Buy article online - an online subscription or single-article purchase is required to access this article.

In the polymeric title complex, [CuCl
2(C
3H
6N
4)
2]
n, there are two ligands in the asymmetric unit. The Cu atom adopts an elongated octahedral geometry, with two 2-ethyltetrazole ligands [Cu—N = 2.0037 (16) and 2.0136 (16) Å] and two Cl atoms [Cu—Cl = 2.2595 (6) and 2.2796 (6) Å] in equatorial positions. A Cl atom and a symmetry-related 2-ethyltetrazole molecule [Cu—Cl = 2.8845 (8) Å and Cu—N = 2.851 (2) Å] lie in the axial positions of the octahedron. One of the two 2-ethyltetrazole ligands of the asymmetric unit exhibits bidentate binding to two Cu atoms through two N atoms of the tetrazole ring, whereas the other ligand is coordinated in a monodentate fashion
via one tetrazole N atom. The Cu-atom octahedra form dimer entities by sharing edges with equatorial and axial Cl atoms. The dimers are linked together through the 2-ethyltetrazole ligands to form one-dimensional polymeric zigzag chains extending along the
b axis. The chains are connected into infinite layers parallel to the (10
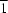
) plane
via the 2-ethyltetrazole ligands.
Supporting information
CCDC reference: 214362
The title complex was prepared by dissolving CuCl2·2H2O (0.0025 mol) in 2-ethyltetrazole (0.025 mol). The fine-grained complex that precipitated immediately was separated by decanting (yield 46%). Two months later, more of the title complex (0.35 g, 42%), in the form of light-green prismatic crystals, was obtained from the decanted solution. Analysis calculated for CuCl2(C3H6N4)2: Cu 19.2, Cl 21.2%; found: Cu 19.3, Cl 21.1 (the results are identical for both obtained fractions).
H atoms were placed in calculated positions, with C—H distances of 0.93–0.97 Å, and were included in the refinement in a riding-model approximation, with Uiso(H) equal to 1.5Ueq(carrier atom) for the methyl group and 1.2Ueq(carrier atom) for other H atoms.
Data collection: R3m software (Nicolet, 1980); cell refinement: R3m software (Nicolet, 1980); data reduction: OMNIBUS (Galdecka, 2002); program(s) used to solve structure: SIR97 (Altomare et al., 1999); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997),
PLATON (Spek, 2003); software used to prepare material for publication: SHELXL97.
poly[[[di-µ-chloro-bis[chloro(2-ethyltetrazole-kN
4)copper(II)]]- di-µ-ethyltetrazole-k
2N
1:
N4]
top
Crystal data top
| [CuCl2(C6H12N8)] | F(000) = 668 |
| Mr = 330.68 | Dx = 1.713 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 25 reflections |
| a = 10.234 (2) Å | θ = 22.0–24.1° |
| b = 13.690 (4) Å | µ = 2.11 mm−1 |
| c = 10.478 (2) Å | T = 293 K |
| β = 119.174 (15)° | Prism, blue |
| V = 1281.9 (5) Å3 | 0.60 × 0.60 × 0.52 mm |
| Z = 4 | |
Data collection top
Nicolet R3m four-circle
diffractometer | 3444 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.019 |
| Graphite monochromator | θmax = 30.1°, θmin = 2.3° |
| ω/2θ scans | h = 0→14 |
Absorption correction: ψ scan
(North et al., 1968) | k = 0→19 |
| Tmin = 0.298, Tmax = 0.333 | l = −14→12 |
| 4079 measured reflections | 3 standard reflections every 100 reflections |
| 3763 independent reflections | intensity decay: none |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.034 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.098 | H-atom parameters constrained |
| S = 1.09 | w = 1/[σ2(Fo2) + (0.0535P)2 + 0.5059P]
where P = (Fo2 + 2Fc2)/3 |
| 3763 reflections | (Δ/σ)max < 0.001 |
| 156 parameters | Δρmax = 0.57 e Å−3 |
| 0 restraints | Δρmin = −0.59 e Å−3 |
Crystal data top
| [CuCl2(C6H12N8)] | V = 1281.9 (5) Å3 |
| Mr = 330.68 | Z = 4 |
| Monoclinic, P21/n | Mo Kα radiation |
| a = 10.234 (2) Å | µ = 2.11 mm−1 |
| b = 13.690 (4) Å | T = 293 K |
| c = 10.478 (2) Å | 0.60 × 0.60 × 0.52 mm |
| β = 119.174 (15)° | |
Data collection top
Nicolet R3m four-circle
diffractometer | 3444 reflections with I > 2σ(I) |
Absorption correction: ψ scan
(North et al., 1968) | Rint = 0.019 |
| Tmin = 0.298, Tmax = 0.333 | 3 standard reflections every 100 reflections |
| 4079 measured reflections | intensity decay: none |
| 3763 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.034 | 0 restraints |
| wR(F2) = 0.098 | H-atom parameters constrained |
| S = 1.09 | Δρmax = 0.57 e Å−3 |
| 3763 reflections | Δρmin = −0.59 e Å−3 |
| 156 parameters | |
Special details top
Experimental. Because of instability in air, the single-crystal was mounted in a glass capillary. |
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Cu1 | 0.57901 (2) | 0.123100 (17) | 0.04226 (2) | 0.03685 (9) | |
| Cl1 | 0.52049 (5) | 0.02443 (3) | 0.18239 (5) | 0.03839 (11) | |
| Cl2 | 0.64953 (6) | 0.23024 (4) | −0.07677 (6) | 0.05315 (14) | |
| N1A | 0.1548 (2) | 0.22734 (17) | −0.24365 (19) | 0.0545 (5) | |
| N2A | 0.15985 (18) | 0.23031 (13) | −0.11470 (18) | 0.0413 (3) | |
| N3A | 0.28670 (18) | 0.20117 (14) | −0.00508 (18) | 0.0431 (4) | |
| N4A | 0.37140 (17) | 0.17808 (12) | −0.06414 (17) | 0.0382 (3) | |
| C5A | 0.2906 (2) | 0.19436 (17) | −0.2072 (2) | 0.0473 (5) | |
| H5A | 0.3251 | 0.1839 | −0.2735 | 0.057* | |
| C6A | 0.0295 (3) | 0.2595 (2) | −0.0991 (3) | 0.0581 (6) | |
| H61A | −0.0540 | 0.2167 | −0.1581 | 0.070* | |
| H62A | 0.0005 | 0.3255 | −0.1355 | 0.070* | |
| C7A | 0.0612 (3) | 0.2552 (3) | 0.0542 (3) | 0.0686 (7) | |
| H71A | 0.0992 | 0.1917 | 0.0938 | 0.103* | |
| H72A | −0.0293 | 0.2671 | 0.0580 | 0.103* | |
| H73A | 0.1341 | 0.3039 | 0.1106 | 0.103* | |
| N1B | 0.9880 (2) | −0.00382 (16) | 0.1502 (2) | 0.0513 (4) | |
| N2B | 1.00320 (17) | 0.02045 (13) | 0.27913 (18) | 0.0412 (3) | |
| N3B | 0.88395 (18) | 0.06117 (15) | 0.27274 (18) | 0.0448 (4) | |
| N4B | 0.78574 (16) | 0.06495 (13) | 0.13237 (17) | 0.0391 (3) | |
| C5B | 0.8505 (2) | 0.02601 (19) | 0.0605 (2) | 0.0502 (5) | |
| H5B | 0.8044 | 0.0205 | −0.0407 | 0.060* | |
| C6B | 1.1409 (2) | 0.0025 (2) | 0.4187 (3) | 0.0603 (6) | |
| H61B | 1.1429 | −0.0651 | 0.4474 | 0.072* | |
| H62B | 1.1409 | 0.0436 | 0.4941 | 0.072* | |
| C7B | 1.2771 (3) | 0.0234 (2) | 0.4061 (3) | 0.0664 (7) | |
| H71B | 1.2830 | −0.0221 | 0.3394 | 0.100* | |
| H72B | 1.3644 | 0.0171 | 0.5005 | 0.100* | |
| H73B | 1.2716 | 0.0887 | 0.3705 | 0.100* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cu1 | 0.02610 (12) | 0.04774 (15) | 0.03535 (13) | 0.00211 (8) | 0.01393 (9) | 0.00712 (8) |
| Cl1 | 0.0366 (2) | 0.0452 (2) | 0.0351 (2) | 0.00032 (16) | 0.01886 (16) | 0.00204 (16) |
| Cl2 | 0.0471 (3) | 0.0618 (3) | 0.0529 (3) | −0.0049 (2) | 0.0263 (2) | 0.0136 (2) |
| N1A | 0.0445 (9) | 0.0771 (13) | 0.0356 (8) | 0.0232 (9) | 0.0147 (7) | 0.0080 (8) |
| N2A | 0.0330 (7) | 0.0501 (9) | 0.0381 (7) | 0.0098 (6) | 0.0152 (6) | 0.0066 (7) |
| N3A | 0.0326 (7) | 0.0573 (10) | 0.0378 (8) | 0.0089 (7) | 0.0160 (6) | 0.0074 (7) |
| N4A | 0.0324 (7) | 0.0448 (8) | 0.0362 (7) | 0.0054 (6) | 0.0157 (6) | 0.0044 (6) |
| C5A | 0.0422 (10) | 0.0610 (12) | 0.0367 (9) | 0.0151 (9) | 0.0176 (8) | 0.0049 (8) |
| C6A | 0.0410 (11) | 0.0791 (16) | 0.0586 (13) | 0.0209 (11) | 0.0276 (10) | 0.0155 (12) |
| C7A | 0.0551 (13) | 0.097 (2) | 0.0678 (16) | 0.0141 (14) | 0.0412 (13) | 0.0123 (14) |
| N1B | 0.0370 (8) | 0.0712 (12) | 0.0446 (9) | 0.0085 (8) | 0.0190 (7) | −0.0025 (8) |
| N2B | 0.0296 (7) | 0.0550 (9) | 0.0378 (8) | 0.0024 (6) | 0.0156 (6) | 0.0071 (7) |
| N3B | 0.0315 (7) | 0.0656 (11) | 0.0364 (8) | 0.0039 (7) | 0.0159 (6) | 0.0030 (7) |
| N4B | 0.0289 (7) | 0.0524 (9) | 0.0351 (7) | 0.0009 (6) | 0.0149 (6) | 0.0032 (6) |
| C5B | 0.0342 (9) | 0.0763 (15) | 0.0376 (9) | 0.0048 (9) | 0.0155 (8) | −0.0043 (9) |
| C6B | 0.0377 (10) | 0.0933 (18) | 0.0407 (10) | 0.0118 (11) | 0.0120 (8) | 0.0189 (11) |
| C7B | 0.0340 (10) | 0.094 (2) | 0.0599 (14) | 0.0102 (11) | 0.0141 (10) | 0.0063 (13) |
Geometric parameters (Å, º) top
| Cu1—N4A | 2.0037 (16) | C7A—H71A | 0.9600 |
| Cu1—N4B | 2.0136 (16) | C7A—H72A | 0.9600 |
| Cu1—Cl2 | 2.2595 (6) | C7A—H73A | 0.9600 |
| Cu1—Cl1 | 2.2796 (6) | N1B—C5B | 1.319 (3) |
| Cu1—N1Ai | 2.851 (2) | N1B—N2B | 1.325 (3) |
| Cu1—Cl1ii | 2.8845 (8) | N2B—N3B | 1.313 (2) |
| N1A—N2A | 1.328 (2) | N2B—C6B | 1.475 (3) |
| N1A—C5A | 1.328 (3) | N3B—N4B | 1.317 (2) |
| N2A—N3A | 1.308 (2) | N4B—C5B | 1.333 (3) |
| N2A—C6A | 1.476 (3) | C5B—H5B | 0.9300 |
| N3A—N4A | 1.326 (2) | C6B—C7B | 1.490 (4) |
| N4A—C5A | 1.331 (2) | C6B—H61B | 0.9700 |
| C5A—H5A | 0.9300 | C6B—H62B | 0.9700 |
| C6A—C7A | 1.477 (4) | C7B—H71B | 0.9600 |
| C6A—H61A | 0.9700 | C7B—H72B | 0.9600 |
| C6A—H62A | 0.9700 | C7B—H73B | 0.9600 |
| | | |
| N4A—Cu1—N4B | 175.10 (6) | H61A—C6A—H62A | 107.9 |
| N4A—Cu1—Cl2 | 89.22 (5) | C6A—C7A—H71A | 109.5 |
| N4B—Cu1—Cl2 | 88.29 (5) | C6A—C7A—H72A | 109.5 |
| N4A—Cu1—Cl1 | 92.33 (5) | H71A—C7A—H72A | 109.5 |
| N4B—Cu1—Cl1 | 90.55 (5) | C6A—C7A—H73A | 109.5 |
| Cl2—Cu1—Cl1 | 174.32 (2) | H71A—C7A—H73A | 109.5 |
| N4A—Cu1—N1Ai | 88.63 (7) | H72A—C7A—H73A | 109.5 |
| N4B—Cu1—N1Ai | 95.40 (7) | C5B—N1B—N2B | 101.72 (17) |
| Cl2—Cu1—N1Ai | 86.01 (5) | N3B—N2B—N1B | 114.20 (16) |
| Cl1—Cu1—N1Ai | 88.56 (5) | N3B—N2B—C6B | 122.44 (18) |
| N4A—Cu1—Cl1ii | 87.26 (5) | N1B—N2B—C6B | 123.36 (18) |
| N4B—Cu1—Cl1ii | 88.74 (5) | N2B—N3B—N4B | 104.94 (16) |
| Cl2—Cu1—Cl1ii | 94.86 (3) | N3B—N4B—C5B | 107.21 (16) |
| Cl1—Cu1—Cl1ii | 90.67 (2) | N3B—N4B—Cu1 | 126.48 (13) |
| N1Ai—Cu1—Cl1ii | 175.79 (4) | C5B—N4B—Cu1 | 126.27 (14) |
| N2A—N1A—C5A | 101.65 (16) | N1B—C5B—N4B | 111.91 (19) |
| N3A—N2A—N1A | 114.45 (16) | N1B—C5B—H5B | 124.0 |
| N3A—N2A—C6A | 123.30 (17) | N4B—C5B—H5B | 124.0 |
| N1A—N2A—C6A | 122.18 (17) | N2B—C6B—C7B | 111.3 (2) |
| N2A—N3A—N4A | 104.94 (15) | N2B—C6B—H61B | 109.4 |
| N3A—N4A—C5A | 107.31 (15) | C7B—C6B—H61B | 109.4 |
| N3A—N4A—Cu1 | 125.96 (12) | N2B—C6B—H62B | 109.4 |
| C5A—N4A—Cu1 | 126.68 (13) | C7B—C6B—H62B | 109.4 |
| N1A—C5A—N4A | 111.65 (18) | H61B—C6B—H62B | 108.0 |
| N1A—C5A—H5A | 124.2 | C6B—C7B—H71B | 109.5 |
| N4A—C5A—H5A | 124.2 | C6B—C7B—H72B | 109.5 |
| N2A—C6A—C7A | 112.28 (19) | H71B—C7B—H72B | 109.5 |
| N2A—C6A—H61A | 109.1 | C6B—C7B—H73B | 109.5 |
| C7A—C6A—H61A | 109.1 | H71B—C7B—H73B | 109.5 |
| N2A—C6A—H62A | 109.1 | H72B—C7B—H73B | 109.5 |
| C7A—C6A—H62A | 109.1 | | |
| | | |
| C5A—N1A—N2A—N3A | −0.5 (3) | C5B—N1B—N2B—N3B | 1.0 (3) |
| C5A—N1A—N2A—C6A | −177.6 (2) | C5B—N1B—N2B—C6B | −179.7 (2) |
| N1A—N2A—N3A—N4A | 0.4 (2) | N1B—N2B—N3B—N4B | −0.7 (2) |
| C6A—N2A—N3A—N4A | 177.4 (2) | C6B—N2B—N3B—N4B | 180.0 (2) |
| N2A—N3A—N4A—C5A | −0.2 (2) | N2B—N3B—N4B—C5B | 0.1 (2) |
| N2A—N3A—N4A—Cu1 | −177.60 (13) | N2B—N3B—N4B—Cu1 | −177.83 (13) |
| Cl2—Cu1—N4A—N3A | −134.61 (17) | Cl2—Cu1—N4B—N3B | 118.58 (17) |
| Cl1—Cu1—N4A—N3A | 39.92 (17) | Cl1—Cu1—N4B—N3B | −55.86 (17) |
| N1Ai—Cu1—N4A—N3A | −48.58 (17) | N1Ai—Cu1—N4B—N3B | 32.75 (18) |
| Cl1ii—Cu1—N4A—N3A | 130.48 (17) | Cl1ii—Cu1—N4B—N3B | −146.51 (17) |
| Cl2—Cu1—N4A—C5A | 48.43 (19) | Cl2—Cu1—N4B—C5B | −59.00 (19) |
| Cl1—Cu1—N4A—C5A | −137.04 (18) | Cl1—Cu1—N4B—C5B | 126.56 (19) |
| N1Ai—Cu1—N4A—C5A | 134.46 (19) | N1Ai—Cu1—N4B—C5B | −144.83 (19) |
| Cl1ii—Cu1—N4A—C5A | −46.48 (18) | Cl1ii—Cu1—N4B—C5B | 35.90 (19) |
| N2A—N1A—C5A—N4A | 0.4 (3) | N2B—N1B—C5B—N4B | −0.9 (3) |
| N3A—N4A—C5A—N1A | −0.2 (3) | N3B—N4B—C5B—N1B | 0.5 (3) |
| Cu1—N4A—C5A—N1A | 177.27 (16) | Cu1—N4B—C5B—N1B | 178.47 (16) |
| N3A—N2A—C6A—C7A | 2.0 (4) | N3B—N2B—C6B—C7B | −140.3 (2) |
| N1A—N2A—C6A—C7A | 178.8 (2) | N1B—N2B—C6B—C7B | 40.5 (3) |
| Symmetry codes: (i) x+1/2, −y+1/2, z+1/2; (ii) −x+1, −y, −z. |
Experimental details
| Crystal data |
| Chemical formula | [CuCl2(C6H12N8)] |
| Mr | 330.68 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 293 |
| a, b, c (Å) | 10.234 (2), 13.690 (4), 10.478 (2) |
| β (°) | 119.174 (15) |
| V (Å3) | 1281.9 (5) |
| Z | 4 |
| Radiation type | Mo Kα |
| µ (mm−1) | 2.11 |
| Crystal size (mm) | 0.60 × 0.60 × 0.52 |
| |
| Data collection |
| Diffractometer | Nicolet R3m four-circle
diffractometer |
| Absorption correction | ψ scan
(North et al., 1968) |
| Tmin, Tmax | 0.298, 0.333 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 4079, 3763, 3444 |
| Rint | 0.019 |
| (sin θ/λ)max (Å−1) | 0.705 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.034, 0.098, 1.09 |
| No. of reflections | 3763 |
| No. of parameters | 156 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.57, −0.59 |
Selected bond lengths (Å) top| Cu1—N4A | 2.0037 (16) | N2A—N3A | 1.308 (2) |
| Cu1—N4B | 2.0136 (16) | N3A—N4A | 1.326 (2) |
| Cu1—Cl2 | 2.2595 (6) | N4A—C5A | 1.331 (2) |
| Cu1—Cl1 | 2.2796 (6) | N1B—C5B | 1.319 (3) |
| Cu1—N1Ai | 2.851 (2) | N1B—N2B | 1.325 (3) |
| Cu1—Cl1ii | 2.8845 (8) | N2B—N3B | 1.313 (2) |
| N1A—N2A | 1.328 (2) | N3B—N4B | 1.317 (2) |
| N1A—C5A | 1.328 (3) | N4B—C5B | 1.333 (3) |
| Symmetry codes: (i) x+1/2, −y+1/2, z+1/2; (ii) −x+1, −y, −z. |

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.
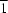 ) plane via the 2-ethyltetrazole ligands.
) plane via the 2-ethyltetrazole ligands.
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2003/06/00/fr1419/fr1419fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2003/06/00/fr1419/fr1419fig2thm.gif)

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry



This work is a part of a project dealing with the synthesis, structures and properties of complexes of copper(II) chloride with 2-monosubstituted tetrazoles. To date, only two such complexes have been structurally characterized, namely CuCl2L, where L is 2-tertbutyltetrazole (Lyakhov, Gaponik, Degtyarik & Ivashkevich, 2003), and Cu3Cl6L4, where L is 2-allyltetrazole (Lyakhov, Gaponik, Degtyarik, Matulis, et al., 2003). Only one complex, [NiL6](BF4)2, where L is 2-methyltetrazole (van den Heuvel et al., 1983), exists in the Cambridge Structural Database (Version 5.24 of November 2002; Allen, 2002) with respect to 2-substituted tetrazole complexes. A new complex of copper(II) chloride, CuCl2L2, where L is 2-ethyltetrazole, (I) (Fig. 1), has been synthesized and investigated in the present work. There are two 2-ethyltetrazole ligands in the asymmetric unit of (I), and these are denoted by A and B.
The geometric parameters of the tetrazole rings of the two 2-ethyltetrazole molecules in (I) are very similar (Table 1). Bond lengths and angles are comparable to those obtained earlier for copper(II) chloride complexes with 2-tertbutyltetrazole (Lyakhov, Gaponik, Degtyarik & Ivashkevich, 2003) and 2-allyltetrazole (Lyakhov, Gaponik, Degtyarik, Matulis et al., 2003), and also for nickel(II) tetrafluoroborate with 2-methyltetrazole (van den Heuvel et al., 1983).
The tetrazole rings in the 2-ethyltetrazole molecules are planar to within 0.002 (3) and 0.003 (3) Å for ligands A and B, respectively.
The Cu atom exhibits axially elongated octahedral coordination, with two 2-ethyltetrazole ligands [Cu1—N4A=2.0037 (16) and Cu1—N4B=2.0136 (16) Å] and two Cl atoms [Cu1—Cl1=2.2796 (6) and Cu1—Cl2=2.2595 (6) Å] in the equatorial positions. Atom Cl1 and a symmetry-related 2-ethyltetrazole molecule lie in the axial positions of the octahedron [Cu1—Cl1ii=2.8845 (8) and Cu1—N1Ai=2.851 (2) Å; symmetry codes: (i) 1/2 + x, 1/2 − y, 1/2 + z; (ii) 1 − x, −y, −z]. Ligand A thus exhibits bidentate binding with the two Cu atoms through atoms N4 and N1 of the tetrazole ring, whereas ligand B is coordinated in a monodentate fashion via atom N4.
In the crystal structure of (I), the Cu-atom octahedra are associated with dimer entities, by sharing edges with symmetry-related Cl1 atoms (Fig. 2). The separation between the Cu atoms in the dimer is 3.6557 (10) Å. The dimer entities are linked by ligand A to form one-dimensional polymeric zigzag chains extended along the b axis. Via ligand A, the chains are connected to infinite layers parallel to the (1 0 1) plane. Only van der Waals interactions exist between the sheets. Ligand B and atom Cl2 do not participate in the formation of the polymeric structure of (I).
Recently, we discussed an example? of bidentate bridge coordination of the 2-allyltetrazole ligand via atoms N3 and N4 of the tetrazole ring in the complex Cu3Cl6L4 (Lyakhov, Gaponik, Degtyarik, Matulis, et al., 2003). The structural data obtained for (I) revealed bidentate bridge coordination of the 2-ethyltetrazole ligand A through atoms N4 and N1. By comparison with structures in the Cambridge Structural Database (Version 5.24, November 2002 release; Allen, 2002) that contain 1-monosubstituted tetrazoles having only N4-coordination, it can be concluded that 2-monosubstituted tetrazoles exhibit more multiform coordination. Further results on complexes of 2-monosubstituted tetrazoles will show how often bidentate coordination takes place for these ligands.
The bidentate bridge coordination of 2-substituted tetrazoles is probably responsible for the difference in the crystal structures of CuII chloride complexes with 1-ethyltetrazole (Virovets et al., 1995) and 2-ethyltetrazole. These compounds are of the same composition; both have octahedrally coordinated Cu atoms and layered polymeric structures. However, whereas all the Cl atoms (and only these atoms) participate in polymeric structure formation in the complex with 1-ethyltetrazole, only one Cl atom, Cl1, plays a bridging role in the polymeric structure of (I), but bidentate bridge coordination of ligand A makes it possible to form a two-dimensional polymeric network.