 (9) | 1.0471 (2) | 0.0243 (5) | |
| H16A | 0.3435 | 0.2907 | 1.0748 | 0.029* | |
| H16B | 0.6250 | 0.3348 | 1.1237 | 0.029* | |
| C14 | 0.5083 (5) | 0.19546 (9) | 0.9534 (2) | 0.0233 (4) | |
| H14A | 0.3877 | 0.1772 | 1.0042 | 0.028* | |
| H14B | 0.3713 | 0.2152 | 0.8571 | 0.028* | |
| C15 | 0.6943 (5) | 0.24964 (9) | 1.0370 (2) | 0.0286 (5) | |
| H15A | 0.8238 | 0.2302 | 1.1349 | 0.034* | |
| H15B | 0.8231 | 0.2660 | 0.9889 | 0.034* | |
| C27 | 0.7073 (5) | 0.85271 (8) | 0.6442 (2) | 0.0198 (4) | |
| C17 | 0.3605 (5) | 0.35270 (9) | 0.9092 (2) | 0.0245 (5) | |
| O11 | 0.8400 (3) | 0.55159 (6) | 0.65415 (15) | 0.0254 (3) | |
| O12 | 0.9335 (4) | 0.58758 (7) | 0.86847 (17) | 0.0301 (4) | |
| N11 | 0.4288 (4) | 0.47027 (7) | 0.66308 (18) | 0.0216 (4) | |
| H11A | 0.3137 | 0.4400 | 0.6832 | 0.032* | |
| H11B | 0.5664 | 0.4489 | 0.6396 | 0.032* | |
| H11C | 0.3131 | 0.4997 | 0.5882 | 0.032* | |
| C12 | 0.5809 (5) | 0.50654 (8) | 0.7909 (2) | 0.0191 (4) | |
| H12A | 0.6858 | 0.4741 | 0.8753 | 0.023* | |
| H12B | 0.4314 | 0.5331 | 0.8108 | 0.023* | |
| C11 | 0.8023 (5) | 0.55239 (8) | 0.7686 (2) | 0.0188 (4) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O23 | 0.0322 (10) | 0.0197 (7) | 0.0298 (9) | −0.0035 (6) | 0.0153 (8) | −0.0067 (6) |
| O25 | 0.0419 (10) | 0.0231 (7) | 0.0283 (9) | −0.0067 (6) | 0.0213 (8) | −0.0080 (6) |
| O26 | 0.0392 (10) | 0.0242 (7) | 0.0295 (9) | −0.0116 (6) | 0.0189 (8) | −0.0074 (6) |
| O21 | 0.0308 (9) | 0.0263 (7) | 0.0239 (8) | 0.0063 (6) | 0.0144 (7) | −0.0009 (6) |
| O22 | 0.0279 (9) | 0.0339 (8) | 0.0268 (9) | 0.0078 (6) | 0.0127 (7) | 0.0068 (6) |
| O13 | 0.0293 (9) | 0.0192 (7) | 0.0264 (9) | 0.0014 (6) | 0.0118 (7) | 0.0003 (5) |
| O24 | 0.0453 (11) | 0.0234 (7) | 0.0428 (10) | −0.0147 (7) | 0.0328 (9) | −0.0115 (6) |
| O16 | 0.0458 (12) | 0.0251 (7) | 0.0305 (9) | 0.0107 (6) | 0.0201 (9) | 0.0021 (6) |
| O14 | 0.0396 (11) | 0.0245 (7) | 0.0308 (9) | 0.0094 (6) | 0.0222 (8) | 0.0043 (6) |
| O15 | 0.0639 (13) | 0.0248 (8) | 0.0372 (10) | 0.0047 (7) | 0.0362 (10) | 0.0024 (6) |
| N21 | 0.0264 (10) | 0.0208 (8) | 0.0225 (9) | 0.0036 (7) | 0.0121 (8) | −0.0029 (6) |
| C22 | 0.0201 (11) | 0.0220 (9) | 0.0195 (10) | −0.0014 (7) | 0.0093 (9) | −0.0033 (7) |
| C13 | 0.0183 (11) | 0.0191 (9) | 0.0221 (11) | −0.0045 (7) | 0.0075 (9) | −0.0062 (7) |
| C25 | 0.0258 (12) | 0.0215 (9) | 0.0215 (11) | −0.0061 (8) | 0.0119 (9) | −0.0043 (7) |
| C23 | 0.0184 (11) | 0.0217 (9) | 0.0203 (11) | −0.0031 (7) | 0.0066 (9) | −0.0026 (7) |
| C21 | 0.0180 (11) | 0.0172 (9) | 0.0241 (11) | −0.0050 (7) | 0.0102 (9) | −0.0036 (7) |
| C26 | 0.0251 (12) | 0.0194 (9) | 0.0201 (11) | −0.0037 (7) | 0.0107 (9) | −0.0035 (7) |
| C24 | 0.0307 (13) | 0.0234 (9) | 0.0218 (11) | −0.0083 (8) | 0.0137 (10) | −0.0056 (7) |
| C16 | 0.0268 (12) | 0.0205 (9) | 0.0235 (11) | 0.0004 (8) | 0.0071 (10) | −0.0055 (7) |
| C14 | 0.0227 (12) | 0.0191 (9) | 0.0300 (12) | −0.0010 (7) | 0.0122 (10) | −0.0037 (7) |
| C15 | 0.0228 (12) | 0.0266 (10) | 0.0280 (12) | 0.0045 (8) | 0.0008 (10) | −0.0066 (8) |
| C27 | 0.0209 (11) | 0.0161 (9) | 0.0211 (11) | 0.0032 (7) | 0.0077 (9) | 0.0004 (7) |
| C17 | 0.0324 (13) | 0.0163 (9) | 0.0298 (12) | −0.0050 (8) | 0.0171 (11) | −0.0032 (8) |
| O11 | 0.0305 (9) | 0.0265 (7) | 0.0249 (8) | −0.0123 (6) | 0.0155 (7) | −0.0069 (6) |
| O12 | 0.0331 (10) | 0.0333 (8) | 0.0306 (9) | −0.0149 (7) | 0.0165 (8) | −0.0163 (6) |
| N11 | 0.0254 (10) | 0.0214 (8) | 0.0218 (9) | −0.0076 (7) | 0.0128 (8) | −0.0028 (6) |
| C12 | 0.0187 (11) | 0.0208 (9) | 0.0190 (10) | −0.0015 (7) | 0.0088 (9) | −0.0020 (7) |
| C11 | 0.0158 (10) | 0.0169 (9) | 0.0238 (11) | 0.0005 (7) | 0.0084 (9) | −0.0009 (7) |
Geometric parameters (Å, º) top
| O23—C23 | 1.225 (2) | C25—H25B | 0.9900 |
| O25—C27 | 1.214 (2) | C23—C24 | 1.512 (3) |
| O26—C27 | 1.326 (2) | C26—C27 | 1.505 (3) |
| O26—H26 | 0.8400 | C26—H26A | 0.9900 |
| O21—C21 | 1.252 (2) | C26—H26B | 0.9900 |
| O22—C21 | 1.256 (2) | C24—H24A | 0.9900 |
| O13—C13 | 1.221 (2) | C24—H24B | 0.9900 |
| O24—C23 | 1.306 (2) | C16—C17 | 1.506 (3) |
| O24—H24 | 0.8400 | C16—C15 | 1.529 (3) |
| O16—C17 | 1.328 (3) | C16—H16A | 0.9900 |
| O16—H16 | 0.8400 | C16—H16B | 0.9900 |
| O14—C13 | 1.313 (2) | C14—C15 | 1.533 (3) |
| O14—H14 | 0.8400 | C14—H14A | 0.9900 |
| O15—C17 | 1.205 (3) | C14—H14B | 0.9900 |
| N21—C22 | 1.482 (2) | C15—H15A | 0.9900 |
| N21—H21A | 0.9100 | C15—H15B | 0.9900 |
| N21—H21B | 0.9100 | O11—C11 | 1.252 (2) |
| N21—H21C | 0.9100 | O12—C11 | 1.257 (2) |
| C22—C21 | 1.520 (3) | N11—C12 | 1.486 (3) |
| C22—H22A | 0.9900 | N11—H11A | 0.9100 |
| C22—H22B | 0.9900 | N11—H11B | 0.9100 |
| C13—C14 | 1.502 (3) | N11—H11C | 0.9100 |
| C25—C26 | 1.524 (2) | C12—C11 | 1.522 (3) |
| C25—C24 | 1.530 (3) | C12—H12A | 0.9900 |
| C25—H25A | 0.9900 | C12—H12B | 0.9900 |
| | | |
| C27—O26—H26 | 109.5 | C25—C24—H24B | 109.1 |
| C23—O24—H24 | 109.5 | H24A—C24—H24B | 107.8 |
| C17—O16—H16 | 109.5 | C17—C16—C15 | 113.30 (18) |
| C13—O14—H14 | 109.5 | C17—C16—H16A | 108.9 |
| C22—N21—H21A | 109.5 | C15—C16—H16A | 108.9 |
| C22—N21—H21B | 109.5 | C17—C16—H16B | 108.9 |
| H21A—N21—H21B | 109.5 | C15—C16—H16B | 108.9 |
| C22—N21—H21C | 109.5 | H16A—C16—H16B | 107.7 |
| H21A—N21—H21C | 109.5 | C13—C14—C15 | 111.21 (18) |
| H21B—N21—H21C | 109.5 | C13—C14—H14A | 109.4 |
| N21—C22—C21 | 112.13 (16) | C15—C14—H14A | 109.4 |
| N21—C22—H22A | 109.2 | C13—C14—H14B | 109.4 |
| C21—C22—H22A | 109.2 | C15—C14—H14B | 109.4 |
| N21—C22—H22B | 109.2 | H14A—C14—H14B | 108.0 |
| C21—C22—H22B | 109.2 | C16—C15—C14 | 112.17 (19) |
| H22A—C22—H22B | 107.9 | C16—C15—H15A | 109.2 |
| O13—C13—O14 | 122.23 (18) | C14—C15—H15A | 109.2 |
| O13—C13—C14 | 124.05 (18) | C16—C15—H15B | 109.2 |
| O14—C13—C14 | 113.72 (16) | C14—C15—H15B | 109.2 |
| C26—C25—C24 | 112.25 (16) | H15A—C15—H15B | 107.9 |
| C26—C25—H25A | 109.2 | O25—C27—O26 | 118.69 (18) |
| C24—C25—H25A | 109.2 | O25—C27—C26 | 123.50 (17) |
| C26—C25—H25B | 109.2 | O26—C27—C26 | 117.78 (17) |
| C24—C25—H25B | 109.2 | O15—C17—O16 | 119.58 (19) |
| H25A—C25—H25B | 107.9 | O15—C17—C16 | 123.7 (2) |
| O23—C23—O24 | 122.74 (17) | O16—C17—C16 | 116.62 (18) |
| O23—C23—C24 | 123.6 (2) | C12—N11—H11A | 109.5 |
| O24—C23—C24 | 113.64 (17) | C12—N11—H11B | 109.5 |
| O21—C21—O22 | 125.30 (19) | H11A—N11—H11B | 109.5 |
| O21—C21—C22 | 117.85 (17) | C12—N11—H11C | 109.5 |
| O22—C21—C22 | 116.85 (17) | H11A—N11—H11C | 109.5 |
| C27—C26—C25 | 111.92 (17) | H11B—N11—H11C | 109.5 |
| C27—C26—H26A | 109.2 | N11—C12—C11 | 111.60 (16) |
| C25—C26—H26A | 109.2 | N11—C12—H12A | 109.3 |
| C27—C26—H26B | 109.2 | C11—C12—H12A | 109.3 |
| C25—C26—H26B | 109.2 | N11—C12—H12B | 109.3 |
| H26A—C26—H26B | 107.9 | C11—C12—H12B | 109.3 |
| C23—C24—C25 | 112.57 (17) | H12A—C12—H12B | 108.0 |
| C23—C24—H24A | 109.1 | O11—C11—O12 | 125.71 (18) |
| C25—C24—H24A | 109.1 | O11—C11—C12 | 117.77 (17) |
| C23—C24—H24B | 109.1 | O12—C11—C12 | 116.51 (18) |
| | | |
| N21—C22—C21—O21 | −4.0 (2) | C17—C16—C15—C14 | −75.0 (2) |
| N21—C22—C21—O22 | 176.98 (16) | C13—C14—C15—C16 | 176.98 (18) |
| C24—C25—C26—C27 | 168.16 (18) | C25—C26—C27—O25 | −6.6 (3) |
| O23—C23—C24—C25 | −142.3 (2) | C25—C26—C27—O26 | 175.47 (18) |
| O24—C23—C24—C25 | 37.5 (3) | C15—C16—C17—O15 | −4.1 (3) |
| C26—C25—C24—C23 | 178.14 (18) | C15—C16—C17—O16 | 178.57 (19) |
| O13—C13—C14—C15 | 99.0 (2) | N11—C12—C11—O11 | −3.6 (2) |
| O14—C13—C14—C15 | −80.7 (2) | N11—C12—C11—O12 | 176.97 (17) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O14—H14···O21i | 0.84 | 1.74 | 2.580 (2) | 176 |
| O16—H16···O12ii | 0.84 | 1.75 | 2.565 (2) | 164 |
| N11—H11A···O15 | 0.91 | 2.12 | 2.827 (2) | 134 |
| N11—H11B···O23iii | 0.91 | 1.96 | 2.860 (2) | 168 |
| N11—H11C···O23 | 0.91 | 2.11 | 2.971 (2) | 158 |
| O24—H24···O11iv | 0.84 | 1.72 | 2.5636 (18) | 176 |
| O26—H26···O22v | 0.84 | 1.73 | 2.555 (2) | 167 |
| N21—H21A···O25 | 0.91 | 2.05 | 2.798 (2) | 139 |
| N21—H21B···O13ii | 0.91 | 1.96 | 2.861 (2) | 172 |
| N21—H21C···O13vi | 0.91 | 2.14 | 2.984 (2) | 155 |
| Symmetry codes: (i) x+1, y−1, z; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+1, −z+1; (iv) x−1, y, z; (v) −x+1, −y+2, −z+1; (vi) x, y+1, z. |
(175) Propane-1,3-dicarboxylic acid – Aminoethanoic acid (1/1)
top
Crystal data top
| C5H8O4·C2H5NO2 | Z = 4 |
| Mr = 207.18 | F(000) = 440 |
| Triclinic, P1 | Dx = 1.490 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 4.9098 (7) Å | Cell parameters from 7461 reflections |
| b = 20.194 (3) Å | θ = 2.1–29.6° |
| c = 10.1777 (15) Å | µ = 0.13 mm−1 |
| α = 85.571 (13)° | T = 175 K |
| β = 113.118 (10)° | Prism, colourless |
| γ = 88.306 (12)° | 0.40 × 0.23 × 0.10 mm |
| V = 923.3 (2) Å3 | |
Data collection top
STOE IPDS 2
diffractometer | 2580 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.052 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 29.2°, θmin = 2.0° |
| rotation method scans | h = −6→5 |
| 9568 measured reflections | k = −27→27 |
| 4319 independent reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.049 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.088 | H-atom parameters constrained |
| S = 0.89 | w = 1/[σ2(Fo2) + (0.0329P)2]
where P = (Fo2 + 2Fc2)/3 |
| 4319 reflections | (Δ/σ)max < 0.001 |
| 259 parameters | Δρmax = 0.22 e Å−3 |
| 0 restraints | Δρmin = −0.23 e Å−3 |
Crystal data top
| C5H8O4·C2H5NO2 | γ = 88.306 (12)° |
| Mr = 207.18 | V = 923.3 (2) Å3 |
| Triclinic, P1 | Z = 4 |
| a = 4.9098 (7) Å | Mo Kα radiation |
| b = 20.194 (3) Å | µ = 0.13 mm−1 |
| c = 10.1777 (15) Å | T = 175 K |
| α = 85.571 (13)° | 0.40 × 0.23 × 0.10 mm |
| β = 113.118 (10)° | |
Data collection top
STOE IPDS 2
diffractometer | 2580 reflections with I > 2σ(I) |
| 9568 measured reflections | Rint = 0.052 |
| 4319 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.049 | 0 restraints |
| wR(F2) = 0.088 | H-atom parameters constrained |
| S = 0.89 | Δρmax = 0.22 e Å−3 |
| 4319 reflections | Δρmin = −0.23 e Å−3 |
| 259 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O23 | 0.1870 (4) | 0.58898 (6) | 0.45114 (16) | 0.0223 (3) | |
| O25 | 0.6532 (4) | 0.84810 (6) | 0.75156 (17) | 0.0248 (3) | |
| O26 | 0.8537 (4) | 0.90432 (6) | 0.62332 (17) | 0.0253 (4) | |
| H26 | 0.8731 | 0.9023 | 0.5453 | 0.038* | |
| O21 | 0.1280 (4) | 1.05536 (6) | 0.82397 (16) | 0.0232 (3) | |
| O22 | 0.0102 (4) | 1.09092 (7) | 0.59471 (17) | 0.0263 (4) | |
| O13 | 0.7721 (3) | 0.08762 (6) | 1.01479 (16) | 0.0206 (3) | |
| O24 | 0.0731 (4) | 0.65463 (6) | 0.59004 (18) | 0.0271 (4) | |
| H24 | 0.0011 | 0.6199 | 0.6096 | 0.041* | |
| O16 | 0.1825 (4) | 0.40414 (7) | 0.90858 (17) | 0.0281 (4) | |
| H16 | 0.1714 | 0.4036 | 0.9889 | 0.042* | |
| O14 | 0.7902 (4) | 0.15354 (7) | 0.83352 (17) | 0.0262 (4) | |
| H14 | 0.8959 | 0.1209 | 0.8296 | 0.039* | |
| O15 | 0.4050 (4) | 0.34553 (7) | 0.80242 (18) | 0.0324 (4) | |
| N21 | 0.5783 (4) | 0.97136 (7) | 0.84576 (19) | 0.0194 (4) | |
| H21A | 0.6999 | 0.9390 | 0.8359 | 0.029* | |
| H21B | 0.4525 | 0.9525 | 0.8815 | 0.029* | |
| H21C | 0.6895 | 1.0000 | 0.9071 | 0.029* | |
| C22 | 0.4044 (5) | 1.00819 (9) | 0.7039 (2) | 0.0174 (4) | |
| H22A | 0.3135 | 0.9760 | 0.6330 | 0.021* | |
| H22B | 0.5395 | 1.0340 | 0.6719 | 0.021* | |
| C13 | 0.6991 (5) | 0.14026 (9) | 0.9372 (2) | 0.0167 (4) | |
| C25 | 0.4744 (5) | 0.74496 (9) | 0.5774 (2) | 0.0189 (4) | |
| H25A | 0.3283 | 0.7612 | 0.6144 | 0.023* | |
| H25B | 0.6304 | 0.7165 | 0.6561 | 0.023* | |
| C23 | 0.1879 (5) | 0.64342 (9) | 0.4973 (2) | 0.0174 (4) | |
| C21 | 0.1618 (5) | 1.05536 (9) | 0.7083 (2) | 0.0172 (4) | |
| C26 | 0.6149 (5) | 0.80440 (9) | 0.5328 (2) | 0.0181 (4) | |
| H26A | 0.7902 | 0.7883 | 0.5163 | 0.022* | |
| H26B | 0.4704 | 0.8274 | 0.4414 | 0.022* | |
| C24 | 0.3189 (5) | 0.70327 (9) | 0.4523 (2) | 0.0204 (4) | |
| H24A | 0.1590 | 0.7314 | 0.3748 | 0.025* | |
| H24B | 0.4636 | 0.6882 | 0.4135 | 0.025* | |
| C16 | 0.4997 (5) | 0.30820 (9) | 1.0471 (2) | 0.0213 (4) | |
| H16A | 0.3413 | 0.2907 | 1.0747 | 0.026* | |
| H16B | 0.6229 | 0.3349 | 1.1238 | 0.026* | |
| C14 | 0.5064 (5) | 0.19536 (9) | 0.9533 (2) | 0.0201 (4) | |
| H14A | 0.3859 | 0.1769 | 1.0042 | 0.024* | |
| H14B | 0.3689 | 0.2153 | 0.8571 | 0.024* | |
| C15 | 0.6933 (5) | 0.24963 (9) | 1.0374 (2) | 0.0251 (5) | |
| H15A | 0.8221 | 0.2302 | 1.1355 | 0.030* | |
| H15B | 0.8229 | 0.2659 | 0.9897 | 0.030* | |
| C27 | 0.7086 (5) | 0.85301 (8) | 0.6451 (2) | 0.0173 (4) | |
| C17 | 0.3590 (5) | 0.35270 (9) | 0.9092 (2) | 0.0202 (4) | |
| O11 | 0.8391 (3) | 0.55165 (6) | 0.65370 (16) | 0.0223 (3) | |
| O12 | 0.9358 (4) | 0.58730 (7) | 0.86881 (17) | 0.0259 (4) | |
| N11 | 0.4277 (4) | 0.47053 (7) | 0.66306 (18) | 0.0187 (4) | |
| H11A | 0.3117 | 0.4404 | 0.6830 | 0.028* | |
| H11B | 0.5652 | 0.4490 | 0.6395 | 0.028* | |
| H11C | 0.3126 | 0.5001 | 0.5882 | 0.028* | |
| C12 | 0.5803 (5) | 0.50660 (9) | 0.7910 (2) | 0.0166 (4) | |
| H12A | 0.6848 | 0.4740 | 0.8753 | 0.020* | |
| H12B | 0.4311 | 0.5333 | 0.8111 | 0.020* | |
| C11 | 0.8031 (5) | 0.55238 (8) | 0.7686 (2) | 0.0168 (4) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O23 | 0.0276 (9) | 0.0155 (7) | 0.0266 (9) | −0.0038 (6) | 0.0128 (7) | −0.0062 (5) |
| O25 | 0.0364 (10) | 0.0207 (7) | 0.0244 (9) | −0.0062 (6) | 0.0183 (8) | −0.0074 (5) |
| O26 | 0.0346 (10) | 0.0213 (7) | 0.0245 (9) | −0.0103 (6) | 0.0151 (8) | −0.0065 (6) |
| O21 | 0.0272 (9) | 0.0230 (7) | 0.0217 (8) | 0.0057 (6) | 0.0130 (7) | −0.0014 (6) |
| O22 | 0.0259 (9) | 0.0286 (8) | 0.0247 (9) | 0.0073 (6) | 0.0125 (7) | 0.0066 (6) |
| O13 | 0.0233 (9) | 0.0159 (6) | 0.0222 (8) | 0.0001 (5) | 0.0089 (7) | −0.0001 (5) |
| O24 | 0.0394 (10) | 0.0194 (7) | 0.0370 (10) | −0.0121 (7) | 0.0286 (9) | −0.0097 (6) |
| O16 | 0.0381 (11) | 0.0225 (7) | 0.0254 (9) | 0.0085 (6) | 0.0160 (8) | 0.0018 (6) |
| O14 | 0.0339 (10) | 0.0221 (7) | 0.0269 (9) | 0.0089 (6) | 0.0187 (8) | 0.0041 (6) |
| O15 | 0.0544 (12) | 0.0215 (7) | 0.0330 (10) | 0.0028 (7) | 0.0309 (9) | 0.0018 (6) |
| N21 | 0.0239 (10) | 0.0165 (8) | 0.0199 (9) | 0.0024 (6) | 0.0113 (8) | −0.0019 (6) |
| C22 | 0.0172 (11) | 0.0188 (9) | 0.0167 (10) | −0.0025 (7) | 0.0070 (8) | −0.0028 (7) |
| C13 | 0.0172 (11) | 0.0159 (8) | 0.0176 (11) | −0.0030 (7) | 0.0071 (9) | −0.0036 (7) |
| C25 | 0.0197 (11) | 0.0187 (9) | 0.0195 (10) | −0.0039 (7) | 0.0088 (9) | −0.0024 (7) |
| C23 | 0.0174 (11) | 0.0184 (9) | 0.0161 (10) | −0.0018 (7) | 0.0065 (9) | −0.0008 (7) |
| C21 | 0.0164 (11) | 0.0158 (9) | 0.0220 (11) | −0.0044 (7) | 0.0100 (9) | −0.0036 (7) |
| C26 | 0.0219 (11) | 0.0166 (9) | 0.0179 (10) | −0.0026 (7) | 0.0094 (9) | −0.0041 (7) |
| C24 | 0.0254 (12) | 0.0201 (9) | 0.0196 (11) | −0.0070 (8) | 0.0120 (10) | −0.0055 (7) |
| C16 | 0.0217 (12) | 0.0185 (9) | 0.0203 (11) | 0.0000 (8) | 0.0043 (9) | −0.0052 (7) |
| C14 | 0.0199 (11) | 0.0173 (9) | 0.0250 (11) | −0.0009 (7) | 0.0108 (9) | −0.0035 (7) |
| C15 | 0.0211 (12) | 0.0223 (10) | 0.0262 (12) | 0.0045 (8) | 0.0031 (10) | −0.0060 (8) |
| C27 | 0.0159 (10) | 0.0147 (8) | 0.0192 (11) | 0.0020 (7) | 0.0052 (9) | −0.0005 (7) |
| C17 | 0.0235 (12) | 0.0155 (9) | 0.0250 (12) | −0.0040 (8) | 0.0128 (10) | −0.0027 (7) |
| O11 | 0.0278 (9) | 0.0226 (7) | 0.0217 (8) | −0.0101 (6) | 0.0144 (7) | −0.0051 (5) |
| O12 | 0.0278 (9) | 0.0294 (8) | 0.0259 (9) | −0.0126 (6) | 0.0136 (7) | −0.0141 (6) |
| N11 | 0.0233 (10) | 0.0171 (8) | 0.0192 (9) | −0.0069 (7) | 0.0116 (8) | −0.0025 (6) |
| C12 | 0.0164 (11) | 0.0186 (9) | 0.0157 (10) | −0.0016 (7) | 0.0075 (8) | −0.0018 (7) |
| C11 | 0.0164 (10) | 0.0143 (8) | 0.0209 (11) | 0.0007 (7) | 0.0091 (9) | −0.0003 (7) |
Geometric parameters (Å, º) top
| O23—C23 | 1.228 (2) | C25—H25B | 0.9900 |
| O25—C27 | 1.214 (3) | C23—C24 | 1.509 (3) |
| O26—C27 | 1.328 (2) | C26—C27 | 1.506 (3) |
| O26—H26 | 0.8400 | C26—H26A | 0.9900 |
| O21—C21 | 1.252 (3) | C26—H26B | 0.9900 |
| O22—C21 | 1.253 (2) | C24—H24A | 0.9900 |
| O13—C13 | 1.228 (2) | C24—H24B | 0.9900 |
| O24—C23 | 1.305 (3) | C16—C17 | 1.504 (3) |
| O24—H24 | 0.8400 | C16—C15 | 1.529 (3) |
| O16—C17 | 1.331 (3) | C16—H16A | 0.9900 |
| O16—H16 | 0.8400 | C16—H16B | 0.9900 |
| O14—C13 | 1.311 (2) | C14—C15 | 1.537 (3) |
| O14—H14 | 0.8400 | C14—H14A | 0.9900 |
| O15—C17 | 1.212 (3) | C14—H14B | 0.9900 |
| N21—C22 | 1.482 (3) | C15—H15A | 0.9900 |
| N21—H21A | 0.9100 | C15—H15B | 0.9900 |
| N21—H21B | 0.9100 | O11—C11 | 1.251 (2) |
| N21—H21C | 0.9100 | O12—C11 | 1.256 (2) |
| C22—C21 | 1.520 (3) | N11—C12 | 1.483 (3) |
| C22—H22A | 0.9900 | N11—H11A | 0.9100 |
| C22—H22B | 0.9900 | N11—H11B | 0.9100 |
| C13—C14 | 1.499 (3) | N11—H11C | 0.9100 |
| C25—C26 | 1.527 (3) | C12—C11 | 1.525 (3) |
| C25—C24 | 1.534 (3) | C12—H12A | 0.9900 |
| C25—H25A | 0.9900 | C12—H12B | 0.9900 |
| | | |
| C27—O26—H26 | 109.5 | C25—C24—H24B | 109.2 |
| C23—O24—H24 | 109.5 | H24A—C24—H24B | 107.9 |
| C17—O16—H16 | 109.5 | C17—C16—C15 | 113.39 (18) |
| C13—O14—H14 | 109.5 | C17—C16—H16A | 108.9 |
| C22—N21—H21A | 109.5 | C15—C16—H16A | 108.9 |
| C22—N21—H21B | 109.5 | C17—C16—H16B | 108.9 |
| H21A—N21—H21B | 109.5 | C15—C16—H16B | 108.9 |
| C22—N21—H21C | 109.5 | H16A—C16—H16B | 107.7 |
| H21A—N21—H21C | 109.5 | C13—C14—C15 | 111.32 (18) |
| H21B—N21—H21C | 109.5 | C13—C14—H14A | 109.4 |
| N21—C22—C21 | 112.08 (16) | C15—C14—H14A | 109.4 |
| N21—C22—H22A | 109.2 | C13—C14—H14B | 109.4 |
| C21—C22—H22A | 109.2 | C15—C14—H14B | 109.4 |
| N21—C22—H22B | 109.2 | H14A—C14—H14B | 108.0 |
| C21—C22—H22B | 109.2 | C16—C15—C14 | 112.01 (18) |
| H22A—C22—H22B | 107.9 | C16—C15—H15A | 109.2 |
| O13—C13—O14 | 122.14 (18) | C14—C15—H15A | 109.2 |
| O13—C13—C14 | 123.87 (18) | C16—C15—H15B | 109.2 |
| O14—C13—C14 | 113.97 (16) | C14—C15—H15B | 109.2 |
| C26—C25—C24 | 112.14 (17) | H15A—C15—H15B | 107.9 |
| C26—C25—H25A | 109.2 | O25—C27—O26 | 119.15 (18) |
| C24—C25—H25A | 109.2 | O25—C27—C26 | 123.38 (17) |
| C26—C25—H25B | 109.2 | O26—C27—C26 | 117.45 (18) |
| C24—C25—H25B | 109.2 | O15—C17—O16 | 119.13 (18) |
| H25A—C25—H25B | 107.9 | O15—C17—C16 | 123.97 (19) |
| O23—C23—O24 | 122.57 (17) | O16—C17—C16 | 116.85 (18) |
| O23—C23—C24 | 123.5 (2) | C12—N11—H11A | 109.5 |
| O24—C23—C24 | 113.88 (17) | C12—N11—H11B | 109.5 |
| O21—C21—O22 | 125.3 (2) | H11A—N11—H11B | 109.5 |
| O21—C21—C22 | 117.76 (18) | C12—N11—H11C | 109.5 |
| O22—C21—C22 | 116.92 (18) | H11A—N11—H11C | 109.5 |
| C27—C26—C25 | 111.87 (17) | H11B—N11—H11C | 109.5 |
| C27—C26—H26A | 109.2 | N11—C12—C11 | 111.48 (16) |
| C25—C26—H26A | 109.2 | N11—C12—H12A | 109.3 |
| C27—C26—H26B | 109.2 | C11—C12—H12A | 109.3 |
| C25—C26—H26B | 109.2 | N11—C12—H12B | 109.3 |
| H26A—C26—H26B | 107.9 | C11—C12—H12B | 109.3 |
| C23—C24—C25 | 112.18 (17) | H12A—C12—H12B | 108.0 |
| C23—C24—H24A | 109.2 | O11—C11—O12 | 125.87 (18) |
| C25—C24—H24A | 109.2 | O11—C11—C12 | 117.66 (17) |
| C23—C24—H24B | 109.2 | O12—C11—C12 | 116.47 (18) |
| | | |
| N21—C22—C21—O21 | −3.4 (2) | C17—C16—C15—C14 | −74.9 (2) |
| N21—C22—C21—O22 | 177.07 (17) | C13—C14—C15—C16 | 176.58 (18) |
| C24—C25—C26—C27 | 167.68 (18) | C25—C26—C27—O25 | −6.3 (3) |
| O23—C23—C24—C25 | −142.6 (2) | C25—C26—C27—O26 | 175.11 (18) |
| O24—C23—C24—C25 | 37.7 (3) | C15—C16—C17—O15 | −3.7 (3) |
| C26—C25—C24—C23 | 178.32 (18) | C15—C16—C17—O16 | 178.74 (18) |
| O13—C13—C14—C15 | 98.0 (2) | N11—C12—C11—O11 | −3.2 (2) |
| O14—C13—C14—C15 | −80.7 (2) | N11—C12—C11—O12 | 177.22 (17) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O14—H14···O21i | 0.84 | 1.74 | 2.579 (2) | 178 |
| O16—H16···O12ii | 0.84 | 1.74 | 2.567 (2) | 166 |
| N11—H11A···O15 | 0.91 | 2.12 | 2.826 (2) | 133 |
| N11—H11B···O23iii | 0.91 | 1.96 | 2.860 (2) | 168 |
| N11—H11C···O23 | 0.91 | 2.10 | 2.967 (2) | 158 |
| O24—H24···O11iv | 0.84 | 1.73 | 2.5655 (18) | 176 |
| O26—H26···O22v | 0.84 | 1.73 | 2.552 (2) | 166 |
| N21—H21A···O25 | 0.91 | 2.07 | 2.800 (2) | 137 |
| N21—H21B···O13ii | 0.91 | 1.95 | 2.859 (2) | 172 |
| N21—H21C···O13vi | 0.91 | 2.12 | 2.974 (2) | 156 |
| Symmetry codes: (i) x+1, y−1, z; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+1, −z+1; (iv) x−1, y, z; (v) −x+1, −y+2, −z+1; (vi) x, y+1, z. |
(150) Propane-1,3-dicarboxylic acid – Aminoethanoic acid (1/1)
top
Crystal data top
| C5H8O4·C2H5NO2 | Z = 4 |
| Mr = 207.18 | F(000) = 440 |
| Triclinic, P1 | Dx = 1.496 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 4.9021 (7) Å | Cell parameters from 7886 reflections |
| b = 20.167 (3) Å | θ = 2.1–29.6° |
| c = 10.1686 (15) Å | µ = 0.13 mm−1 |
| α = 85.540 (11)° | T = 150 K |
| β = 113.122 (10)° | Prism, colourless |
| γ = 88.270 (11)° | 0.40 × 0.23 × 0.10 mm |
| V = 919.7 (2) Å3 | |
Data collection top
STOE IPDS 2
diffractometer | 2840 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.040 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 29.2°, θmin = 2.0° |
| rotation method scans | h = −6→5 |
| 10066 measured reflections | k = −27→27 |
| 4417 independent reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.077 | H-atom parameters constrained |
| S = 0.88 | w = 1/[σ2(Fo2) + (0.0327P)2]
where P = (Fo2 + 2Fc2)/3 |
| 4417 reflections | (Δ/σ)max = 0.001 |
| 259 parameters | Δρmax = 0.21 e Å−3 |
| 0 restraints | Δρmin = −0.20 e Å−3 |
Crystal data top
| C5H8O4·C2H5NO2 | γ = 88.270 (11)° |
| Mr = 207.18 | V = 919.7 (2) Å3 |
| Triclinic, P1 | Z = 4 |
| a = 4.9021 (7) Å | Mo Kα radiation |
| b = 20.167 (3) Å | µ = 0.13 mm−1 |
| c = 10.1686 (15) Å | T = 150 K |
| α = 85.540 (11)° | 0.40 × 0.23 × 0.10 mm |
| β = 113.122 (10)° | |
Data collection top
STOE IPDS 2
diffractometer | 2840 reflections with I > 2σ(I) |
| 10066 measured reflections | Rint = 0.040 |
| 4417 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.039 | 0 restraints |
| wR(F2) = 0.077 | H-atom parameters constrained |
| S = 0.88 | Δρmax = 0.21 e Å−3 |
| 4417 reflections | Δρmin = −0.20 e Å−3 |
| 259 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O23 | 0.1874 (3) | 0.58914 (5) | 0.45078 (12) | 0.0203 (3) | |
| O25 | 0.6546 (3) | 0.84812 (5) | 0.75242 (13) | 0.0223 (3) | |
| O26 | 0.8549 (3) | 0.90476 (5) | 0.62412 (13) | 0.0228 (3) | |
| H26 | 0.8755 | 0.9028 | 0.5463 | 0.034* | |
| O21 | 0.1276 (3) | 1.05551 (5) | 0.82410 (12) | 0.0206 (2) | |
| O22 | 0.0071 (3) | 1.09063 (5) | 0.59387 (13) | 0.0232 (3) | |
| O13 | 0.7716 (3) | 0.08766 (5) | 1.01507 (12) | 0.0190 (2) | |
| O24 | 0.0737 (3) | 0.65465 (5) | 0.59016 (14) | 0.0243 (3) | |
| H24 | −0.0057 | 0.6204 | 0.6065 | 0.036* | |
| O16 | 0.1809 (3) | 0.40439 (5) | 0.90802 (13) | 0.0255 (3) | |
| H16 | 0.1707 | 0.4040 | 0.9886 | 0.038* | |
| O14 | 0.7880 (3) | 0.15347 (5) | 0.83294 (13) | 0.0232 (3) | |
| H14 | 0.8872 | 0.1201 | 0.8266 | 0.035* | |
| O15 | 0.4039 (3) | 0.34547 (5) | 0.80201 (14) | 0.0290 (3) | |
| N21 | 0.5793 (3) | 0.97142 (6) | 0.84584 (14) | 0.0179 (3) | |
| H21A | 0.7037 | 0.9394 | 0.8364 | 0.027* | |
| H21B | 0.4531 | 0.9520 | 0.8807 | 0.027* | |
| H21C | 0.6879 | 1.0002 | 0.9076 | 0.027* | |
| C22 | 0.4054 (4) | 1.00833 (7) | 0.70355 (17) | 0.0160 (3) | |
| H22A | 0.3151 | 0.9761 | 0.6325 | 0.019* | |
| H22B | 0.5407 | 1.0343 | 0.6718 | 0.019* | |
| C13 | 0.6982 (4) | 0.14032 (7) | 0.93729 (16) | 0.0157 (3) | |
| C25 | 0.4768 (4) | 0.74507 (7) | 0.57750 (17) | 0.0168 (3) | |
| H25A | 0.3307 | 0.7613 | 0.6146 | 0.020* | |
| H25B | 0.6328 | 0.7164 | 0.6562 | 0.020* | |
| C23 | 0.1886 (4) | 0.64362 (7) | 0.49715 (17) | 0.0160 (3) | |
| C21 | 0.1611 (4) | 1.05547 (7) | 0.70778 (17) | 0.0155 (3) | |
| C26 | 0.6178 (4) | 0.80449 (7) | 0.53373 (17) | 0.0173 (3) | |
| H26A | 0.7941 | 0.7884 | 0.5178 | 0.021* | |
| H26B | 0.4738 | 0.8275 | 0.4419 | 0.021* | |
| C24 | 0.3205 (4) | 0.70337 (7) | 0.45289 (18) | 0.0197 (3) | |
| H24A | 0.1606 | 0.7317 | 0.3753 | 0.024* | |
| H24B | 0.4653 | 0.6882 | 0.4140 | 0.024* | |
| C16 | 0.4986 (4) | 0.30835 (7) | 1.04724 (18) | 0.0202 (3) | |
| H16A | 0.3402 | 0.2911 | 1.0752 | 0.024* | |
| H16B | 0.6230 | 0.3351 | 1.1237 | 0.024* | |
| C14 | 0.5041 (4) | 0.19555 (7) | 0.95314 (18) | 0.0186 (3) | |
| H14A | 0.3831 | 0.1771 | 1.0038 | 0.022* | |
| H14B | 0.3668 | 0.2155 | 0.8567 | 0.022* | |
| C15 | 0.6904 (4) | 0.24967 (8) | 1.03731 (19) | 0.0233 (4) | |
| H15A | 0.8191 | 0.2301 | 1.1355 | 0.028* | |
| H15B | 0.8208 | 0.2659 | 0.9898 | 0.028* | |
| C27 | 0.7102 (4) | 0.85318 (7) | 0.64558 (17) | 0.0159 (3) | |
| C17 | 0.3573 (4) | 0.35294 (7) | 0.90864 (18) | 0.0196 (3) | |
| O11 | 0.8392 (3) | 0.55172 (5) | 0.65368 (12) | 0.0202 (2) | |
| O12 | 0.9381 (3) | 0.58701 (5) | 0.86944 (13) | 0.0231 (3) | |
| N11 | 0.4275 (3) | 0.47064 (6) | 0.66303 (14) | 0.0176 (3) | |
| H11A | 0.3137 | 0.4399 | 0.6834 | 0.026* | |
| H11B | 0.5653 | 0.4497 | 0.6386 | 0.026* | |
| H11C | 0.3097 | 0.5002 | 0.5885 | 0.026* | |
| C12 | 0.5797 (4) | 0.50657 (7) | 0.79090 (17) | 0.0161 (3) | |
| H12A | 0.6837 | 0.4738 | 0.8753 | 0.019* | |
| H12B | 0.4301 | 0.5334 | 0.8108 | 0.019* | |
| C11 | 0.8035 (3) | 0.55227 (7) | 0.76915 (17) | 0.0149 (3) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O23 | 0.0243 (7) | 0.0161 (5) | 0.0228 (6) | −0.0029 (4) | 0.0112 (5) | −0.0050 (4) |
| O25 | 0.0321 (7) | 0.0192 (5) | 0.0213 (6) | −0.0043 (5) | 0.0160 (6) | −0.0054 (4) |
| O26 | 0.0321 (7) | 0.0192 (5) | 0.0223 (6) | −0.0088 (5) | 0.0151 (6) | −0.0058 (4) |
| O21 | 0.0226 (6) | 0.0212 (5) | 0.0190 (6) | 0.0041 (4) | 0.0100 (5) | −0.0014 (4) |
| O22 | 0.0222 (7) | 0.0267 (6) | 0.0207 (6) | 0.0051 (5) | 0.0102 (5) | 0.0047 (4) |
| O13 | 0.0216 (6) | 0.0154 (5) | 0.0199 (6) | 0.0002 (4) | 0.0083 (5) | −0.0004 (4) |
| O24 | 0.0339 (7) | 0.0184 (5) | 0.0329 (7) | −0.0106 (5) | 0.0247 (6) | −0.0090 (5) |
| O16 | 0.0354 (8) | 0.0208 (6) | 0.0229 (7) | 0.0077 (5) | 0.0155 (6) | 0.0011 (4) |
| O14 | 0.0309 (7) | 0.0200 (5) | 0.0229 (6) | 0.0071 (5) | 0.0168 (6) | 0.0035 (4) |
| O15 | 0.0480 (9) | 0.0201 (6) | 0.0300 (7) | 0.0020 (5) | 0.0281 (7) | 0.0010 (5) |
| N21 | 0.0205 (7) | 0.0163 (6) | 0.0179 (7) | 0.0030 (5) | 0.0091 (6) | −0.0017 (5) |
| C22 | 0.0165 (8) | 0.0170 (7) | 0.0159 (7) | −0.0020 (5) | 0.0076 (6) | −0.0025 (5) |
| C13 | 0.0153 (8) | 0.0151 (7) | 0.0159 (8) | −0.0027 (6) | 0.0047 (7) | −0.0038 (5) |
| C25 | 0.0205 (8) | 0.0165 (7) | 0.0163 (8) | −0.0041 (6) | 0.0096 (7) | −0.0037 (5) |
| C23 | 0.0145 (8) | 0.0174 (7) | 0.0150 (8) | −0.0013 (5) | 0.0046 (6) | −0.0017 (5) |
| C21 | 0.0151 (8) | 0.0146 (7) | 0.0182 (8) | −0.0037 (5) | 0.0078 (6) | −0.0029 (5) |
| C26 | 0.0213 (8) | 0.0161 (7) | 0.0168 (8) | −0.0031 (6) | 0.0095 (7) | −0.0030 (5) |
| C24 | 0.0249 (9) | 0.0193 (7) | 0.0178 (8) | −0.0065 (6) | 0.0108 (7) | −0.0047 (6) |
| C16 | 0.0221 (9) | 0.0162 (7) | 0.0213 (8) | −0.0001 (6) | 0.0072 (7) | −0.0045 (6) |
| C14 | 0.0190 (8) | 0.0155 (7) | 0.0223 (8) | −0.0009 (6) | 0.0091 (7) | −0.0027 (6) |
| C15 | 0.0185 (9) | 0.0206 (8) | 0.0252 (9) | 0.0031 (6) | 0.0025 (7) | −0.0051 (6) |
| C27 | 0.0153 (8) | 0.0128 (7) | 0.0184 (8) | 0.0013 (5) | 0.0056 (6) | −0.0003 (5) |
| C17 | 0.0247 (9) | 0.0143 (7) | 0.0241 (9) | −0.0038 (6) | 0.0139 (7) | −0.0031 (6) |
| O11 | 0.0243 (7) | 0.0207 (5) | 0.0199 (6) | −0.0086 (4) | 0.0124 (5) | −0.0043 (4) |
| O12 | 0.0247 (7) | 0.0261 (6) | 0.0233 (6) | −0.0103 (5) | 0.0123 (5) | −0.0117 (4) |
| N11 | 0.0214 (7) | 0.0163 (6) | 0.0185 (7) | −0.0063 (5) | 0.0110 (6) | −0.0022 (5) |
| C12 | 0.0157 (8) | 0.0176 (7) | 0.0164 (7) | −0.0015 (6) | 0.0078 (6) | −0.0023 (5) |
| C11 | 0.0136 (8) | 0.0136 (7) | 0.0185 (8) | 0.0011 (5) | 0.0077 (6) | −0.0009 (5) |
Geometric parameters (Å, º) top
| O23—C23 | 1.2280 (18) | C25—H25B | 0.9900 |
| O25—C27 | 1.2176 (19) | C23—C24 | 1.505 (2) |
| O26—C27 | 1.3279 (18) | C26—C27 | 1.503 (2) |
| O26—H26 | 0.8400 | C26—H26A | 0.9900 |
| O21—C21 | 1.2563 (19) | C26—H26B | 0.9900 |
| O22—C21 | 1.2518 (19) | C24—H24A | 0.9900 |
| O13—C13 | 1.2271 (17) | C24—H24B | 0.9900 |
| O24—C23 | 1.3056 (19) | C16—C17 | 1.508 (2) |
| O24—H24 | 0.8400 | C16—C15 | 1.523 (2) |
| O16—C17 | 1.328 (2) | C16—H16A | 0.9900 |
| O16—H16 | 0.8400 | C16—H16B | 0.9900 |
| O14—C13 | 1.3117 (19) | C14—C15 | 1.533 (2) |
| O14—H14 | 0.8400 | C14—H14A | 0.9900 |
| O15—C17 | 1.212 (2) | C14—H14B | 0.9900 |
| N21—C22 | 1.483 (2) | C15—H15A | 0.9900 |
| N21—H21A | 0.9100 | C15—H15B | 0.9900 |
| N21—H21B | 0.9100 | O11—C11 | 1.2541 (19) |
| N21—H21C | 0.9100 | O12—C11 | 1.2557 (19) |
| C22—C21 | 1.523 (2) | N11—C12 | 1.480 (2) |
| C22—H22A | 0.9900 | N11—H11A | 0.9100 |
| C22—H22B | 0.9900 | N11—H11B | 0.9100 |
| C13—C14 | 1.502 (2) | N11—H11C | 0.9100 |
| C25—C26 | 1.523 (2) | C12—C11 | 1.524 (2) |
| C25—C24 | 1.530 (2) | C12—H12A | 0.9900 |
| C25—H25A | 0.9900 | C12—H12B | 0.9900 |
| | | |
| C27—O26—H26 | 109.5 | C25—C24—H24B | 109.1 |
| C23—O24—H24 | 109.5 | H24A—C24—H24B | 107.8 |
| C17—O16—H16 | 109.5 | C17—C16—C15 | 113.27 (14) |
| C13—O14—H14 | 109.5 | C17—C16—H16A | 108.9 |
| C22—N21—H21A | 109.5 | C15—C16—H16A | 108.9 |
| C22—N21—H21B | 109.5 | C17—C16—H16B | 108.9 |
| H21A—N21—H21B | 109.5 | C15—C16—H16B | 108.9 |
| C22—N21—H21C | 109.5 | H16A—C16—H16B | 107.7 |
| H21A—N21—H21C | 109.5 | C13—C14—C15 | 111.24 (14) |
| H21B—N21—H21C | 109.5 | C13—C14—H14A | 109.4 |
| N21—C22—C21 | 111.96 (13) | C15—C14—H14A | 109.4 |
| N21—C22—H22A | 109.2 | C13—C14—H14B | 109.4 |
| C21—C22—H22A | 109.2 | C15—C14—H14B | 109.4 |
| N21—C22—H22B | 109.2 | H14A—C14—H14B | 108.0 |
| C21—C22—H22B | 109.2 | C16—C15—C14 | 112.36 (14) |
| H22A—C22—H22B | 107.9 | C16—C15—H15A | 109.1 |
| O13—C13—O14 | 122.15 (14) | C14—C15—H15A | 109.1 |
| O13—C13—C14 | 123.87 (14) | C16—C15—H15B | 109.1 |
| O14—C13—C14 | 113.97 (12) | C14—C15—H15B | 109.1 |
| C26—C25—C24 | 112.64 (13) | H15A—C15—H15B | 107.9 |
| C26—C25—H25A | 109.1 | O25—C27—O26 | 119.01 (14) |
| C24—C25—H25A | 109.1 | O25—C27—C26 | 123.29 (13) |
| C26—C25—H25B | 109.1 | O26—C27—C26 | 117.69 (14) |
| C24—C25—H25B | 109.1 | O15—C17—O16 | 119.42 (14) |
| H25A—C25—H25B | 107.8 | O15—C17—C16 | 123.78 (15) |
| O23—C23—O24 | 122.42 (13) | O16—C17—C16 | 116.75 (14) |
| O23—C23—C24 | 123.66 (15) | C12—N11—H11A | 109.5 |
| O24—C23—C24 | 113.92 (13) | C12—N11—H11B | 109.5 |
| O22—C21—O21 | 125.27 (15) | H11A—N11—H11B | 109.5 |
| O22—C21—C22 | 117.00 (14) | C12—N11—H11C | 109.5 |
| O21—C21—C22 | 117.72 (13) | H11A—N11—H11C | 109.5 |
| C27—C26—C25 | 112.15 (13) | H11B—N11—H11C | 109.5 |
| C27—C26—H26A | 109.2 | N11—C12—C11 | 111.57 (13) |
| C25—C26—H26A | 109.2 | N11—C12—H12A | 109.3 |
| C27—C26—H26B | 109.2 | C11—C12—H12A | 109.3 |
| C25—C26—H26B | 109.2 | N11—C12—H12B | 109.3 |
| H26A—C26—H26B | 107.9 | C11—C12—H12B | 109.3 |
| C23—C24—C25 | 112.62 (13) | H12A—C12—H12B | 108.0 |
| C23—C24—H24A | 109.1 | O11—C11—O12 | 125.60 (14) |
| C25—C24—H24A | 109.1 | O11—C11—C12 | 117.52 (13) |
| C23—C24—H24B | 109.1 | O12—C11—C12 | 116.87 (14) |
| | | |
| N21—C22—C21—O22 | 177.79 (13) | C17—C16—C15—C14 | −74.51 (18) |
| N21—C22—C21—O21 | −2.99 (18) | C13—C14—C15—C16 | 176.54 (14) |
| C24—C25—C26—C27 | 167.31 (14) | C25—C26—C27—O25 | −5.9 (2) |
| O23—C23—C24—C25 | −142.48 (16) | C25—C26—C27—O26 | 175.38 (14) |
| O24—C23—C24—C25 | 37.6 (2) | C15—C16—C17—O15 | −4.0 (2) |
| C26—C25—C24—C23 | 178.41 (14) | C15—C16—C17—O16 | 178.47 (14) |
| O13—C13—C14—C15 | 97.87 (18) | N11—C12—C11—O11 | −2.93 (19) |
| O14—C13—C14—C15 | −81.22 (17) | N11—C12—C11—O12 | 177.57 (13) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O14—H14···O21i | 0.84 | 1.74 | 2.5745 (16) | 174 |
| O16—H16···O12ii | 0.84 | 1.74 | 2.5658 (17) | 166 |
| N11—H11A···O15 | 0.91 | 2.11 | 2.8238 (16) | 135 |
| N11—H11B···O23iii | 0.91 | 1.96 | 2.8554 (18) | 169 |
| N11—H11C···O23 | 0.91 | 2.10 | 2.9644 (17) | 158 |
| O24—H24···O11iv | 0.84 | 1.72 | 2.5626 (14) | 176 |
| O26—H26···O22v | 0.84 | 1.73 | 2.5550 (16) | 166 |
| N21—H21A···O25 | 0.91 | 2.07 | 2.7944 (17) | 136 |
| N21—H21B···O13ii | 0.91 | 1.95 | 2.8551 (17) | 171 |
| N21—H21C···O13vi | 0.91 | 2.11 | 2.9719 (18) | 157 |
| Symmetry codes: (i) x+1, y−1, z; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+1, −z+1; (iv) x−1, y, z; (v) −x+1, −y+2, −z+1; (vi) x, y+1, z. |
(125) Propane-1,3-dicarboxylic acid – Aminoethanoic acid (1/1)
top
Crystal data top
| C5H8O4·C2H5NO2 | Z = 4 |
| Mr = 207.18 | F(000) = 440 |
| Triclinic, P1 | Dx = 1.501 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 4.8952 (6) Å | Cell parameters from 8370 reflections |
| b = 20.151 (3) Å | θ = 2.1–29.6° |
| c = 10.1585 (13) Å | µ = 0.13 mm−1 |
| α = 85.518 (11)° | T = 125 K |
| β = 113.088 (9)° | Prism, colourless |
| γ = 88.264 (11)° | 0.40 × 0.23 × 0.10 mm |
| V = 916.9 (2) Å3 | |
Data collection top
STOE IPDS 2
diffractometer | 2935 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.038 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 29.2°, θmin = 2.0° |
| rotation method scans | h = −6→5 |
| 9997 measured reflections | k = −27→27 |
| 4391 independent reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.075 | H-atom parameters constrained |
| S = 0.90 | w = 1/[σ2(Fo2) + (0.0331P)2]
where P = (Fo2 + 2Fc2)/3 |
| 4391 reflections | (Δ/σ)max = 0.001 |
| 259 parameters | Δρmax = 0.23 e Å−3 |
| 0 restraints | Δρmin = −0.20 e Å−3 |
Crystal data top
| C5H8O4·C2H5NO2 | γ = 88.264 (11)° |
| Mr = 207.18 | V = 916.9 (2) Å3 |
| Triclinic, P1 | Z = 4 |
| a = 4.8952 (6) Å | Mo Kα radiation |
| b = 20.151 (3) Å | µ = 0.13 mm−1 |
| c = 10.1585 (13) Å | T = 125 K |
| α = 85.518 (11)° | 0.40 × 0.23 × 0.10 mm |
| β = 113.088 (9)° | |
Data collection top
STOE IPDS 2
diffractometer | 2935 reflections with I > 2σ(I) |
| 9997 measured reflections | Rint = 0.038 |
| 4391 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.039 | 0 restraints |
| wR(F2) = 0.075 | H-atom parameters constrained |
| S = 0.90 | Δρmax = 0.23 e Å−3 |
| 4391 reflections | Δρmin = −0.20 e Å−3 |
| 259 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O23 | 0.1879 (3) | 0.58911 (5) | 0.45029 (12) | 0.0173 (2) | |
| O25 | 0.6557 (3) | 0.84815 (5) | 0.75302 (12) | 0.0188 (2) | |
| O26 | 0.8564 (3) | 0.90507 (5) | 0.62477 (12) | 0.0192 (2) | |
| H26 | 0.8812 | 0.9026 | 0.5480 | 0.029* | |
| O21 | 0.1277 (3) | 1.05564 (5) | 0.82429 (12) | 0.0176 (2) | |
| O22 | 0.0047 (3) | 1.09042 (5) | 0.59311 (12) | 0.0196 (2) | |
| O13 | 0.7715 (3) | 0.08758 (5) | 1.01545 (12) | 0.0163 (2) | |
| O24 | 0.0747 (3) | 0.65473 (5) | 0.59045 (13) | 0.0208 (3) | |
| H24 | −0.0037 | 0.6204 | 0.6074 | 0.031* | |
| O16 | 0.1792 (3) | 0.40479 (5) | 0.90748 (13) | 0.0218 (3) | |
| H16 | 0.1693 | 0.4046 | 0.9882 | 0.033* | |
| O14 | 0.7857 (3) | 0.15346 (5) | 0.83223 (12) | 0.0199 (2) | |
| H14 | 0.8885 | 0.1204 | 0.8271 | 0.030* | |
| O15 | 0.4030 (3) | 0.34543 (5) | 0.80176 (13) | 0.0248 (3) | |
| N21 | 0.5804 (3) | 0.97171 (6) | 0.84609 (14) | 0.0152 (3) | |
| H21A | 0.7024 | 0.9391 | 0.8366 | 0.023* | |
| H21B | 0.4542 | 0.9530 | 0.8821 | 0.023* | |
| H21C | 0.6917 | 1.0005 | 0.9071 | 0.023* | |
| C22 | 0.4059 (3) | 1.00839 (7) | 0.70358 (16) | 0.0137 (3) | |
| H22A | 0.3164 | 0.9760 | 0.6327 | 0.016* | |
| H22B | 0.5408 | 1.0345 | 0.6717 | 0.016* | |
| C13 | 0.6967 (3) | 0.14032 (7) | 0.93711 (16) | 0.0133 (3) | |
| C25 | 0.4781 (4) | 0.74509 (7) | 0.57783 (16) | 0.0151 (3) | |
| H25A | 0.3315 | 0.7613 | 0.6149 | 0.018* | |
| H25B | 0.6344 | 0.7164 | 0.6566 | 0.018* | |
| C23 | 0.1894 (3) | 0.64365 (7) | 0.49710 (16) | 0.0142 (3) | |
| C21 | 0.1601 (3) | 1.05532 (7) | 0.70766 (16) | 0.0136 (3) | |
| C26 | 0.6191 (4) | 0.80469 (7) | 0.53377 (16) | 0.0149 (3) | |
| H26A | 0.7955 | 0.7886 | 0.5176 | 0.018* | |
| H26B | 0.4748 | 0.8278 | 0.4421 | 0.018* | |
| C24 | 0.3224 (4) | 0.70350 (7) | 0.45288 (17) | 0.0168 (3) | |
| H24A | 0.1628 | 0.7319 | 0.3751 | 0.020* | |
| H24B | 0.4678 | 0.6882 | 0.4142 | 0.020* | |
| C16 | 0.4975 (4) | 0.30849 (7) | 1.04730 (17) | 0.0172 (3) | |
| H16A | 0.3387 | 0.2914 | 1.0754 | 0.021* | |
| H16B | 0.6223 | 0.3352 | 1.1237 | 0.021* | |
| C14 | 0.5022 (4) | 0.19550 (7) | 0.95298 (17) | 0.0158 (3) | |
| H14A | 0.3807 | 0.1770 | 1.0035 | 0.019* | |
| H14B | 0.3651 | 0.2156 | 0.8565 | 0.019* | |
| C15 | 0.6889 (4) | 0.24962 (7) | 1.03760 (18) | 0.0198 (3) | |
| H15A | 0.8168 | 0.2299 | 1.1359 | 0.024* | |
| H15B | 0.8204 | 0.2658 | 0.9904 | 0.024* | |
| C27 | 0.7117 (3) | 0.85331 (7) | 0.64638 (16) | 0.0139 (3) | |
| C17 | 0.3562 (4) | 0.35311 (7) | 0.90847 (17) | 0.0166 (3) | |
| O11 | 0.8392 (3) | 0.55171 (5) | 0.65348 (12) | 0.0172 (2) | |
| O12 | 0.9398 (3) | 0.58671 (5) | 0.86977 (12) | 0.0197 (2) | |
| N11 | 0.4250 (3) | 0.47085 (6) | 0.66249 (14) | 0.0150 (3) | |
| H11A | 0.3073 | 0.4409 | 0.6821 | 0.023* | |
| H11B | 0.5625 | 0.4490 | 0.6390 | 0.023* | |
| H11C | 0.3109 | 0.5007 | 0.5875 | 0.023* | |
| C12 | 0.5791 (3) | 0.50673 (7) | 0.79119 (16) | 0.0136 (3) | |
| H12A | 0.6823 | 0.4739 | 0.8756 | 0.016* | |
| H12B | 0.4301 | 0.5338 | 0.8111 | 0.016* | |
| C11 | 0.8048 (3) | 0.55207 (7) | 0.76937 (16) | 0.0135 (3) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O23 | 0.0199 (6) | 0.0139 (5) | 0.0194 (6) | −0.0023 (4) | 0.0087 (5) | −0.0043 (4) |
| O25 | 0.0263 (6) | 0.0168 (5) | 0.0176 (6) | −0.0036 (4) | 0.0126 (5) | −0.0041 (4) |
| O26 | 0.0264 (7) | 0.0163 (5) | 0.0197 (6) | −0.0069 (4) | 0.0133 (5) | −0.0050 (4) |
| O21 | 0.0199 (6) | 0.0188 (5) | 0.0159 (6) | 0.0028 (4) | 0.0092 (5) | −0.0012 (4) |
| O22 | 0.0188 (6) | 0.0216 (5) | 0.0180 (6) | 0.0037 (4) | 0.0085 (5) | 0.0041 (4) |
| O13 | 0.0182 (6) | 0.0130 (5) | 0.0170 (6) | 0.0001 (4) | 0.0066 (5) | −0.0007 (4) |
| O24 | 0.0291 (7) | 0.0156 (5) | 0.0282 (6) | −0.0091 (5) | 0.0210 (6) | −0.0072 (4) |
| O16 | 0.0301 (7) | 0.0175 (5) | 0.0195 (6) | 0.0063 (4) | 0.0126 (6) | 0.0007 (4) |
| O14 | 0.0268 (7) | 0.0164 (5) | 0.0201 (6) | 0.0056 (4) | 0.0144 (5) | 0.0023 (4) |
| O15 | 0.0412 (8) | 0.0178 (5) | 0.0246 (6) | 0.0015 (5) | 0.0238 (6) | 0.0006 (4) |
| N21 | 0.0173 (7) | 0.0144 (6) | 0.0148 (6) | 0.0023 (5) | 0.0075 (5) | −0.0014 (4) |
| C22 | 0.0138 (7) | 0.0151 (6) | 0.0123 (7) | −0.0007 (5) | 0.0052 (6) | −0.0016 (5) |
| C13 | 0.0129 (7) | 0.0128 (6) | 0.0141 (7) | −0.0027 (5) | 0.0049 (6) | −0.0033 (5) |
| C25 | 0.0183 (8) | 0.0149 (7) | 0.0152 (7) | −0.0040 (5) | 0.0094 (6) | −0.0035 (5) |
| C23 | 0.0117 (7) | 0.0160 (7) | 0.0135 (7) | −0.0008 (5) | 0.0036 (6) | −0.0014 (5) |
| C21 | 0.0129 (7) | 0.0127 (6) | 0.0168 (7) | −0.0040 (5) | 0.0069 (6) | −0.0032 (5) |
| C26 | 0.0178 (8) | 0.0144 (7) | 0.0141 (7) | −0.0022 (5) | 0.0080 (6) | −0.0016 (5) |
| C24 | 0.0210 (8) | 0.0157 (7) | 0.0164 (7) | −0.0050 (6) | 0.0099 (7) | −0.0033 (5) |
| C16 | 0.0186 (8) | 0.0147 (7) | 0.0172 (8) | −0.0004 (5) | 0.0054 (6) | −0.0047 (5) |
| C14 | 0.0159 (8) | 0.0140 (7) | 0.0182 (8) | −0.0001 (5) | 0.0077 (6) | −0.0016 (5) |
| C15 | 0.0161 (8) | 0.0180 (7) | 0.0206 (8) | 0.0020 (6) | 0.0019 (7) | −0.0048 (6) |
| C27 | 0.0124 (7) | 0.0115 (6) | 0.0152 (7) | 0.0012 (5) | 0.0031 (6) | −0.0007 (5) |
| C17 | 0.0206 (8) | 0.0120 (6) | 0.0204 (8) | −0.0046 (5) | 0.0109 (7) | −0.0033 (5) |
| O11 | 0.0210 (6) | 0.0179 (5) | 0.0169 (6) | −0.0071 (4) | 0.0111 (5) | −0.0041 (4) |
| O12 | 0.0200 (6) | 0.0229 (5) | 0.0197 (6) | −0.0079 (4) | 0.0097 (5) | −0.0094 (4) |
| N11 | 0.0182 (7) | 0.0139 (6) | 0.0156 (6) | −0.0048 (5) | 0.0091 (5) | −0.0016 (4) |
| C12 | 0.0131 (7) | 0.0145 (6) | 0.0137 (7) | −0.0016 (5) | 0.0058 (6) | −0.0021 (5) |
| C11 | 0.0130 (7) | 0.0119 (6) | 0.0161 (7) | 0.0004 (5) | 0.0064 (6) | −0.0010 (5) |
Geometric parameters (Å, º) top
| O23—C23 | 1.2303 (18) | C25—H25B | 0.9900 |
| O25—C27 | 1.2150 (19) | C23—C24 | 1.5073 (19) |
| O26—C27 | 1.3299 (17) | C26—C27 | 1.506 (2) |
| O26—H26 | 0.8400 | C26—H26A | 0.9900 |
| O21—C21 | 1.2561 (19) | C26—H26B | 0.9900 |
| O22—C21 | 1.2561 (18) | C24—H24A | 0.9900 |
| O13—C13 | 1.2297 (17) | C24—H24B | 0.9900 |
| O24—C23 | 1.3069 (19) | C16—C17 | 1.508 (2) |
| O24—H24 | 0.8400 | C16—C15 | 1.522 (2) |
| O16—C17 | 1.3315 (19) | C16—H16A | 0.9900 |
| O16—H16 | 0.8400 | C16—H16B | 0.9900 |
| O14—C13 | 1.3122 (18) | C14—C15 | 1.534 (2) |
| O14—H14 | 0.8400 | C14—H14A | 0.9900 |
| O15—C17 | 1.213 (2) | C14—H14B | 0.9900 |
| N21—C22 | 1.4820 (19) | C15—H15A | 0.9900 |
| N21—H21A | 0.9100 | C15—H15B | 0.9900 |
| N21—H21B | 0.9100 | O11—C11 | 1.2535 (18) |
| N21—H21C | 0.9100 | O12—C11 | 1.2549 (18) |
| C22—C21 | 1.523 (2) | N11—C12 | 1.4860 (19) |
| C22—H22A | 0.9900 | N11—H11A | 0.9100 |
| C22—H22B | 0.9900 | N11—H11B | 0.9100 |
| C13—C14 | 1.501 (2) | N11—H11C | 0.9100 |
| C25—C26 | 1.5256 (19) | C12—C11 | 1.525 (2) |
| C25—C24 | 1.530 (2) | C12—H12A | 0.9900 |
| C25—H25A | 0.9900 | C12—H12B | 0.9900 |
| | | |
| C27—O26—H26 | 109.5 | C25—C24—H24B | 109.1 |
| C23—O24—H24 | 109.5 | H24A—C24—H24B | 107.8 |
| C17—O16—H16 | 109.5 | C17—C16—C15 | 113.34 (13) |
| C13—O14—H14 | 109.5 | C17—C16—H16A | 108.9 |
| C22—N21—H21A | 109.5 | C15—C16—H16A | 108.9 |
| C22—N21—H21B | 109.5 | C17—C16—H16B | 108.9 |
| H21A—N21—H21B | 109.5 | C15—C16—H16B | 108.9 |
| C22—N21—H21C | 109.5 | H16A—C16—H16B | 107.7 |
| H21A—N21—H21C | 109.5 | C13—C14—C15 | 111.18 (13) |
| H21B—N21—H21C | 109.5 | C13—C14—H14A | 109.4 |
| N21—C22—C21 | 111.89 (12) | C15—C14—H14A | 109.4 |
| N21—C22—H22A | 109.2 | C13—C14—H14B | 109.4 |
| C21—C22—H22A | 109.2 | C15—C14—H14B | 109.4 |
| N21—C22—H22B | 109.2 | H14A—C14—H14B | 108.0 |
| C21—C22—H22B | 109.2 | C16—C15—C14 | 112.47 (13) |
| H22A—C22—H22B | 107.9 | C16—C15—H15A | 109.1 |
| O13—C13—O14 | 122.16 (14) | C14—C15—H15A | 109.1 |
| O13—C13—C14 | 123.89 (13) | C16—C15—H15B | 109.1 |
| O14—C13—C14 | 113.94 (12) | C14—C15—H15B | 109.1 |
| C26—C25—C24 | 112.40 (12) | H15A—C15—H15B | 107.8 |
| C26—C25—H25A | 109.1 | O25—C27—O26 | 119.09 (13) |
| C24—C25—H25A | 109.1 | O25—C27—C26 | 123.40 (13) |
| C26—C25—H25B | 109.1 | O26—C27—C26 | 117.50 (13) |
| C24—C25—H25B | 109.1 | O15—C17—O16 | 119.43 (14) |
| H25A—C25—H25B | 107.9 | O15—C17—C16 | 123.68 (14) |
| O23—C23—O24 | 122.54 (13) | O16—C17—C16 | 116.84 (13) |
| O23—C23—C24 | 123.56 (15) | C12—N11—H11A | 109.5 |
| O24—C23—C24 | 113.90 (13) | C12—N11—H11B | 109.5 |
| O22—C21—O21 | 125.25 (15) | H11A—N11—H11B | 109.5 |
| O22—C21—C22 | 117.00 (13) | C12—N11—H11C | 109.5 |
| O21—C21—C22 | 117.75 (13) | H11A—N11—H11C | 109.5 |
| C27—C26—C25 | 111.91 (12) | H11B—N11—H11C | 109.5 |
| C27—C26—H26A | 109.2 | N11—C12—C11 | 111.46 (12) |
| C25—C26—H26A | 109.2 | N11—C12—H12A | 109.3 |
| C27—C26—H26B | 109.2 | C11—C12—H12A | 109.3 |
| C25—C26—H26B | 109.2 | N11—C12—H12B | 109.3 |
| H26A—C26—H26B | 107.9 | C11—C12—H12B | 109.3 |
| C23—C24—C25 | 112.47 (13) | H12A—C12—H12B | 108.0 |
| C23—C24—H24A | 109.1 | O11—C11—O12 | 125.71 (14) |
| C25—C24—H24A | 109.1 | O11—C11—C12 | 117.59 (13) |
| C23—C24—H24B | 109.1 | O12—C11—C12 | 116.70 (13) |
| | | |
| N21—C22—C21—O22 | 177.84 (12) | C17—C16—C15—C14 | −74.23 (17) |
| N21—C22—C21—O21 | −2.58 (18) | C13—C14—C15—C16 | 176.32 (13) |
| C24—C25—C26—C27 | 167.46 (13) | C25—C26—C27—O25 | −6.1 (2) |
| O23—C23—C24—C25 | −142.69 (16) | C25—C26—C27—O26 | 175.42 (13) |
| O24—C23—C24—C25 | 37.30 (19) | C15—C16—C17—O15 | −4.1 (2) |
| C26—C25—C24—C23 | 178.60 (13) | C15—C16—C17—O16 | 178.32 (13) |
| O13—C13—C14—C15 | 97.45 (17) | N11—C12—C11—O11 | −2.49 (18) |
| O14—C13—C14—C15 | −81.62 (16) | N11—C12—C11—O12 | 177.39 (13) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O14—H14···O21i | 0.84 | 1.74 | 2.5739 (16) | 175 |
| O16—H16···O12ii | 0.84 | 1.74 | 2.5644 (16) | 166 |
| N11—H11A···O15 | 0.91 | 2.13 | 2.8245 (15) | 133 |
| N11—H11B···O23iii | 0.91 | 1.96 | 2.8537 (18) | 168 |
| N11—H11C···O23 | 0.91 | 2.09 | 2.9564 (17) | 159 |
| O24—H24···O11iv | 0.84 | 1.72 | 2.5623 (14) | 176 |
| O26—H26···O22v | 0.84 | 1.73 | 2.5539 (16) | 167 |
| N21—H21A···O25 | 0.91 | 2.06 | 2.7956 (17) | 137 |
| N21—H21B···O13ii | 0.91 | 1.95 | 2.8522 (17) | 172 |
| N21—H21C···O13vi | 0.91 | 2.11 | 2.9639 (17) | 156 |
| Symmetry codes: (i) x+1, y−1, z; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+1, −z+1; (iv) x−1, y, z; (v) −x+1, −y+2, −z+1; (vi) x, y+1, z. |
(100) Propane-1,3-dicarboxylic acid – Aminoethanoic acid (1/1)
top
Crystal data top
| C5H8O4·C2H5NO2 | Z = 4 |
| Mr = 207.18 | F(000) = 440 |
| Triclinic, P1 | Dx = 1.505 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 4.8884 (7) Å | Cell parameters from 8917 reflections |
| b = 20.136 (3) Å | θ = 2.1–29.6° |
| c = 10.1518 (15) Å | µ = 0.13 mm−1 |
| α = 85.486 (13)° | T = 100 K |
| β = 113.065 (10)° | Prism, colourless |
| γ = 88.263 (12)° | 0.40 × 0.23 × 0.10 mm |
| V = 914.5 (2) Å3 | |
Data collection top
STOE IPDS 2
diffractometer | 2766 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.052 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 29.2°, θmin = 2.0° |
| rotation method scans | h = −6→5 |
| 8816 measured reflections | k = −27→27 |
| 4247 independent reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.053 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.097 | H-atom parameters constrained |
| S = 0.98 | w = 1/[σ2(Fo2) + (0.0367P)2]
where P = (Fo2 + 2Fc2)/3 |
| 4247 reflections | (Δ/σ)max = 0.001 |
| 259 parameters | Δρmax = 0.31 e Å−3 |
| 0 restraints | Δρmin = −0.29 e Å−3 |
Crystal data top
| C5H8O4·C2H5NO2 | γ = 88.263 (12)° |
| Mr = 207.18 | V = 914.5 (2) Å3 |
| Triclinic, P1 | Z = 4 |
| a = 4.8884 (7) Å | Mo Kα radiation |
| b = 20.136 (3) Å | µ = 0.13 mm−1 |
| c = 10.1518 (15) Å | T = 100 K |
| α = 85.486 (13)° | 0.40 × 0.23 × 0.10 mm |
| β = 113.065 (10)° | |
Data collection top
STOE IPDS 2
diffractometer | 2766 reflections with I > 2σ(I) |
| 8816 measured reflections | Rint = 0.052 |
| 4247 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.053 | 0 restraints |
| wR(F2) = 0.097 | H-atom parameters constrained |
| S = 0.98 | Δρmax = 0.31 e Å−3 |
| 4247 reflections | Δρmin = −0.29 e Å−3 |
| 259 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O23 | 0.1875 (4) | 0.58907 (6) | 0.44968 (16) | 0.0138 (3) | |
| O25 | 0.6555 (4) | 0.84827 (7) | 0.75344 (17) | 0.0154 (3) | |
| O26 | 0.8581 (4) | 0.90525 (7) | 0.62556 (17) | 0.0151 (3) | |
| H26 | 0.8961 | 0.9012 | 0.5527 | 0.023* | |
| O21 | 0.1280 (4) | 1.05580 (7) | 0.82434 (16) | 0.0145 (3) | |
| O22 | 0.0018 (4) | 1.09019 (7) | 0.59227 (17) | 0.0161 (3) | |
| O13 | 0.7712 (3) | 0.08755 (6) | 1.01603 (16) | 0.0129 (3) | |
| O24 | 0.0754 (4) | 0.65477 (7) | 0.59062 (18) | 0.0164 (3) | |
| H24 | 0.0083 | 0.6196 | 0.6125 | 0.025* | |
| O16 | 0.1782 (4) | 0.40501 (7) | 0.90676 (17) | 0.0170 (3) | |
| H16 | 0.1737 | 0.4056 | 0.9885 | 0.025* | |
| O14 | 0.7839 (4) | 0.15336 (7) | 0.83151 (17) | 0.0159 (3) | |
| H14 | 0.8916 | 0.1208 | 0.8282 | 0.024* | |
| O15 | 0.4030 (4) | 0.34543 (7) | 0.80174 (18) | 0.0197 (4) | |
| N21 | 0.5811 (4) | 0.97199 (8) | 0.84592 (19) | 0.0124 (4) | |
| H21A | 0.7074 | 0.9402 | 0.8369 | 0.019* | |
| H21B | 0.4552 | 0.9522 | 0.8809 | 0.019* | |
| H21C | 0.6881 | 1.0011 | 0.9075 | 0.019* | |
| C22 | 0.4058 (5) | 1.00861 (9) | 0.7031 (2) | 0.0107 (4) | |
| H22A | 0.3168 | 0.9761 | 0.6324 | 0.013* | |
| H22B | 0.5406 | 1.0348 | 0.6710 | 0.013* | |
| C13 | 0.6952 (5) | 0.14024 (9) | 0.9368 (2) | 0.0105 (4) | |
| C25 | 0.4806 (5) | 0.74504 (9) | 0.5787 (2) | 0.0114 (4) | |
| H25A | 0.3343 | 0.7610 | 0.6162 | 0.014* | |
| H25B | 0.6376 | 0.7163 | 0.6570 | 0.014* | |
| C23 | 0.1899 (5) | 0.64358 (9) | 0.4969 (2) | 0.0111 (4) | |
| C21 | 0.1586 (5) | 1.05554 (9) | 0.7067 (2) | 0.0104 (4) | |
| C26 | 0.6206 (5) | 0.80486 (9) | 0.5342 (2) | 0.0117 (4) | |
| H26A | 0.7977 | 0.7890 | 0.5182 | 0.014* | |
| H26B | 0.4758 | 0.8279 | 0.4424 | 0.014* | |
| C24 | 0.3238 (5) | 0.70367 (9) | 0.4524 (2) | 0.0129 (4) | |
| H24A | 0.1643 | 0.7322 | 0.3748 | 0.015* | |
| H24B | 0.4693 | 0.6884 | 0.4136 | 0.015* | |
| C16 | 0.4952 (5) | 0.30859 (9) | 1.0473 (2) | 0.0133 (4) | |
| H16A | 0.3355 | 0.2915 | 1.0749 | 0.016* | |
| H16B | 0.6194 | 0.3354 | 1.1238 | 0.016* | |
| C14 | 0.5003 (5) | 0.19553 (9) | 0.9527 (2) | 0.0120 (4) | |
| H14A | 0.3781 | 0.1770 | 1.0030 | 0.014* | |
| H14B | 0.3636 | 0.2158 | 0.8562 | 0.014* | |
| C15 | 0.6883 (5) | 0.24948 (10) | 1.0379 (2) | 0.0158 (4) | |
| H15A | 0.8156 | 0.2296 | 1.1363 | 0.019* | |
| H15B | 0.8206 | 0.2656 | 0.9911 | 0.019* | |
| C27 | 0.7122 (5) | 0.85358 (9) | 0.6466 (2) | 0.0106 (4) | |
| C17 | 0.3553 (5) | 0.35312 (9) | 0.9077 (2) | 0.0123 (4) | |
| O11 | 0.8386 (4) | 0.55177 (7) | 0.65326 (16) | 0.0139 (3) | |
| O12 | 0.9420 (4) | 0.58647 (7) | 0.87026 (17) | 0.0158 (3) | |
| N11 | 0.4247 (4) | 0.47105 (8) | 0.66252 (19) | 0.0123 (4) | |
| H11A | 0.3045 | 0.4416 | 0.6817 | 0.018* | |
| H11B | 0.5623 | 0.4486 | 0.6400 | 0.018* | |
| H11C | 0.3129 | 0.5010 | 0.5871 | 0.018* | |
| C12 | 0.5787 (5) | 0.50689 (9) | 0.7909 (2) | 0.0106 (4) | |
| H12A | 0.6816 | 0.4740 | 0.8754 | 0.013* | |
| H12B | 0.4295 | 0.5341 | 0.8107 | 0.013* | |
| C11 | 0.8053 (5) | 0.55222 (9) | 0.7695 (2) | 0.0102 (4) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O23 | 0.0166 (8) | 0.0109 (7) | 0.0153 (8) | −0.0032 (5) | 0.0071 (7) | −0.0045 (5) |
| O25 | 0.0215 (9) | 0.0136 (7) | 0.0153 (8) | −0.0030 (6) | 0.0113 (7) | −0.0037 (5) |
| O26 | 0.0203 (9) | 0.0134 (7) | 0.0149 (8) | −0.0055 (6) | 0.0099 (7) | −0.0037 (5) |
| O21 | 0.0166 (8) | 0.0154 (7) | 0.0132 (8) | 0.0024 (6) | 0.0080 (7) | −0.0008 (5) |
| O22 | 0.0156 (8) | 0.0177 (7) | 0.0153 (8) | 0.0030 (6) | 0.0074 (7) | 0.0022 (5) |
| O13 | 0.0139 (8) | 0.0104 (6) | 0.0135 (8) | 0.0001 (5) | 0.0046 (7) | −0.0004 (5) |
| O24 | 0.0226 (9) | 0.0125 (7) | 0.0226 (9) | −0.0079 (6) | 0.0166 (7) | −0.0061 (6) |
| O16 | 0.0244 (10) | 0.0142 (7) | 0.0147 (8) | 0.0048 (6) | 0.0109 (7) | 0.0001 (5) |
| O14 | 0.0214 (9) | 0.0119 (7) | 0.0172 (8) | 0.0045 (6) | 0.0118 (7) | 0.0027 (5) |
| O15 | 0.0315 (10) | 0.0133 (7) | 0.0207 (9) | 0.0019 (6) | 0.0180 (8) | 0.0007 (6) |
| N21 | 0.0146 (9) | 0.0100 (7) | 0.0131 (9) | 0.0019 (6) | 0.0065 (8) | −0.0006 (6) |
| C22 | 0.0121 (10) | 0.0113 (9) | 0.0108 (10) | −0.0008 (7) | 0.0066 (8) | −0.0014 (7) |
| C13 | 0.0096 (10) | 0.0101 (8) | 0.0116 (10) | −0.0024 (7) | 0.0035 (9) | −0.0035 (7) |
| C25 | 0.0100 (10) | 0.0134 (9) | 0.0110 (10) | −0.0022 (7) | 0.0042 (8) | −0.0020 (7) |
| C23 | 0.0090 (10) | 0.0135 (9) | 0.0098 (10) | 0.0002 (7) | 0.0028 (8) | −0.0011 (7) |
| C21 | 0.0090 (10) | 0.0095 (8) | 0.0142 (10) | −0.0033 (7) | 0.0057 (9) | −0.0017 (7) |
| C26 | 0.0145 (11) | 0.0097 (8) | 0.0130 (10) | −0.0018 (7) | 0.0075 (9) | −0.0025 (7) |
| C24 | 0.0157 (11) | 0.0130 (9) | 0.0120 (10) | −0.0038 (7) | 0.0070 (9) | −0.0030 (7) |
| C16 | 0.0135 (11) | 0.0112 (9) | 0.0128 (10) | −0.0002 (7) | 0.0024 (9) | −0.0030 (7) |
| C14 | 0.0144 (11) | 0.0101 (9) | 0.0132 (10) | −0.0001 (7) | 0.0074 (9) | −0.0012 (7) |
| C15 | 0.0128 (11) | 0.0155 (9) | 0.0157 (11) | 0.0020 (8) | 0.0018 (9) | −0.0038 (7) |
| C27 | 0.0087 (10) | 0.0091 (8) | 0.0115 (10) | 0.0016 (7) | 0.0018 (8) | 0.0002 (7) |
| C17 | 0.0127 (11) | 0.0094 (9) | 0.0155 (11) | −0.0032 (7) | 0.0063 (9) | −0.0017 (7) |
| O11 | 0.0169 (9) | 0.0139 (7) | 0.0136 (8) | −0.0050 (6) | 0.0083 (7) | −0.0033 (5) |
| O12 | 0.0162 (8) | 0.0182 (7) | 0.0156 (8) | −0.0067 (6) | 0.0075 (7) | −0.0079 (6) |
| N11 | 0.0159 (9) | 0.0116 (8) | 0.0123 (9) | −0.0040 (6) | 0.0082 (8) | −0.0016 (6) |
| C12 | 0.0121 (11) | 0.0117 (9) | 0.0097 (10) | −0.0017 (7) | 0.0058 (8) | −0.0020 (7) |
| C11 | 0.0092 (10) | 0.0092 (8) | 0.0137 (10) | 0.0011 (7) | 0.0061 (8) | −0.0004 (7) |
Geometric parameters (Å, º) top
| O23—C23 | 1.230 (2) | C25—H25B | 0.9900 |
| O25—C27 | 1.218 (3) | C23—C24 | 1.514 (3) |
| O26—C27 | 1.329 (2) | C26—C27 | 1.506 (3) |
| O26—H26 | 0.8400 | C26—H26A | 0.9900 |
| O21—C21 | 1.261 (3) | C26—H26B | 0.9900 |
| O22—C21 | 1.251 (3) | C24—H24A | 0.9900 |
| O13—C13 | 1.232 (2) | C24—H24B | 0.9900 |
| O24—C23 | 1.309 (3) | C16—C17 | 1.511 (3) |
| O24—H24 | 0.8400 | C16—C15 | 1.528 (3) |
| O16—C17 | 1.334 (3) | C16—H16A | 0.9900 |
| O16—H16 | 0.8400 | C16—H16B | 0.9900 |
| O14—C13 | 1.313 (3) | C14—C15 | 1.536 (3) |
| O14—H14 | 0.8400 | C14—H14A | 0.9900 |
| O15—C17 | 1.206 (3) | C14—H14B | 0.9900 |
| N21—C22 | 1.483 (3) | C15—H15A | 0.9900 |
| N21—H21A | 0.9100 | C15—H15B | 0.9900 |
| N21—H21B | 0.9100 | O11—C11 | 1.254 (3) |
| N21—H21C | 0.9100 | O12—C11 | 1.254 (3) |
| C22—C21 | 1.525 (3) | N11—C12 | 1.483 (3) |
| C22—H22A | 0.9900 | N11—H11A | 0.9100 |
| C22—H22B | 0.9900 | N11—H11B | 0.9100 |
| C13—C14 | 1.502 (3) | N11—H11C | 0.9100 |
| C25—C26 | 1.526 (3) | C12—C11 | 1.525 (3) |
| C25—C24 | 1.537 (3) | C12—H12A | 0.9900 |
| C25—H25A | 0.9900 | C12—H12B | 0.9900 |
| | | |
| C27—O26—H26 | 109.5 | C25—C24—H24B | 109.2 |
| C23—O24—H24 | 109.5 | H24A—C24—H24B | 107.9 |
| C17—O16—H16 | 109.5 | C17—C16—C15 | 113.01 (18) |
| C13—O14—H14 | 109.5 | C17—C16—H16A | 109.0 |
| C22—N21—H21A | 109.5 | C15—C16—H16A | 109.0 |
| C22—N21—H21B | 109.5 | C17—C16—H16B | 109.0 |
| H21A—N21—H21B | 109.5 | C15—C16—H16B | 109.0 |
| C22—N21—H21C | 109.5 | H16A—C16—H16B | 107.8 |
| H21A—N21—H21C | 109.5 | C13—C14—C15 | 111.04 (18) |
| H21B—N21—H21C | 109.5 | C13—C14—H14A | 109.4 |
| N21—C22—C21 | 112.03 (17) | C15—C14—H14A | 109.4 |
| N21—C22—H22A | 109.2 | C13—C14—H14B | 109.4 |
| C21—C22—H22A | 109.2 | C15—C14—H14B | 109.4 |
| N21—C22—H22B | 109.2 | H14A—C14—H14B | 108.0 |
| C21—C22—H22B | 109.2 | C16—C15—C14 | 112.16 (18) |
| H22A—C22—H22B | 107.9 | C16—C15—H15A | 109.2 |
| O13—C13—O14 | 122.31 (19) | C14—C15—H15A | 109.2 |
| O13—C13—C14 | 123.72 (19) | C16—C15—H15B | 109.2 |
| O14—C13—C14 | 113.96 (16) | C14—C15—H15B | 109.2 |
| C26—C25—C24 | 111.99 (17) | H15A—C15—H15B | 107.9 |
| C26—C25—H25A | 109.2 | O25—C27—O26 | 119.10 (18) |
| C24—C25—H25A | 109.2 | O25—C27—C26 | 123.30 (17) |
| C26—C25—H25B | 109.2 | O26—C27—C26 | 117.60 (18) |
| C24—C25—H25B | 109.2 | O15—C17—O16 | 119.60 (19) |
| H25A—C25—H25B | 107.9 | O15—C17—C16 | 124.0 (2) |
| O23—C23—O24 | 122.71 (18) | O16—C17—C16 | 116.40 (19) |
| O23—C23—C24 | 123.5 (2) | C12—N11—H11A | 109.5 |
| O24—C23—C24 | 113.84 (17) | C12—N11—H11B | 109.5 |
| O22—C21—O21 | 125.5 (2) | H11A—N11—H11B | 109.5 |
| O22—C21—C22 | 117.20 (18) | C12—N11—H11C | 109.5 |
| O21—C21—C22 | 117.27 (18) | H11A—N11—H11C | 109.5 |
| C27—C26—C25 | 111.88 (17) | H11B—N11—H11C | 109.5 |
| C27—C26—H26A | 109.2 | N11—C12—C11 | 111.66 (17) |
| C25—C26—H26A | 109.2 | N11—C12—H12A | 109.3 |
| C27—C26—H26B | 109.2 | C11—C12—H12A | 109.3 |
| C25—C26—H26B | 109.2 | N11—C12—H12B | 109.3 |
| H26A—C26—H26B | 107.9 | C11—C12—H12B | 109.3 |
| C23—C24—C25 | 112.07 (17) | H12A—C12—H12B | 107.9 |
| C23—C24—H24A | 109.2 | O12—C11—O11 | 125.93 (19) |
| C25—C24—H24A | 109.2 | O12—C11—C12 | 116.87 (18) |
| C23—C24—H24B | 109.2 | O11—C11—C12 | 117.20 (18) |
| | | |
| N21—C22—C21—O22 | 178.23 (17) | C17—C16—C15—C14 | −74.2 (2) |
| N21—C22—C21—O21 | −2.3 (2) | C13—C14—C15—C16 | 176.23 (18) |
| C24—C25—C26—C27 | 167.06 (18) | C25—C26—C27—O25 | −6.2 (3) |
| O23—C23—C24—C25 | −142.8 (2) | C25—C26—C27—O26 | 174.87 (18) |
| O24—C23—C24—C25 | 37.3 (3) | C15—C16—C17—O15 | −4.0 (3) |
| C26—C25—C24—C23 | 178.91 (18) | C15—C16—C17—O16 | 178.57 (17) |
| O13—C13—C14—C15 | 96.8 (2) | N11—C12—C11—O12 | 177.91 (18) |
| O14—C13—C14—C15 | −82.0 (2) | N11—C12—C11—O11 | −2.3 (3) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O14—H14···O21i | 0.84 | 1.73 | 2.571 (2) | 177 |
| O16—H16···O12ii | 0.84 | 1.75 | 2.568 (2) | 164 |
| N11—H11A···O15 | 0.91 | 2.14 | 2.824 (2) | 131 |
| N11—H11B···O23iii | 0.91 | 1.96 | 2.851 (2) | 168 |
| N11—H11C···O23 | 0.91 | 2.08 | 2.953 (2) | 160 |
| O24—H24···O11iv | 0.84 | 1.72 | 2.5611 (19) | 175 |
| O26—H26···O22v | 0.84 | 1.73 | 2.555 (2) | 168 |
| N21—H21A···O25 | 0.91 | 2.08 | 2.793 (2) | 135 |
| N21—H21B···O13ii | 0.91 | 1.95 | 2.849 (2) | 171 |
| N21—H21C···O13vi | 0.91 | 2.10 | 2.961 (2) | 158 |
| Symmetry codes: (i) x+1, y−1, z; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+1, −z+1; (iv) x−1, y, z; (v) −x+1, −y+2, −z+1; (vi) x, y+1, z. |
Experimental details
| | (300) | (275) | (250) | (225) |
| Crystal data |
| Chemical formula | C5H8O4·C2H5NO2 | C5H8O4·C2H5NO2 | C5H8O4·C2H5NO2 | C5H8O4·C2H5NO2 |
| Mr | 207.18 | 207.18 | 207.18 | 207.18 |
| Crystal system, space group | Monoclinic, P21/c | Monoclinic, P21/c | Monoclinic, P21/c | Monoclinic, P21/c |
| Temperature (K) | 300 | 275 | 250 | 225 |
| a, b, c (Å) | 4.9110 (7), 20.922 (3), 10.2744 (15) | 4.9022 (7), 20.901 (3), 10.2730 (15) | 4.8960 (6), 20.878 (2), 10.2701 (13) | 4.8888 (6), 20.859 (2), 10.2664 (13) |
| α, β, γ (°) | 90, 114.586 (10), 90 | 90, 114.60 (1), 90 | 90, 114.634 (9), 90 | 90, 114.648 (9), 90 |
| V (Å3) | 959.9 (2) | 957.0 (2) | 954.3 (2) | 951.6 (2) |
| Z | 4 | 4 | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα | Mo Kα |
| µ (mm−1) | 0.13 | 0.13 | 0.13 | 0.13 |
| Crystal size (mm) | 0.32 × 0.17 × 0.10 | 0.32 × 0.17 × 0.10 | 0.32 × 0.17 × 0.10 | 0.32 × 0.17 × 0.10 |
| |
| Data collection |
| Diffractometer | STOE IPDS 2
diffractometer | STOE IPDS 2
diffractometer | STOE IPDS 2
diffractometer | STOE IPDS 2
diffractometer |
| Absorption correction | – | – | – | – |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 6828, 2578, 1421 | 6792, 2570, 1461 | 6775, 2562, 1535 | 6775, 2558, 1594 |
| Rint | 0.046 | 0.047 | 0.045 | 0.043 |
| (sin θ/λ)max (Å−1) | 0.686 | 0.687 | 0.686 | 0.686 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.048, 0.096, 0.89 | 0.043, 0.085, 0.87 | 0.044, 0.086, 0.88 | 0.042, 0.081, 0.88 |
| No. of reflections | 2578 | 2570 | 2562 | 2558 |
| No. of parameters | 130 | 130 | 130 | 130 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.19, −0.17 | 0.16, −0.18 | 0.16, −0.20 | 0.21, −0.18 |
| | (200) | (175) | (150) | (125) |
| Crystal data |
| Chemical formula | C5H8O4·C2H5NO2 | C5H8O4·C2H5NO2 | C5H8O4·C2H5NO2 | C5H8O4·C2H5NO2 |
| Mr | 207.18 | 207.18 | 207.18 | 207.18 |
| Crystal system, space group | Triclinic, P1 | Triclinic, P1 | Triclinic, P1 | Triclinic, P1 |
| Temperature (K) | 200 | 175 | 150 | 125 |
| a, b, c (Å) | 4.9155 (7), 20.215 (3), 10.1862 (16) | 4.9098 (7), 20.194 (3), 10.1777 (15) | 4.9021 (7), 20.167 (3), 10.1686 (15) | 4.8952 (6), 20.151 (3), 10.1585 (13) |
| α, β, γ (°) | 85.578 (14), 113.133 (11), 88.301 (13) | 85.571 (13), 113.118 (10), 88.306 (12) | 85.540 (11), 113.122 (10), 88.270 (11) | 85.518 (11), 113.088 (9), 88.264 (11) |
| V (Å3) | 926.1 (2) | 923.3 (2) | 919.7 (2) | 916.9 (2) |
| Z | 4 | 4 | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα | Mo Kα |
| µ (mm−1) | 0.13 | 0.13 | 0.13 | 0.13 |
| Crystal size (mm) | 0.40 × 0.23 × 0.10 | 0.40 × 0.23 × 0.10 | 0.40 × 0.23 × 0.10 | 0.40 × 0.23 × 0.10 |
| |
| Data collection |
| Diffractometer | STOE IPDS 2
diffractometer | STOE IPDS 2
diffractometer | STOE IPDS 2
diffractometer | STOE IPDS 2
diffractometer |
| Absorption correction | – | – | – | – |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 9663, 4312, 2491 | 9568, 4319, 2580 | 10066, 4417, 2840 | 9997, 4391, 2935 |
| Rint | 0.055 | 0.052 | 0.040 | 0.038 |
| (sin θ/λ)max (Å−1) | 0.686 | 0.687 | 0.686 | 0.686 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.049, 0.081, 0.89 | 0.049, 0.088, 0.89 | 0.039, 0.077, 0.88 | 0.039, 0.075, 0.90 |
| No. of reflections | 4312 | 4319 | 4417 | 4391 |
| No. of parameters | 259 | 259 | 259 | 259 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.19, −0.21 | 0.22, −0.23 | 0.21, −0.20 | 0.23, −0.20 |
| | (100) |
| Crystal data |
| Chemical formula | C5H8O4·C2H5NO2 |
| Mr | 207.18 |
| Crystal system, space group | Triclinic, P1 |
| Temperature (K) | 100 |
| a, b, c (Å) | 4.8884 (7), 20.136 (3), 10.1518 (15) |
| α, β, γ (°) | 85.486 (13), 113.065 (10), 88.263 (12) |
| V (Å3) | 914.5 (2) |
| Z | 4 |
| Radiation type | Mo Kα |
| µ (mm−1) | 0.13 |
| Crystal size (mm) | 0.40 × 0.23 × 0.10 |
| |
| Data collection |
| Diffractometer | STOE IPDS 2
diffractometer |
| Absorption correction | – |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 8816, 4247, 2766 |
| Rint | 0.052 |
| (sin θ/λ)max (Å−1) | 0.686 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.053, 0.097, 0.98 |
| No. of reflections | 4247 |
| No. of parameters | 259 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.31, −0.29 |
Hydrogen-bond geometry (Å, º) for (300) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1B···O3i | 0.89 | 2.14 | 2.9671 (19) | 154.2 |
| N1—H1C···O3ii | 0.89 | 2.00 | 2.8824 (19) | 169.4 |
| N1—H1A···O5 | 0.89 | 2.00 | 2.7665 (18) | 143.2 |
| O4—H4···O1iii | 0.82 | 1.75 | 2.5706 (18) | 177.5 |
| O6—H6···O2iv | 0.82 | 1.75 | 2.5444 (18) | 164.4 |
| Symmetry codes: (i) −x+1, y−1/2, −z+1/2; (ii) x, −y+3/2, z+1/2; (iii) −x, y+1/2, −z+1/2; (iv) −x+1, −y+1, −z. |
Hydrogen-bond geometry (Å, º) for (275) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1B···O3i | 0.89 | 2.13 | 2.9564 (18) | 154.2 |
| N1—H1C···O3ii | 0.89 | 2.00 | 2.8789 (18) | 169.4 |
| N1—H1A···O5 | 0.89 | 2.00 | 2.7658 (17) | 143.2 |
| O4—H4···O1iii | 0.82 | 1.75 | 2.5680 (17) | 176.4 |
| O6—H6···O2iv | 0.82 | 1.74 | 2.5425 (17) | 164.5 |
| Symmetry codes: (i) −x+1, y−1/2, −z+1/2; (ii) x, −y+3/2, z+1/2; (iii) −x, y+1/2, −z+1/2; (iv) −x+1, −y+1, −z. |
Hydrogen-bond geometry (Å, º) for (250) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1B···O3i | 0.90 | 2.12 | 2.9504 (17) | 153.4 |
| N1—H1C···O3ii | 0.90 | 1.99 | 2.8760 (18) | 169.5 |
| N1—H1A···O5 | 0.90 | 1.98 | 2.7621 (17) | 143.8 |
| O4—H4···O1iii | 0.83 | 1.74 | 2.5676 (16) | 177.9 |
| O6—H6···O2iv | 0.83 | 1.73 | 2.5414 (16) | 164.6 |
| Symmetry codes: (i) −x+1, y−1/2, −z+1/2; (ii) x, −y+3/2, z+1/2; (iii) −x, y+1/2, −z+1/2; (iv) −x+1, −y+1, −z. |
Hydrogen-bond geometry (Å, º) for (225) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1B···O3i | 0.90 | 2.11 | 2.9420 (16) | 154.0 |
| N1—H1C···O3ii | 0.90 | 1.98 | 2.8716 (16) | 169.5 |
| N1—H1A···O5 | 0.90 | 1.99 | 2.7641 (15) | 143.2 |
| O4—H4···O1iii | 0.83 | 1.74 | 2.5650 (15) | 174.7 |
| O6—H6···O2iv | 0.83 | 1.73 | 2.5417 (15) | 164.9 |
| Symmetry codes: (i) −x+1, y−1/2, −z+1/2; (ii) x, −y+3/2, z+1/2; (iii) −x, y+1/2, −z+1/2; (iv) −x+1, −y+1, −z. |
Hydrogen-bond geometry (Å, º) for (200) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O14—H14···O21i | 0.84 | 1.74 | 2.580 (2) | 176.1 |
| O16—H16···O12ii | 0.84 | 1.75 | 2.565 (2) | 163.5 |
| N11—H11A···O15 | 0.91 | 2.12 | 2.827 (2) | 133.5 |
| N11—H11B···O23iii | 0.91 | 1.96 | 2.860 (2) | 168.1 |
| N11—H11C···O23 | 0.91 | 2.11 | 2.971 (2) | 158.0 |
| O24—H24···O11iv | 0.84 | 1.72 | 2.5636 (18) | 176.0 |
| O26—H26···O22v | 0.84 | 1.73 | 2.555 (2) | 167.3 |
| N21—H21A···O25 | 0.91 | 2.05 | 2.798 (2) | 138.5 |
| N21—H21B···O13ii | 0.91 | 1.96 | 2.861 (2) | 172.2 |
| N21—H21C···O13vi | 0.91 | 2.14 | 2.984 (2) | 154.5 |
| Symmetry codes: (i) x+1, y−1, z; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+1, −z+1; (iv) x−1, y, z; (v) −x+1, −y+2, −z+1; (vi) x, y+1, z. |
Hydrogen-bond geometry (Å, º) for (175) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O14—H14···O21i | 0.84 | 1.74 | 2.579 (2) | 177.5 |
| O16—H16···O12ii | 0.84 | 1.74 | 2.567 (2) | 165.9 |
| N11—H11A···O15 | 0.91 | 2.12 | 2.826 (2) | 133.2 |
| N11—H11B···O23iii | 0.91 | 1.96 | 2.860 (2) | 168.2 |
| N11—H11C···O23 | 0.91 | 2.10 | 2.967 (2) | 158.3 |
| O24—H24···O11iv | 0.84 | 1.73 | 2.5655 (18) | 176.3 |
| O26—H26···O22v | 0.84 | 1.73 | 2.552 (2) | 165.7 |
| N21—H21A···O25 | 0.91 | 2.07 | 2.800 (2) | 136.8 |
| N21—H21B···O13ii | 0.91 | 1.95 | 2.859 (2) | 171.8 |
| N21—H21C···O13vi | 0.91 | 2.12 | 2.974 (2) | 156.1 |
| Symmetry codes: (i) x+1, y−1, z; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+1, −z+1; (iv) x−1, y, z; (v) −x+1, −y+2, −z+1; (vi) x, y+1, z. |
Hydrogen-bond geometry (Å, º) for (150) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O14—H14···O21i | 0.84 | 1.74 | 2.5745 (16) | 173.6 |
| O16—H16···O12ii | 0.84 | 1.74 | 2.5658 (17) | 165.6 |
| N11—H11A···O15 | 0.91 | 2.11 | 2.8238 (16) | 134.5 |
| N11—H11B···O23iii | 0.91 | 1.96 | 2.8554 (18) | 169.3 |
| N11—H11C···O23 | 0.91 | 2.10 | 2.9644 (17) | 157.5 |
| O24—H24···O11iv | 0.84 | 1.72 | 2.5626 (14) | 175.5 |
| O26—H26···O22v | 0.84 | 1.73 | 2.5550 (16) | 166.1 |
| N21—H21A···O25 | 0.91 | 2.07 | 2.7944 (17) | 135.6 |
| N21—H21B···O13ii | 0.91 | 1.95 | 2.8551 (17) | 171.1 |
| N21—H21C···O13vi | 0.91 | 2.11 | 2.9719 (18) | 157.0 |
| Symmetry codes: (i) x+1, y−1, z; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+1, −z+1; (iv) x−1, y, z; (v) −x+1, −y+2, −z+1; (vi) x, y+1, z. |
Hydrogen-bond geometry (Å, º) for (125) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O14—H14···O21i | 0.84 | 1.74 | 2.5739 (16) | 175.1 |
| O16—H16···O12ii | 0.84 | 1.74 | 2.5644 (16) | 165.7 |
| N11—H11A···O15 | 0.91 | 2.13 | 2.8245 (15) | 132.5 |
| N11—H11B···O23iii | 0.91 | 1.96 | 2.8537 (18) | 168.4 |
| N11—H11C···O23 | 0.91 | 2.09 | 2.9564 (17) | 158.6 |
| O24—H24···O11iv | 0.84 | 1.72 | 2.5623 (14) | 176.1 |
| O26—H26···O22v | 0.84 | 1.73 | 2.5539 (16) | 167.0 |
| N21—H21A···O25 | 0.91 | 2.06 | 2.7956 (17) | 136.8 |
| N21—H21B···O13ii | 0.91 | 1.95 | 2.8522 (17) | 172.2 |
| N21—H21C···O13vi | 0.91 | 2.11 | 2.9639 (17) | 156.0 |
| Symmetry codes: (i) x+1, y−1, z; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+1, −z+1; (iv) x−1, y, z; (v) −x+1, −y+2, −z+1; (vi) x, y+1, z. |
Hydrogen-bond geometry (Å, º) for (100) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O14—H14···O21i | 0.84 | 1.73 | 2.571 (2) | 177.3 |
| O16—H16···O12ii | 0.84 | 1.75 | 2.568 (2) | 163.8 |
| N11—H11A···O15 | 0.91 | 2.14 | 2.824 (2) | 131.3 |
| N11—H11B···O23iii | 0.91 | 1.96 | 2.851 (2) | 167.5 |
| N11—H11C···O23 | 0.91 | 2.08 | 2.953 (2) | 159.5 |
| O24—H24···O11iv | 0.84 | 1.72 | 2.5611 (19) | 174.5 |
| O26—H26···O22v | 0.84 | 1.73 | 2.555 (2) | 167.8 |
| N21—H21A···O25 | 0.91 | 2.08 | 2.793 (2) | 134.6 |
| N21—H21B···O13ii | 0.91 | 1.95 | 2.849 (2) | 171.4 |
| N21—H21C···O13vi | 0.91 | 2.10 | 2.961 (2) | 157.7 |
| Symmetry codes: (i) x+1, y−1, z; (ii) −x+1, −y+1, −z+2; (iii) −x+1, −y+1, −z+1; (iv) x−1, y, z; (v) −x+1, −y+2, −z+1; (vi) x, y+1, z. |

 Subscribe to Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials
Subscribe to Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.

 Subscribe to Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials
Subscribe to Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/b/issues/2012/03/00/gp5050/gp5050fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/b/issues/2012/03/00/gp5050/gp5050fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/b/issues/2012/03/00/gp5050/gp5050fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/b/issues/2012/03/00/gp5050/gp5050fig4thm.gif)
![[Figure 5]](http://journals.iucr.org/b/issues/2012/03/00/gp5050/gp5050fig5thm.gif)
![[Figure 6]](http://journals.iucr.org/b/issues/2012/03/00/gp5050/gp5050fig6thm.gif)
![[Figure 7]](http://journals.iucr.org/b/issues/2012/03/00/gp5050/gp5050fig7thm.gif)
![[Figure 8]](http://journals.iucr.org/b/issues/2012/03/00/gp5050/gp5050fig8thm.gif)

 Subscribe to Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials
Subscribe to Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials


