Tartronic acid forms a hydrogen-bonded complex, C
5H
5NO·C
3H
4O
5, (I), with 2-pyridone, while it forms acid salts, namely 3-hydroxypyridinium hydrogen tartronate, (II), and 4-hydroxypyridinium hydrogen tartronate, (III), both C
5H
6NO
+·C
3H
3O
5−, with 3-hydroxypyridine and 4-hydroxypyridine, respectively. In (I), the pyridone molecules and the acid molecules form
R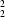
(8) and
R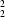
(10) hydrogen-bonded rings, respectively, around the inversion centres. In (II) and (III), the cations and anions are linked by N—H

O and O—H

O hydrogen bonds to form a hydrogen-bonded chain. In each of (I), (II) and (III), an intermolecular hydrogen bond is formed between a carboxyl group and the hydroxyl group attached to the central C atom, and in (I), the hydroxyl group participates in an intramolecular hydrogen bond with a carbonyl group. No intermolecular hydrogen bond is formed between the carboxyl groups in (I), or between the carboxyl and carboxylate groups in (II) and (III).
Supporting information
CCDC references: 254923; 254924; 254925
Crystals of (I), (II) and (III) were grown by slow evaporation at room temperature of ethanol solutions of tartronic acid and the respective hydroxypyridines, in a molar ratio of 1:1.
For all compounds, H atoms attached to C atoms were treated as riding, with C—H = 0.98 (methine H) or 0.93 Å (aromatic H), and with Uiso(H) = 1.2Ueq(C). For (I) and (II), H atoms involved in O—H···O and N—H···O hydrogen bonds were refined. For (III), the O—H distances and the C—O—H angles were fixed to 0.82 Å and 109.5°, respectively, while the torsion angles around the C—O bonds were refined, with Uiso(H) = 1.5Ueq(O). H atoms attached to N atoms were constrained in the riding model, with N—H = 0.86 Å and Uiso(H) = 1.2Ueq(N). For (II) and (III), Friedel opposites were merged.
For all compounds, data collection: MSC/AFC Diffractometer Control Software (Molecular Structure Corporation, 1990); cell refinement: MSC/AFC Diffractometer Control Software; data reduction: TEXSAN for Windows (Molecular Structure Corporation, 1997-1999); program(s) used to solve structure: SIR92 (Altomare et al., 1994); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: ORTEP-3 (Farrugia, 1997); software used to prepare material for publication: TEXSAN for Windows.
Crystal data top
| C5H5NO·C3H4O5 | Z = 2 |
| Mr = 215.16 | F(000) = 224 |
| Triclinic, P1 | Dx = 1.549 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 5.879 (4) Å | Cell parameters from 25 reflections |
| b = 6.859 (4) Å | θ = 10.3–11.5° |
| c = 12.588 (8) Å | µ = 0.14 mm−1 |
| α = 74.61 (5)° | T = 296 K |
| β = 80.94 (6)° | Plate, colourless |
| γ = 70.95 (5)° | 0.30 × 0.20 × 0.08 mm |
| V = 461.2 (5) Å3 | |
Data collection top
Rigaku AFC-5R
diffractometer | Rint = 0.032 |
| Radiation source: Rigaku rotating anode | θmax = 30.0°, θmin = 3.2° |
| Graphite monochromator | h = −8→8 |
| ω/2θ scans | k = −9→1 |
| 3319 measured reflections | l = −17→17 |
| 2698 independent reflections | 3 standard reflections every 97 reflections |
| 1323 reflections with I > 2σ(I) | intensity decay: 0.0% |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.065 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.191 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.08P)2]
where P = (Fo2 + 2Fc2)/3 |
| 2698 reflections | (Δ/σ)max = 0.001 |
| 153 parameters | Δρmax = 0.26 e Å−3 |
| 0 restraints | Δρmin = −0.24 e Å−3 |
| 0 constraints | |
Crystal data top
| C5H5NO·C3H4O5 | γ = 70.95 (5)° |
| Mr = 215.16 | V = 461.2 (5) Å3 |
| Triclinic, P1 | Z = 2 |
| a = 5.879 (4) Å | Mo Kα radiation |
| b = 6.859 (4) Å | µ = 0.14 mm−1 |
| c = 12.588 (8) Å | T = 296 K |
| α = 74.61 (5)° | 0.30 × 0.20 × 0.08 mm |
| β = 80.94 (6)° | |
Data collection top
Rigaku AFC-5R
diffractometer | Rint = 0.032 |
| 3319 measured reflections | 3 standard reflections every 97 reflections |
| 2698 independent reflections | intensity decay: 0.0% |
| 1323 reflections with I > 2σ(I) | |
Refinement top
| R[F2 > 2σ(F2)] = 0.065 | 0 restraints |
| wR(F2) = 0.191 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.00 | Δρmax = 0.26 e Å−3 |
| 2698 reflections | Δρmin = −0.24 e Å−3 |
| 153 parameters | |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.5760 (4) | 0.2370 (4) | 0.44393 (17) | 0.0441 (6) | |
| O2 | 0.4022 (4) | 0.7641 (4) | 0.01997 (18) | 0.0366 (6) | |
| O3 | 0.8635 (4) | 0.7274 (4) | −0.05312 (16) | 0.0403 (6) | |
| O4 | 0.9596 (4) | 0.7379 (4) | 0.10989 (19) | 0.0426 (7) | |
| O5 | 0.5145 (4) | 0.3892 (4) | 0.17429 (18) | 0.0455 (7) | |
| O6 | 0.6360 (5) | 0.5405 (4) | 0.28236 (19) | 0.0421 (6) | |
| N1 | 0.6855 (5) | 0.0014 (4) | 0.6052 (2) | 0.0349 (6) | |
| C2 | 0.6995 (6) | 0.1784 (5) | 0.5281 (2) | 0.0331 (7) | |
| C3 | 0.8526 (6) | 0.2838 (6) | 0.5480 (3) | 0.0422 (8) | |
| C4 | 0.9782 (7) | 0.2080 (6) | 0.6388 (3) | 0.0477 (9) | |
| C5 | 0.9599 (6) | 0.0227 (6) | 0.7161 (3) | 0.0414 (8) | |
| C6 | 0.8131 (6) | −0.0764 (6) | 0.6969 (2) | 0.0392 (8) | |
| C7 | 0.5646 (5) | 0.7342 (5) | 0.0989 (2) | 0.0282 (6) | |
| C8 | 0.8142 (5) | 0.7321 (5) | 0.0429 (2) | 0.0274 (6) | |
| C9 | 0.5710 (5) | 0.5335 (5) | 0.1894 (2) | 0.0310 (7) | |
| H1 | 0.586 (5) | −0.074 (5) | 0.590 (3) | 0.033 (8)* | |
| H2 | 0.867 | 0.407 | 0.498 | 0.051* | |
| H3 | 1.079 | 0.280 | 0.650 | 0.057* | |
| H4 | 1.047 | −0.030 | 0.779 | 0.050* | |
| H5 | 0.798 | −0.200 | 0.747 | 0.047* | |
| H6 | 0.505 | 0.855 | 0.134 | 0.034* | |
| H7 | 0.477 (7) | 0.712 (6) | −0.042 (3) | 0.058 (12)* | |
| H8 | 1.081 (7) | 0.733 (6) | 0.077 (3) | 0.054 (13)* | |
| H9 | 0.614 (9) | 0.444 (8) | 0.330 (4) | 0.083 (16)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0575 (15) | 0.0457 (15) | 0.0328 (12) | −0.0280 (13) | −0.0139 (10) | 0.0069 (11) |
| O2 | 0.0284 (11) | 0.0495 (15) | 0.0319 (11) | −0.0128 (11) | −0.0086 (9) | −0.0038 (11) |
| O3 | 0.0419 (13) | 0.0536 (15) | 0.0254 (11) | −0.0181 (12) | −0.0009 (9) | −0.0055 (10) |
| O4 | 0.0300 (13) | 0.0721 (19) | 0.0351 (12) | −0.0244 (13) | −0.0032 (10) | −0.0158 (12) |
| O5 | 0.0609 (16) | 0.0472 (15) | 0.0382 (12) | −0.0334 (13) | −0.0081 (11) | −0.0024 (11) |
| O6 | 0.0614 (16) | 0.0451 (15) | 0.0274 (11) | −0.0306 (13) | −0.0129 (10) | 0.0029 (11) |
| N1 | 0.0461 (16) | 0.0363 (15) | 0.0277 (12) | −0.0233 (13) | −0.0080 (11) | 0.0007 (11) |
| C2 | 0.0380 (17) | 0.0341 (18) | 0.0273 (14) | −0.0152 (14) | −0.0010 (12) | −0.0028 (13) |
| C3 | 0.055 (2) | 0.042 (2) | 0.0351 (17) | −0.0297 (18) | −0.0065 (15) | 0.0029 (15) |
| C4 | 0.054 (2) | 0.056 (2) | 0.0438 (19) | −0.0329 (19) | −0.0122 (16) | −0.0036 (17) |
| C5 | 0.049 (2) | 0.047 (2) | 0.0305 (16) | −0.0212 (17) | −0.0111 (14) | 0.0016 (15) |
| C6 | 0.052 (2) | 0.0400 (19) | 0.0268 (15) | −0.0212 (17) | −0.0080 (14) | 0.0032 (14) |
| C7 | 0.0263 (14) | 0.0325 (16) | 0.0263 (13) | −0.0126 (13) | −0.0062 (11) | −0.0004 (12) |
| C8 | 0.0265 (14) | 0.0253 (15) | 0.0279 (14) | −0.0084 (12) | −0.0013 (11) | −0.0016 (11) |
| C9 | 0.0272 (15) | 0.0387 (18) | 0.0299 (15) | −0.0166 (14) | −0.0008 (12) | −0.0049 (13) |
Geometric parameters (Å, º) top
| O1—C2 | 1.279 (7) | C2—C3 | 1.409 (10) |
| O2—C7 | 1.416 (5) | C3—C4 | 1.350 (9) |
| O2—H7 | 0.92 (4) | C3—H2 | 0.93 |
| O3—C8 | 1.204 (3) | C4—C5 | 1.404 (11) |
| O4—C8 | 1.308 (4) | C4—H3 | 0.93 |
| O4—H8 | 0.76 (4) | C5—C6 | 1.342 (10) |
| O5—C9 | 1.208 (7) | C5—H4 | 0.93 |
| O6—C9 | 1.304 (4) | C6—H5 | 0.93 |
| O6—H9 | 0.79 (5) | C7—C8 | 1.522 (4) |
| N1—C2 | 1.355 (4) | C7—C9 | 1.530 (4) |
| N1—C6 | 1.365 (8) | C7—H6 | 0.98 |
| N1—H1 | 0.97 (3) | | |
| | | |
| O1···O6 | 2.569 (4) | O4···H4vii | 2.72 |
| O1···N1i | 2.809 (4) | O4···C5vii | 3.234 (5) |
| O2···H8ii | 1.97 (4) | O4···C4vii | 3.370 (5) |
| O2···O4ii | 2.712 (4) | O5···H7iii | 1.96 (4) |
| O2···O5iii | 2.825 (4) | O5···H5i | 2.52 |
| O2···C8iv | 3.192 (5) | O5···C6i | 3.286 (5) |
| O3···H5v | 2.50 | O6···H3vii | 2.72 |
| O3···H4v | 2.64 | O6···C3viii | 3.383 (5) |
| O3···O3vi | 3.049 (4) | O6···C2 | 3.399 (4) |
| O3···C8vi | 3.084 (5) | C2···H9 | 2.68 (5) |
| O3···C6v | 3.096 (4) | C2···H1i | 2.77 (3) |
| O3···C5v | 3.157 (5) | C7···H8ii | 2.90 (4) |
| | | |
| C7—O2—H7 | 113 (2) | C4—C5—H4 | 121.1 |
| C8—O4—H8 | 107 (3) | C5—C6—N1 | 120.8 (3) |
| C9—O6—H9 | 110 (3) | C5—C6—H5 | 119.6 |
| C2—N1—C6 | 123.3 (3) | N1—C6—H5 | 119.6 |
| C2—N1—H1 | 116.3 (19) | O2—C7—C8 | 110.6 (2) |
| C6—N1—H1 | 120.3 (19) | O2—C7—C9 | 110.9 (3) |
| O1—C2—N1 | 118.7 (3) | C8—C7—C9 | 112.1 (3) |
| O1—C2—C3 | 125.1 (3) | O2—C7—H6 | 107.7 |
| N1—C2—C3 | 116.2 (6) | C8—C7—H6 | 107.7 |
| C4—C3—C2 | 120.7 (4) | C9—C7—H6 | 107.7 |
| C4—C3—H2 | 119.6 | O3—C8—O4 | 125.8 (3) |
| C2—C3—H2 | 119.6 | O3—C8—C7 | 121.6 (3) |
| C3—C4—C5 | 121.1 (7) | O4—C8—C7 | 112.5 (2) |
| C3—C4—H3 | 119.4 | O5—C9—O6 | 125.2 (6) |
| C5—C4—H3 | 119.4 | O5—C9—C7 | 122.0 (3) |
| C6—C5—C4 | 117.9 (6) | O6—C9—C7 | 112.8 (3) |
| C6—C5—H4 | 121.1 | | |
| | | |
| C6—N1—C2—O1 | 178.8 (3) | O2—C7—C8—O3 | −9.4 (4) |
| C6—N1—C2—C3 | −1.0 (5) | C9—C7—C8—O3 | 114.9 (6) |
| O1—C2—C3—C4 | −179.2 (4) | O2—C7—C8—O4 | 170.1 (3) |
| N1—C2—C3—C4 | 0.7 (5) | C9—C7—C8—O4 | −65.7 (6) |
| C2—C3—C4—C5 | −0.1 (6) | O2—C7—C9—O5 | 23.5 (4) |
| C3—C4—C5—C6 | −0.1 (6) | C8—C7—C9—O5 | −100.6 (4) |
| C4—C5—C6—N1 | −0.2 (5) | O2—C7—C9—O6 | −154.7 (3) |
| C2—N1—C6—C5 | 0.8 (5) | C8—C7—C9—O6 | 81.2 (6) |
| Symmetry codes: (i) −x+1, −y, −z+1; (ii) x−1, y, z; (iii) −x+1, −y+1, −z; (iv) −x+1, −y+2, −z; (v) x, y+1, z−1; (vi) −x+2, −y+1, −z; (vii) −x+2, −y+1, −z+1; (viii) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1i | 0.96 (3) | 1.85 (3) | 2.809 (4) | 174 (4) |
| O2—H7···O5iii | 0.93 (4) | 1.96 (4) | 2.825 (4) | 155 (4) |
| O4—H8···O2ix | 0.76 (4) | 1.97 (4) | 2.712 (4) | 164 (4) |
| O6—H9···O1 | 0.80 (5) | 1.77 (5) | 2.569 (4) | 175 (3) |
| O2—H7···O3 | 0.93 (4) | 2.29 (4) | 2.675 (4) | 104 (3) |
| C6—H5···O3x | 0.93 | 2.50 | 3.096 (4) | 122 |
| C6—H5···O5i | 0.93 | 2.52 | 3.286 (5) | 140 |
| Symmetry codes: (i) −x+1, −y, −z+1; (iii) −x+1, −y+1, −z; (ix) x+1, y, z; (x) x, y−1, z+1. |
Crystal data top
| C5H6NO+·C3H3O5− | F(000) = 224 |
| Mr = 215.16 | Dx = 1.467 Mg m−3 |
| Monoclinic, Pc | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P -2yc | Cell parameters from 25 reflections |
| a = 5.7563 (11) Å | θ = 11.0–11.5° |
| b = 5.1871 (7) Å | µ = 0.13 mm−1 |
| c = 16.3206 (19) Å | T = 298 K |
| β = 92.186 (13)° | Plate, colourless |
| V = 486.95 (13) Å3 | 0.50 × 0.50 × 0.10 mm |
| Z = 2 | |
Data collection top
Rigaku AFC-5R
diffractometer | Rint = 0.016 |
| Radiation source: Rigaku rotating anode | θmax = 31.0°, θmin = 2.5° |
| Graphite monochromator | h = −1→8 |
| ω/2θ scans | k = −1→7 |
| 2353 measured reflections | l = −23→23 |
| 1562 independent reflections | 3 standard reflections every 97 reflections |
| 1394 reflections with I > 2σ(I) | intensity decay: 0.2% |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.042 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.119 | w = 1/[σ2(Fo2) + (0.0945P)2]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.00 | (Δ/σ)max = 0.001 |
| 1562 reflections | Δρmax = 0.43 e Å−3 |
| 153 parameters | Δρmin = −0.18 e Å−3 |
| 2 restraints | Extinction correction: SHELXL97 (Sheldrick, 1997), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.115 (19) |
Crystal data top
| C5H6NO+·C3H3O5− | V = 486.95 (13) Å3 |
| Mr = 215.16 | Z = 2 |
| Monoclinic, Pc | Mo Kα radiation |
| a = 5.7563 (11) Å | µ = 0.13 mm−1 |
| b = 5.1871 (7) Å | T = 298 K |
| c = 16.3206 (19) Å | 0.50 × 0.50 × 0.10 mm |
| β = 92.186 (13)° | |
Data collection top
Rigaku AFC-5R
diffractometer | Rint = 0.016 |
| 2353 measured reflections | 3 standard reflections every 97 reflections |
| 1562 independent reflections | intensity decay: 0.2% |
| 1394 reflections with I > 2σ(I) | |
Refinement top
| R[F2 > 2σ(F2)] = 0.042 | 2 restraints |
| wR(F2) = 0.119 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.00 | Δρmax = 0.43 e Å−3 |
| 1562 reflections | Δρmin = −0.18 e Å−3 |
| 153 parameters | |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.9138 (5) | −0.0907 (5) | 0.08939 (14) | 0.0655 (6) | |
| O2 | 0.2852 (2) | 0.4262 (2) | 0.34707 (11) | 0.0335 (3) | |
| O3 | −0.1582 (3) | 0.4706 (4) | 0.28672 (12) | 0.0503 (5) | |
| O4 | −0.2977 (3) | 0.3127 (4) | 0.40281 (12) | 0.0473 (5) | |
| O5 | 0.0705 (4) | −0.0990 (3) | 0.45448 (12) | 0.0529 (5) | |
| O6 | 0.2425 (3) | −0.0767 (3) | 0.33621 (11) | 0.0442 (5) | |
| N1 | 0.4292 (4) | 0.1192 (5) | 0.20162 (13) | 0.0466 (5) | |
| C2 | 0.6141 (5) | −0.0148 (5) | 0.17989 (14) | 0.0466 (6) | |
| C3 | 0.7280 (5) | 0.0497 (5) | 0.10891 (14) | 0.0462 (5) | |
| C4 | 0.6415 (6) | 0.2591 (7) | 0.06331 (15) | 0.0555 (6) | |
| C5 | 0.4496 (6) | 0.3944 (7) | 0.08893 (17) | 0.0597 (7) | |
| C6 | 0.3427 (5) | 0.3205 (7) | 0.15909 (18) | 0.0556 (7) | |
| C7 | 0.1089 (3) | 0.3168 (3) | 0.39297 (12) | 0.0253 (3) | |
| C8 | −0.1301 (3) | 0.3763 (3) | 0.35368 (13) | 0.0286 (3) | |
| C9 | 0.1436 (3) | 0.0219 (3) | 0.39551 (13) | 0.0300 (3) | |
| H1 | 0.344 (8) | 0.059 (9) | 0.237 (3) | 0.073 (13)* | |
| H2 | 0.667 | −0.152 | 0.212 | 0.056* | |
| H3 | 0.968 (9) | −0.025 (10) | 0.043 (3) | 0.082 (15)* | |
| H4 | 0.713 | 0.308 | 0.016 | 0.067* | |
| H5 | 0.393 | 0.535 | 0.059 | 0.072* | |
| H6 | 0.213 | 0.409 | 0.177 | 0.067* | |
| H7 | 0.118 | 0.386 | 0.449 | 0.030* | |
| H8 | 0.271 (6) | 0.591 (7) | 0.344 (2) | 0.042 (7)* | |
| H9 | −0.421 (8) | 0.375 (8) | 0.383 (3) | 0.062 (10)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0825 (16) | 0.0663 (13) | 0.0497 (11) | 0.0027 (12) | 0.0306 (11) | 0.0030 (10) |
| O2 | 0.0223 (5) | 0.0221 (5) | 0.0570 (9) | −0.0003 (5) | 0.0118 (5) | 0.0016 (5) |
| O3 | 0.0352 (8) | 0.0702 (12) | 0.0451 (9) | −0.0014 (8) | −0.0030 (7) | 0.0192 (8) |
| O4 | 0.0211 (6) | 0.0715 (12) | 0.0498 (9) | 0.0010 (7) | 0.0090 (6) | 0.0114 (9) |
| O5 | 0.0744 (13) | 0.0364 (8) | 0.0501 (10) | 0.0122 (8) | 0.0326 (9) | 0.0128 (7) |
| O6 | 0.0635 (11) | 0.0224 (6) | 0.0488 (9) | 0.0011 (6) | 0.0326 (8) | 0.0010 (6) |
| N1 | 0.0561 (12) | 0.0503 (11) | 0.0343 (8) | −0.0167 (9) | 0.0141 (8) | −0.0013 (8) |
| C2 | 0.0677 (15) | 0.0407 (10) | 0.0325 (10) | −0.0113 (10) | 0.0171 (10) | 0.0022 (9) |
| C3 | 0.0594 (14) | 0.0481 (12) | 0.0321 (9) | −0.0122 (11) | 0.0142 (9) | −0.0013 (9) |
| C4 | 0.0677 (16) | 0.0665 (15) | 0.0331 (10) | −0.0093 (14) | 0.0114 (10) | 0.0112 (11) |
| C5 | 0.0651 (17) | 0.0722 (18) | 0.0415 (12) | −0.0028 (15) | −0.0016 (12) | 0.0160 (13) |
| C6 | 0.0459 (14) | 0.0736 (19) | 0.0475 (13) | −0.0042 (12) | 0.0030 (10) | −0.0020 (13) |
| C7 | 0.0195 (6) | 0.0227 (6) | 0.0339 (7) | 0.0020 (5) | 0.0037 (5) | −0.0024 (6) |
| C8 | 0.0229 (6) | 0.0274 (7) | 0.0357 (8) | 0.0012 (6) | 0.0038 (6) | −0.0001 (7) |
| C9 | 0.0291 (8) | 0.0240 (6) | 0.0377 (8) | 0.0036 (6) | 0.0111 (6) | 0.0038 (7) |
Geometric parameters (Å, º) top
| O1—C3 | 1.342 (4) | C2—C3 | 1.394 (3) |
| O1—H3 | 0.90 (5) | C2—H2 | 0.93 |
| O2—C7 | 1.404 (2) | C3—C4 | 1.397 (4) |
| O2—H8 | 0.86 (4) | C4—C5 | 1.387 (5) |
| O3—C8 | 1.203 (3) | C4—H4 | 0.93 |
| O4—C8 | 1.320 (2) | C5—C6 | 1.375 (4) |
| O4—H9 | 0.83 (5) | C5—H5 | 0.93 |
| O5—C9 | 1.236 (3) | C6—H6 | 0.93 |
| O6—C9 | 1.251 (2) | C7—C8 | 1.527 (2) |
| N1—C2 | 1.330 (4) | C7—C9 | 1.543 (2) |
| N1—C6 | 1.339 (4) | C7—H7 | 0.98 |
| N1—H1 | 0.84 (5) | | |
| | | |
| O2···H1 | 2.62 (5) | O5···H7vii | 2.69 |
| O2···H6 | 2.79 | O5···H8vii | 2.71 (3) |
| O2···N1 | 3.004 (3) | O5···C7vii | 3.203 (2) |
| O2···C6 | 3.146 (3) | O5···C4vi | 3.206 (4) |
| O2···C9i | 3.299 (2) | O5···C3vi | 3.269 (3) |
| O2···C8ii | 3.373 (2) | O5···C8vii | 3.362 (3) |
| O3···H2iii | 2.50 | O6···C7vii | 3.377 (2) |
| O3···H6 | 2.86 | C7···H9ii | 2.73 (5) |
| O3···N1iv | 3.262 (3) | C9···H8vii | 2.51 (4) |
| O3···C2iv | 3.305 (3) | C9···H3vi | 2.65 (5) |
| O4···H4v | 2.70 | C9···H1 | 2.88 (5) |
| O5···H4vi | 2.56 | | |
| | | |
| C3—O1—H3 | 107 (3) | C4—C5—H5 | 120.1 |
| C7—O2—H8 | 111 (2) | N1—C6—C5 | 118.7 (3) |
| C8—O4—H9 | 108 (3) | N1—C6—H6 | 120.7 |
| C2—N1—C6 | 123.7 (2) | C5—C6—H6 | 120.7 |
| C2—N1—H1 | 119 (3) | O2—C7—C8 | 110.62 (15) |
| C6—N1—H1 | 116 (3) | O2—C7—C9 | 108.66 (13) |
| N1—C2—C3 | 120.2 (2) | C8—C7—C9 | 108.96 (13) |
| N1—C2—H2 | 119.9 | O2—C7—H7 | 109.5 |
| C3—C2—H2 | 119.9 | C8—C7—H7 | 109.5 |
| O1—C3—C2 | 118.1 (2) | C9—C7—H7 | 109.5 |
| O1—C3—C4 | 124.5 (2) | O3—C8—O4 | 125.29 (19) |
| C2—C3—C4 | 117.4 (3) | O3—C8—C7 | 123.47 (17) |
| C5—C4—C3 | 120.3 (2) | O4—C8—C7 | 111.24 (17) |
| C5—C4—H4 | 119.9 | O5—C9—O6 | 125.01 (17) |
| C3—C4—H4 | 119.9 | O5—C9—C7 | 118.45 (17) |
| C6—C5—C4 | 119.8 (3) | O6—C9—C7 | 116.53 (16) |
| C6—C5—H5 | 120.1 | | |
| | | |
| C6—N1—C2—C3 | −0.6 (4) | O2—C7—C8—O3 | 10.1 (3) |
| N1—C2—C3—O1 | 180.0 (3) | C9—C7—C8—O3 | −109.3 (2) |
| N1—C2—C3—C4 | 0.6 (4) | O2—C7—C8—O4 | −169.49 (16) |
| O1—C3—C4—C5 | −179.3 (3) | C9—C7—C8—O4 | 71.1 (2) |
| C2—C3—C4—C5 | 0.0 (4) | O2—C7—C9—O5 | 153.1 (2) |
| C3—C4—C5—C6 | −0.7 (5) | C8—C7—C9—O5 | −86.3 (3) |
| C2—N1—C6—C5 | 0.0 (4) | O2—C7—C9—O6 | −27.8 (2) |
| C4—C5—C6—N1 | 0.6 (5) | C8—C7—C9—O6 | 92.8 (2) |
| Symmetry codes: (i) x, y+1, z; (ii) x+1, y, z; (iii) x−1, y+1, z; (iv) x−1, y, z; (v) x−1, −y+1, z+1/2; (vi) x−1, −y, z+1/2; (vii) x, y−1, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H3···O5viii | 0.90 (5) | 1.71 (5) | 2.604 (3) | 180 (7) |
| O2—H8···O6i | 0.86 (4) | 1.74 (4) | 2.5956 (19) | 179 (4) |
| O4—H9···O2iv | 0.83 (5) | 1.79 (5) | 2.602 (2) | 165 (4) |
| N1—H1···O6 | 0.83 (5) | 1.88 (5) | 2.682 (3) | 162 (5) |
| C2—H2···O3ix | 0.93 | 2.50 | 3.422 (3) | 174 |
| C4—H4···O5viii | 0.93 | 2.56 | 3.206 (4) | 127 |
| Symmetry codes: (i) x, y+1, z; (iv) x−1, y, z; (viii) x+1, −y, z−1/2; (ix) x+1, y−1, z. |
Crystal data top
| C5H6NO+·C3H3O5− | F(000) = 224 |
| Mr = 215.16 | Dx = 1.574 Mg m−3 |
| Monoclinic, Pn | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P -2yac | Cell parameters from 25 reflections |
| a = 4.0607 (13) Å | θ = 10.8–11.4° |
| b = 13.144 (3) Å | µ = 0.14 mm−1 |
| c = 8.696 (2) Å | T = 298 K |
| β = 101.99 (2)° | Plate, colourless |
| V = 454.0 (2) Å3 | 0.46 × 0.26 × 0.17 mm |
| Z = 2 | |
Data collection top
Rigaku AFC-5R
diffractometer | 861 reflections with I > 2σ(I) |
| Radiation source: Rigaku rotating anode | Rint = 0.038 |
| Graphite monochromator | θmax = 30.0°, θmin = 2.9° |
| ω/2θ scans | h = −1→5 |
Absorption correction: ψ scan
(North et al., 1968) | k = −1→18 |
| Tmin = 0.942, Tmax = 0.977 | l = −12→12 |
| 1985 measured reflections | 3 standard reflections every 97 reflections |
| 1341 independent reflections | intensity decay: 2.0% |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.051 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.133 | H-atom parameters constrained |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.0521P)2]
where P = (Fo2 + 2Fc2)/3 |
| 1341 reflections | (Δ/σ)max = 0.001 |
| 136 parameters | Δρmax = 0.21 e Å−3 |
| 2 restraints | Δρmin = −0.19 e Å−3 |
Crystal data top
| C5H6NO+·C3H3O5− | V = 454.0 (2) Å3 |
| Mr = 215.16 | Z = 2 |
| Monoclinic, Pn | Mo Kα radiation |
| a = 4.0607 (13) Å | µ = 0.14 mm−1 |
| b = 13.144 (3) Å | T = 298 K |
| c = 8.696 (2) Å | 0.46 × 0.26 × 0.17 mm |
| β = 101.99 (2)° | |
Data collection top
Rigaku AFC-5R
diffractometer | 861 reflections with I > 2σ(I) |
Absorption correction: ψ scan
(North et al., 1968) | Rint = 0.038 |
| Tmin = 0.942, Tmax = 0.977 | 3 standard reflections every 97 reflections |
| 1985 measured reflections | intensity decay: 2.0% |
| 1341 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.051 | 2 restraints |
| wR(F2) = 0.133 | H-atom parameters constrained |
| S = 1.00 | Δρmax = 0.21 e Å−3 |
| 1341 reflections | Δρmin = −0.19 e Å−3 |
| 136 parameters | |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.0432 (12) | 0.8918 (2) | 0.9464 (4) | 0.0558 (9) | |
| O2 | 0.5687 (8) | 0.6265 (2) | 0.4054 (3) | 0.0357 (7) | |
| O3 | 0.3569 (10) | 0.4882 (2) | 0.1718 (4) | 0.0526 (9) | |
| O4 | 0.6132 (9) | 0.5659 (2) | 0.0077 (3) | 0.0494 (9) | |
| O5 | 0.8864 (9) | 0.7360 (2) | 0.0931 (4) | 0.0510 (9) | |
| O6 | 0.7017 (9) | 0.8064 (2) | 0.2937 (4) | 0.0458 (8) | |
| N1 | 0.3879 (11) | 0.8035 (3) | 0.5608 (4) | 0.0401 (9) | |
| C2 | 0.3392 (13) | 0.9020 (3) | 0.5858 (5) | 0.0426 (10) | |
| C3 | 0.2261 (13) | 0.9332 (3) | 0.7144 (5) | 0.0454 (12) | |
| C4 | 0.1519 (12) | 0.8613 (3) | 0.8191 (5) | 0.0392 (10) | |
| C5 | 0.1983 (12) | 0.7588 (3) | 0.7885 (5) | 0.0374 (9) | |
| C6 | 0.3127 (13) | 0.7327 (3) | 0.6582 (5) | 0.0432 (11) | |
| C7 | 0.7267 (11) | 0.6245 (3) | 0.2768 (4) | 0.0301 (8) | |
| C8 | 0.5441 (12) | 0.5539 (3) | 0.1482 (4) | 0.0348 (9) | |
| C9 | 0.7685 (11) | 0.7335 (3) | 0.2199 (4) | 0.0351 (10) | |
| H1 | 0.469 | 0.785 | 0.481 | 0.048* | |
| H2 | 0.383 | 0.950 | 0.514 | 0.051* | |
| H3 | 0.199 | 1.002 | 0.732 | 0.054* | |
| H4 | 0.001 | 0.842 | 0.996 | 0.084* | |
| H5 | 0.151 | 0.709 | 0.857 | 0.045* | |
| H6 | 0.339 | 0.664 | 0.636 | 0.052* | |
| H7 | 0.953 | 0.597 | 0.314 | 0.036* | |
| H8 | 0.630 | 0.577 | 0.462 | 0.053* | |
| H9 | 0.720 | 0.619 | 0.006 | 0.074* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.093 (3) | 0.0359 (15) | 0.0512 (19) | 0.004 (2) | 0.0448 (19) | −0.0015 (15) |
| O2 | 0.0484 (17) | 0.0332 (13) | 0.0305 (15) | 0.0042 (13) | 0.0203 (14) | 0.0028 (11) |
| O3 | 0.067 (3) | 0.0397 (15) | 0.0571 (18) | −0.0144 (17) | 0.0257 (18) | −0.0135 (14) |
| O4 | 0.070 (3) | 0.0503 (17) | 0.0333 (14) | −0.0062 (18) | 0.0236 (16) | −0.0125 (14) |
| O5 | 0.077 (3) | 0.0431 (16) | 0.0428 (17) | −0.0120 (18) | 0.0340 (18) | −0.0003 (14) |
| O6 | 0.069 (2) | 0.0318 (13) | 0.0411 (15) | −0.0061 (16) | 0.0218 (16) | −0.0010 (13) |
| N1 | 0.052 (3) | 0.0410 (18) | 0.0321 (17) | −0.0068 (18) | 0.0204 (17) | −0.0014 (15) |
| C2 | 0.060 (3) | 0.0318 (18) | 0.042 (2) | −0.003 (2) | 0.024 (2) | 0.0086 (17) |
| C3 | 0.069 (3) | 0.0207 (16) | 0.054 (3) | 0.0034 (19) | 0.030 (3) | 0.0045 (17) |
| C4 | 0.056 (3) | 0.033 (2) | 0.033 (2) | 0.000 (2) | 0.019 (2) | 0.0013 (16) |
| C5 | 0.053 (3) | 0.0297 (17) | 0.0334 (18) | 0.003 (2) | 0.0185 (19) | 0.0030 (16) |
| C6 | 0.064 (3) | 0.0331 (18) | 0.039 (2) | −0.002 (2) | 0.026 (2) | −0.0009 (18) |
| C7 | 0.037 (2) | 0.0311 (16) | 0.0245 (17) | 0.0060 (18) | 0.0120 (16) | 0.0000 (15) |
| C8 | 0.044 (3) | 0.0267 (16) | 0.037 (2) | 0.0047 (18) | 0.0161 (19) | −0.0048 (15) |
| C9 | 0.043 (3) | 0.034 (2) | 0.0306 (19) | −0.0057 (18) | 0.0139 (19) | −0.0002 (16) |
Geometric parameters (Å, º) top
| O1—C4 | 1.336 (5) | C2—C3 | 1.358 (6) |
| O1—H4 | 0.82 | C2—H2 | 0.93 |
| O2—C7 | 1.400 (4) | C3—C4 | 1.389 (5) |
| O2—H8 | 0.82 | C3—H3 | 0.93 |
| O3—C8 | 1.196 (5) | C4—C5 | 1.394 (5) |
| O4—C8 | 1.318 (5) | C5—C6 | 1.356 (6) |
| O4—H9 | 0.82 | C5—H5 | 0.93 |
| O5—C9 | 1.291 (5) | C6—H6 | 0.93 |
| O6—C9 | 1.215 (5) | C7—C8 | 1.522 (6) |
| N1—C2 | 1.334 (5) | C7—C9 | 1.537 (5) |
| N1—C6 | 1.336 (5) | C7—H7 | 0.98 |
| N1—H1 | 0.86 | | |
| | | |
| O2···H6 | 2.43 | O4···C7ii | 3.385 (5) |
| O2···O3 | 2.729 (4) | O4···C5iii | 3.401 (6) |
| O2···C6 | 2.968 (5) | O5···H5iv | 2.53 |
| O2···C7i | 3.370 (6) | O5···C5iv | 3.178 (6) |
| O3···H7i | 2.67 | O5···C4iv | 3.258 (6) |
| O3···O4i | 3.226 (5) | O6···H3v | 2.57 |
| O3···C8i | 3.376 (6) | O6···N1vi | 3.235 (5) |
| O3···C7i | 3.401 (6) | C2···C4vi | 3.537 (7) |
| O4···H8ii | 2.69 | C2···C3vi | 3.566 (8) |
| O4···H7ii | 2.72 | C5···C6i | 3.554 (7) |
| O4···H5iii | 2.78 | | |
| | | |
| C4—O1—H4 | 109.5 | C4—C5—H5 | 120.4 |
| C7—O2—H8 | 109.5 | N1—C6—C5 | 121.1 (4) |
| C8—O4—H9 | 109.5 | N1—C6—H6 | 119.4 |
| C2—N1—C6 | 120.8 (4) | C5—C6—H6 | 119.4 |
| C2—N1—H1 | 119.6 | O2—C7—C8 | 111.3 (3) |
| C6—N1—H1 | 119.6 | O2—C7—C9 | 109.8 (3) |
| N1—C2—C3 | 121.0 (4) | C8—C7—C9 | 113.7 (3) |
| N1—C2—H2 | 119.5 | O2—C7—H7 | 107.2 |
| C3—C2—H2 | 119.5 | C8—C7—H7 | 107.2 |
| C2—C3—C4 | 119.4 (3) | C9—C7—H7 | 107.2 |
| C2—C3—H3 | 120.3 | O3—C8—O4 | 120.7 (4) |
| C4—C3—H3 | 120.3 | O3—C8—C7 | 123.0 (3) |
| O1—C4—C3 | 119.5 (3) | O4—C8—C7 | 116.3 (4) |
| O1—C4—C5 | 122.0 (3) | O6—C9—O5 | 126.5 (4) |
| C3—C4—C5 | 118.5 (4) | O6—C9—C7 | 120.9 (3) |
| C6—C5—C4 | 119.2 (4) | O5—C9—C7 | 112.6 (4) |
| C6—C5—H5 | 120.4 | | |
| | | |
| C6—N1—C2—C3 | 2.9 (9) | O2—C7—C8—O3 | −19.9 (6) |
| N1—C2—C3—C4 | −1.7 (8) | C9—C7—C8—O3 | −144.6 (4) |
| C2—C3—C4—O1 | 179.5 (5) | O2—C7—C8—O4 | 162.7 (3) |
| C2—C3—C4—C5 | 0.3 (8) | C9—C7—C8—O4 | 38.0 (5) |
| O1—C4—C5—C6 | −179.3 (5) | O2—C7—C9—O6 | 7.4 (6) |
| C3—C4—C5—C6 | −0.2 (8) | C8—C7—C9—O6 | 132.9 (4) |
| C2—N1—C6—C5 | −2.8 (8) | O2—C7—C9—O5 | −174.4 (4) |
| C4—C5—C6—N1 | 1.4 (8) | C8—C7—C9—O5 | −48.9 (5) |
| Symmetry codes: (i) x−1, y, z; (ii) x−1/2, −y+1, z−1/2; (iii) x, y, z−1; (iv) x+1, y, z−1; (v) x+1/2, −y+2, z−1/2; (vi) x+1, y, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H4···O5vii | 0.82 | 1.74 | 2.562 (4) | 177 |
| O2—H8···O3viii | 0.82 | 2.05 | 2.808 (4) | 153 |
| O4—H9···O5 | 0.82 | 1.79 | 2.538 (4) | 151 |
| N1—H1···O2 | 0.86 | 2.25 | 2.862 (5) | 128 |
| N1—H1···O6 | 0.86 | 2.07 | 2.872 (5) | 156 |
| C2—H2···O1v | 0.93 | 2.29 | 3.149 (5) | 153 |
| C3—H3···O6ix | 0.93 | 2.57 | 3.497 (5) | 173 |
| C5—H5···O5vii | 0.93 | 2.54 | 3.179 (6) | 127 |
| C7—H7···O2vi | 0.98 | 2.49 | 3.369 (6) | 149 |
| Symmetry codes: (v) x+1/2, −y+2, z−1/2; (vi) x+1, y, z; (vii) x−1, y, z+1; (viii) x+1/2, −y+1, z+1/2; (ix) x−1/2, −y+2, z+1/2. |
Experimental details
| | (I) | (II) | (III) |
| Crystal data |
| Chemical formula | C5H5NO·C3H4O5 | C5H6NO+·C3H3O5− | C5H6NO+·C3H3O5− |
| Mr | 215.16 | 215.16 | 215.16 |
| Crystal system, space group | Triclinic, P1 | Monoclinic, Pc | Monoclinic, Pn |
| Temperature (K) | 296 | 298 | 298 |
| a, b, c (Å) | 5.879 (4), 6.859 (4), 12.588 (8) | 5.7563 (11), 5.1871 (7), 16.3206 (19) | 4.0607 (13), 13.144 (3), 8.696 (2) |
| α, β, γ (°) | 74.61 (5), 80.94 (6), 70.95 (5) | 90, 92.186 (13), 90 | 90, 101.99 (2), 90 |
| V (Å3) | 461.2 (5) | 486.95 (13) | 454.0 (2) |
| Z | 2 | 2 | 2 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα |
| µ (mm−1) | 0.14 | 0.13 | 0.14 |
| Crystal size (mm) | 0.30 × 0.20 × 0.08 | 0.50 × 0.50 × 0.10 | 0.46 × 0.26 × 0.17 |
| |
| Data collection |
| Diffractometer | Rigaku AFC-5R
diffractometer | Rigaku AFC-5R
diffractometer | Rigaku AFC-5R
diffractometer |
| Absorption correction | – | – | ψ scan
(North et al., 1968) |
| Tmin, Tmax | – | – | 0.942, 0.977 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 3319, 2698, 1323 | 2353, 1562, 1394 | 1985, 1341, 861 |
| Rint | 0.032 | 0.016 | 0.038 |
| (sin θ/λ)max (Å−1) | 0.703 | 0.725 | 0.704 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.065, 0.191, 1.00 | 0.042, 0.119, 1.00 | 0.051, 0.133, 1.00 |
| No. of reflections | 2698 | 1562 | 1341 |
| No. of parameters | 153 | 153 | 136 |
| No. of restraints | 0 | 2 | 2 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.26, −0.24 | 0.43, −0.18 | 0.21, −0.19 |
Selected geometric parameters (Å, º) for (I) top| O1—C2 | 1.279 (7) | O4—C8 | 1.308 (4) |
| O2—C7 | 1.416 (5) | O5—C9 | 1.208 (7) |
| O3—C8 | 1.204 (3) | O6—C9 | 1.304 (4) |
| | | |
| O1—C2—N1 | 118.7 (3) | O3—C8—C7 | 121.6 (3) |
| O1—C2—C3 | 125.1 (3) | O4—C8—C7 | 112.5 (2) |
| O2—C7—C8 | 110.6 (2) | O5—C9—C7 | 122.0 (3) |
| O2—C7—C9 | 110.9 (3) | O6—C9—C7 | 112.8 (3) |
| C8—C7—C9 | 112.1 (3) | | |
Hydrogen-bond geometry (Å, º) for (I) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1i | 0.96 (3) | 1.85 (3) | 2.809 (4) | 174 (4) |
| O2—H7···O5ii | 0.93 (4) | 1.96 (4) | 2.825 (4) | 155 (4) |
| O4—H8···O2iii | 0.76 (4) | 1.97 (4) | 2.712 (4) | 164 (4) |
| O6—H9···O1 | 0.80 (5) | 1.77 (5) | 2.569 (4) | 175 (3) |
| O2—H7···O3 | 0.93 (4) | 2.29 (4) | 2.675 (4) | 104 (3) |
| C6—H5···O3iv | 0.93 | 2.50 | 3.096 (4) | 122 |
| C6—H5···O5i | 0.93 | 2.52 | 3.286 (5) | 140 |
| Symmetry codes: (i) −x+1, −y, −z+1; (ii) −x+1, −y+1, −z; (iii) x+1, y, z; (iv) x, y−1, z+1. |
Selected geometric parameters (Å, º) for (II) top| O1—C3 | 1.342 (4) | O4—C8 | 1.320 (2) |
| O2—C7 | 1.404 (2) | O5—C9 | 1.236 (3) |
| O3—C8 | 1.203 (3) | O6—C9 | 1.251 (2) |
| | | |
| O1—C3—C2 | 118.1 (2) | O3—C8—C7 | 123.47 (17) |
| O1—C3—C4 | 124.5 (2) | O4—C8—C7 | 111.24 (17) |
| O2—C7—C8 | 110.62 (15) | O5—C9—C7 | 118.45 (17) |
| O2—C7—C9 | 108.66 (13) | O6—C9—C7 | 116.53 (16) |
| C8—C7—C9 | 108.96 (13) | | |
Hydrogen-bond geometry (Å, º) for (II) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H3···O5i | 0.90 (5) | 1.71 (5) | 2.604 (3) | 180 (7) |
| O2—H8···O6ii | 0.86 (4) | 1.74 (4) | 2.5956 (19) | 179 (4) |
| O4—H9···O2iii | 0.83 (5) | 1.79 (5) | 2.602 (2) | 165 (4) |
| N1—H1···O6 | 0.83 (5) | 1.88 (5) | 2.682 (3) | 162 (5) |
| C2—H2···O3iv | 0.93 | 2.50 | 3.422 (3) | 174 |
| C4—H4···O5i | 0.93 | 2.56 | 3.206 (4) | 127 |
| Symmetry codes: (i) x+1, −y, z−1/2; (ii) x, y+1, z; (iii) x−1, y, z; (iv) x+1, y−1, z. |
Selected geometric parameters (Å, º) for (III) top| O1—C4 | 1.336 (5) | O4—C8 | 1.318 (5) |
| O2—C7 | 1.400 (4) | O5—C9 | 1.291 (5) |
| O3—C8 | 1.196 (5) | O6—C9 | 1.215 (5) |
| | | |
| O1—C4—C3 | 119.5 (3) | O3—C8—C7 | 123.0 (3) |
| O1—C4—C5 | 122.0 (3) | O4—C8—C7 | 116.3 (4) |
| O2—C7—C8 | 111.3 (3) | O6—C9—C7 | 120.9 (3) |
| O2—C7—C9 | 109.8 (3) | O5—C9—C7 | 112.6 (4) |
| C8—C7—C9 | 113.7 (3) | | |
Hydrogen-bond geometry (Å, º) for (III) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H4···O5i | 0.82 | 1.74 | 2.562 (4) | 177 |
| O2—H8···O3ii | 0.82 | 2.05 | 2.808 (4) | 153 |
| O4—H9···O5 | 0.82 | 1.79 | 2.538 (4) | 151 |
| N1—H1···O2 | 0.86 | 2.25 | 2.862 (5) | 128 |
| N1—H1···O6 | 0.86 | 2.07 | 2.872 (5) | 156 |
| C2—H2···O1iii | 0.93 | 2.29 | 3.149 (5) | 153 |
| C3—H3···O6iv | 0.93 | 2.57 | 3.497 (5) | 173 |
| C5—H5···O5i | 0.93 | 2.54 | 3.179 (6) | 127 |
| C7—H7···O2v | 0.98 | 2.49 | 3.369 (6) | 149 |
| Symmetry codes: (i) x−1, y, z+1; (ii) x+1/2, −y+1, z+1/2; (iii) x+1/2, −y+2, z−1/2; (iv) x−1/2, −y+2, z+1/2; (v) x+1, y, z. |
 O and O—H
O and O—H O hydrogen bonds to form a hydrogen-bonded chain. In each of (I), (II) and (III), an intermolecular hydrogen bond is formed between a carboxyl group and the hydroxyl group attached to the central C atom, and in (I), the hydroxyl group participates in an intramolecular hydrogen bond with a carbonyl group. No intermolecular hydrogen bond is formed between the carboxyl groups in (I), or between the carboxyl and carboxylate groups in (II) and (III).
O hydrogen bonds to form a hydrogen-bonded chain. In each of (I), (II) and (III), an intermolecular hydrogen bond is formed between a carboxyl group and the hydroxyl group attached to the central C atom, and in (I), the hydroxyl group participates in an intramolecular hydrogen bond with a carbonyl group. No intermolecular hydrogen bond is formed between the carboxyl groups in (I), or between the carboxyl and carboxylate groups in (II) and (III).
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2004/10/00/fr1496/fr1496fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2004/10/00/fr1496/fr1496fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2004/10/00/fr1496/fr1496fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/c/issues/2004/10/00/fr1496/fr1496fig4thm.gif)




Tartronic acid, a member of a family of hydroxycarboxylic acids, is of potential interest in crystal engineering for the formation of two- and three-dimensional aggregates with organic bases, as shown in several organic salts of tartaric acid (Aakeröy et al., 1992; Aakeröy & Hitchcock, 1993; Aakeröy Bahra & Niewenhuzen, 1996), malonic acid (Aakeröy & Niewenhuzen, 1994, 1996), dihydroxymalonic acid (Aakeröy Niewenhuzen & Robinson, 1998) and oxamic acid (Aakeröy Hughes & Niewenhuyzen, 1996). Furthermore, tartronic acid is an interesting candidate for the selective synthesis and crystallization of optically active substances, because its acidic anion, the hydrogen tartronate ion, is optically active. Previously, we have reported the three-dimensional hydrogen-bonded structure of imidazolium hydrogen tartronate, where the anions form a homochiral infinite chain but the chains aggregate to form a heterochiral layer (Fukunaga & Ishida, 2003). In the present study, we have selected hydroxypyridines as the counter-base in order to extend the previous examination further, and report here the structures of the title compounds, (I)-(III). Fig. 1 shows the asymmetric units of compounds (I), (II) and (III). \sch
In (I), the base molecule is in the lactam form (Yang & Craven, 1998), and no acid-base interaction involving H-atom transfer is observed. Two pyridone molecules are held together by N—H···O hydrogen bonds, forming a centrosymmetric hydrogen-bonded ring with graph-set discriptor R44(8) (Bernstein et al., 1995), and two molecules of tartronic acid are held together by O—H···O hydrogen bonds between the hydroxyl and carbonyl groups, forming a centrosymmetric R22(10) ring (Fig. 2a and Table 2). The dimeric unit composed of pyridone molecules is connected to that composed of acid molecules via an O6—H9···O1 hydrogen bond on one side, and by C6—H5···O3iv and C6—H5···O5i bifurcated hydrogen bonds on the other side, to afford a molecular tape running along [011] [symmetry codes: (i) 1 − x, −y, 1 − z; (iv) x, y − 1, 1 + z; Fig. 2a and Table 2]. The molecular tapes are stacked parallel to the c direction through an O4—H8···O2iii hydrogen bond, resulting in a layer parallel to (202) [symmetry code: (iii) 1 + x, y, z; Figs. 2 b and 2c, and Table 2]. The closest separation between the pyridone rings in the layer is 3.428 Å, and the centre-to-centre separation is 3.735 (3) Å.
The hydrogen-bonding motif formed between the pyridone dimer and the acid moiety is similar to that reported for 2:1 co-crystals of 2-pyridone and several dicarboxylic acids, namely oxalic acid, succinic acid, adipic acid dihydrate, suberic acid and sebacic acid (Aakeröy, Beatty & Zou, 1998), fumaric acid and meso-2,3-dibromosuccinic acid (Aakeröy et al., 2000), oxalic acid and trans-β-hydromuconic acid (Kashino et al., 2001), and malonic acid and pimeric acid (Edwards et al., 2002). The pyridone dimer is also formed in 1:1 co-crystals of 2-pyridone and some non-centrosymmetric dicarboxylic acids, namely glutaric acid and azelaic acid (Edwards et al., 2002). The pyridone dimer is not formed in the 1:1 co-crystals of 2-pyridone with trans-glutaconic acid and L-tartaric acid (Kashino et al., 2001).
In (II) and (III), H-atom transfer occurs from the acid to the base moiety and a chiral hydrogen tartronate anion is induced in these crystals. In (II), a cation is linked to two neighbouring anions related by a glide plane through the N1—H1···O6, N1—H1···O2, O1—H3···O5v and C4—H4···O5v hydrogen bonds, to form a zigzag chain running along [102] [symmetry code: (v) 1 + x, −y, z − 1/2; Fig. 3a and Table 4]. On the other hand, the anions aggregate through O—H···O hydrogen bonds between the hydroxyl and carboxylate groups (O2—H8···O6vi) and between the carboxyl and hydroxyl groups (O4—H9···O2vii) to form a homochiral plane parallel to (001) [symmetry codes: (vi) x, 1 + y, z; (vii) x − 1, y, z; Fig. 3 b]. The planes related by a glide plane are bridged by cations through the N—H···O, O—H···O and C—H···O hydrogen bonds mentioned above, resulting in a three-dimensional hydrogen-bonded network. It is noted that such a homochiral plane has also been found in some chiral crystals, namely 3-hydroxypyridinium hydrogen L-tartarate (Tafeenko et al., 1990) and 3-hydroxypyridinium hydrogen L-maleate (Aakeröy & Niewenhuyzen, 1994). In these crystals, planes with the same chirality were bridged by the cations.
In (III), an intramolecular O—H···O hydrogen bond is observed between the carboxyl and carboxylate groups in the anion (Fig. 1 and Table 6). A cation is linked to two neighbouring anions, which are related to each other by a translation along [101], through the O1—H4···O5ix, C5—H5···O5ix and bifurcated N—H···O hydrogen bonds to form a zigzag chain running along [121] [symmetry codes: (ix) x − 1, y, 1 + z; Fig. 4a and Table 6). The chains are linked by an O2—H8···O3x hydrogen bond between the hydroxyl and carboxyl groups, resulting in a heterochiral layer parallel to (101) [symmetry code: (x) 1/2 + x, 1 − y, 1/2 + z; Fig. 4 b]. The layers are connected via an C7—H7···O2xiii hydrogen bond [symmetry code: (xiii) 1 + x, y, z; Table 6].
In the crystal of tartronic acid, neighbouring molecules related by a 21 axis form a molecular chain through O—H···O hydrogen bonds between the carboxyl groups (van Eijck et al., 1965). In the crystal of ammonium hydrogen tartronate, hydrogen tartronate anions related by a 21 axis form a very short O—H···O hydrogen bond between the carboxyl and carboxylate groups (Taka et al., 1998; Moritani et al., 2001). This is an indication that a strong intermolecular hydrogen bond is formed between the carboxyl and carboxylate groups of the anions. However, in the crystals of (I), (II) and (III), no such type of hydrogen-bonding is observed. This fact shows that the hydroxyl group attached to the central C atom of tartronic acid or the hydrogen tartronate anion can form stable intermolecular hydrogen bond(s) in place of the carboxyl group.