The title compound, [Cu(C
9H
8BrClNO
2)
2], is a square-planar complex. The potentially tridentate dibasic 2-bromo-4-chloro-6-{[(2-hydroxyethyl)imino]methyl}phenolate ligand coordinates in a
trans-bis fashion to the Cu
II centre
via the imine N and phenolate O atoms. The Cu
II atom lies on the centre of inversion of the molecule. The potentially coordinating hydroxyethyl group remains protonated and uncoordinated, taking part in intermolecular hydrogen bonds with vicinal groups, leading to the formation of a two-dimensional hydrogen-bond network with sheets parallel to the (10
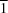
) plane. Substituent effects on the crystal packing and coordination modes of the ligand are discussed.
Supporting information
CCDC reference: 1005486
Data collection: CrysAlis PRO (Agilent, 2010); cell refinement: CrysAlis PRO (Agilent, 2010); data reduction: CrysAlis PRO (Agilent, 2010); program(s) used to solve structure: SIR97 (Altomare et al., 1999); program(s) used to refine structure: SHELXL2012 (Sheldrick, 2008); molecular graphics: Mercury (Macrae et al., 2006); software used to prepare material for publication: WinGX (Farrugia, 2012).
Bis(2-bromo-4-chloro-6-{[(2-hydroxyethyl)imino]methyl}phenolato-
κ2N,
O)copper(II)
top
Crystal data top
| [Cu(C9H8BrClNO2)2] | F(000) = 606 |
| Mr = 618.59 | Dx = 1.985 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.5418 Å |
| Hall symbol: -P 2yn | Cell parameters from 6191 reflections |
| a = 13.1298 (6) Å | θ = 4.2–70.7° |
| b = 4.6502 (1) Å | µ = 8.64 mm−1 |
| c = 16.957 (2) Å | T = 293 K |
| β = 90.977 (9)° | Needle, green |
| V = 1035.20 (13) Å3 | 0.45 × 0.09 × 0.05 mm |
| Z = 2 | |
Data collection top
Agilent SuperNova (Dual, Cu at zero, Atlas)
diffractometer | 1998 independent reflections |
| Mirror monochromator | 1774 reflections with I > 2σ(I) |
| Detector resolution: 10.3756 pixels mm-1 | Rint = 0.046 |
| ω scans | θmax = 71.1°, θmin = 4.2° |
Absorption correction: multi-scan
(CrysAlis PRO; Agilent, 2010) | h = −15→15 |
| Tmin = 0.266, Tmax = 1 | k = −5→5 |
| 12608 measured reflections | l = −20→20 |
Refinement top
| Refinement on F2 | 1 restraint |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.029 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.076 | w = 1/[σ2(Fo2) + (0.0412P)2 + 0.3308P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.11 | (Δ/σ)max = 0.001 |
| 1998 reflections | Δρmax = 0.30 e Å−3 |
| 137 parameters | Δρmin = −0.59 e Å−3 |
Special details top
Experimental. Absorption correction: CrysAlisPro, Version 1.171.36.24
(Agilent Technologies, 2010)
Empirical absorption correction using spherical harmonics,
implemented in SCALE3 ABSPACK scaling algorithm. |
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Cu1 | 0 | 0.5 | 0.5 | 0.03380 (14) | |
| Cl1 | 0.51349 (5) | 0.0906 (2) | 0.38758 (5) | 0.0743 (3) | |
| Br1 | 0.11786 (2) | −0.17824 (8) | 0.30816 (2) | 0.05847 (13) | |
| N1 | 0.12873 (15) | 0.6497 (4) | 0.55269 (11) | 0.0358 (4) | |
| C2 | 0.21922 (18) | 0.5765 (6) | 0.53272 (14) | 0.0390 (5) | |
| H2 | 0.2725 | 0.6697 | 0.559 | 0.047* | |
| C3 | 0.24752 (18) | 0.3676 (6) | 0.47457 (14) | 0.0394 (5) | |
| C4 | 0.17325 (18) | 0.2105 (5) | 0.43017 (13) | 0.0367 (5) | |
| C5 | 0.21223 (19) | 0.0212 (5) | 0.37315 (14) | 0.0393 (5) | |
| C6 | 0.3142 (2) | −0.0192 (6) | 0.36112 (15) | 0.0451 (6) | |
| H6 | 0.3363 | −0.1492 | 0.3234 | 0.054* | |
| C7 | 0.38386 (19) | 0.1368 (7) | 0.40601 (16) | 0.0477 (6) | |
| C8 | 0.35205 (19) | 0.3276 (6) | 0.46146 (15) | 0.0461 (6) | |
| H8 | 0.3998 | 0.4319 | 0.4908 | 0.055* | |
| O9 | 0.07575 (13) | 0.2348 (4) | 0.44007 (10) | 0.0437 (4) | |
| C10 | 0.12363 (19) | 0.8628 (5) | 0.61661 (14) | 0.0391 (5) | |
| H10A | 0.0652 | 0.9871 | 0.6077 | 0.047* | |
| H10B | 0.1844 | 0.9813 | 0.6164 | 0.047* | |
| C11 | 0.1148 (2) | 0.7177 (6) | 0.69618 (15) | 0.0446 (6) | |
| H11A | 0.0952 | 0.8595 | 0.7351 | 0.053* | |
| H11B | 0.0615 | 0.5733 | 0.6932 | 0.053* | |
| O12 | 0.20696 (17) | 0.5858 (5) | 0.72096 (13) | 0.0584 (5) | |
| H12 | 0.247 (3) | 0.715 (7) | 0.733 (2) | 0.072 (12)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cu1 | 0.0316 (2) | 0.0383 (3) | 0.0313 (2) | 0.00047 (18) | −0.00502 (17) | −0.00030 (19) |
| Cl1 | 0.0372 (3) | 0.1123 (7) | 0.0734 (5) | 0.0143 (4) | 0.0035 (3) | −0.0158 (5) |
| Br1 | 0.0540 (2) | 0.0644 (2) | 0.0569 (2) | −0.00257 (13) | −0.00077 (14) | −0.02250 (14) |
| N1 | 0.0371 (10) | 0.0360 (11) | 0.0341 (9) | −0.0002 (8) | −0.0061 (8) | 0.0005 (8) |
| C2 | 0.0340 (11) | 0.0430 (13) | 0.0396 (12) | −0.0047 (10) | −0.0072 (9) | 0.0034 (10) |
| C3 | 0.0368 (12) | 0.0436 (14) | 0.0377 (11) | 0.0020 (10) | −0.0045 (9) | 0.0038 (10) |
| C4 | 0.0361 (11) | 0.0398 (13) | 0.0341 (11) | 0.0006 (9) | −0.0016 (9) | 0.0047 (10) |
| C5 | 0.0441 (13) | 0.0395 (13) | 0.0341 (11) | −0.0005 (10) | −0.0010 (10) | 0.0019 (10) |
| C6 | 0.0492 (14) | 0.0491 (15) | 0.0371 (12) | 0.0083 (12) | 0.0038 (10) | 0.0049 (11) |
| C7 | 0.0350 (12) | 0.0610 (17) | 0.0473 (14) | 0.0095 (11) | 0.0015 (10) | 0.0077 (12) |
| C8 | 0.0378 (13) | 0.0559 (17) | 0.0444 (13) | 0.0005 (11) | −0.0049 (10) | 0.0006 (12) |
| O9 | 0.0337 (9) | 0.0526 (10) | 0.0448 (9) | −0.0005 (7) | −0.0011 (7) | −0.0110 (8) |
| C10 | 0.0379 (12) | 0.0377 (13) | 0.0413 (12) | 0.0001 (9) | −0.0092 (10) | −0.0029 (10) |
| C11 | 0.0466 (14) | 0.0478 (15) | 0.0390 (12) | −0.0039 (11) | −0.0079 (10) | −0.0018 (11) |
| O12 | 0.0664 (13) | 0.0446 (11) | 0.0631 (12) | 0.0024 (10) | −0.0310 (10) | 0.0025 (10) |
Geometric parameters (Å, º) top
| Cu1—N1 | 2.0219 (19) | C4—C5 | 1.410 (3) |
| Cu1—O9 | 1.8913 (18) | C5—C6 | 1.371 (4) |
| Cu1—O9i | 1.8913 (18) | C6—C7 | 1.385 (4) |
| Cu1—N1i | 2.0219 (19) | C6—H6 | 0.93 |
| Cl1—C7 | 1.749 (3) | C7—C8 | 1.363 (4) |
| Br1—C5 | 1.888 (2) | C8—H8 | 0.93 |
| N1—C2 | 1.287 (3) | C10—C11 | 1.515 (3) |
| N1—C10 | 1.471 (3) | C10—H10A | 0.97 |
| C2—C3 | 1.438 (4) | C10—H10B | 0.97 |
| C2—H2 | 0.93 | C11—O12 | 1.414 (3) |
| C3—C8 | 1.406 (4) | C11—H11A | 0.97 |
| C3—C4 | 1.423 (3) | C11—H11B | 0.97 |
| C4—O9 | 1.299 (3) | O12—H12 | 0.821 (19) |
| | | |
| O9—Cu1—O9i | 180 | C5—C6—H6 | 120.5 |
| O9—Cu1—N1i | 88.93 (8) | C7—C6—H6 | 120.5 |
| O9i—Cu1—N1i | 91.07 (8) | C8—C7—C6 | 120.9 (2) |
| O9—Cu1—N1 | 91.07 (8) | C8—C7—Cl1 | 120.9 (2) |
| O9i—Cu1—N1 | 88.93 (8) | C6—C7—Cl1 | 118.2 (2) |
| N1i—Cu1—N1 | 180 | C7—C8—C3 | 120.4 (2) |
| C2—N1—C10 | 115.2 (2) | C7—C8—H8 | 119.8 |
| C2—N1—Cu1 | 124.15 (17) | C3—C8—H8 | 119.8 |
| C10—N1—Cu1 | 120.62 (15) | C4—O9—Cu1 | 130.89 (17) |
| N1—C2—C3 | 127.6 (2) | N1—C10—C11 | 111.2 (2) |
| N1—C2—H2 | 116.2 | N1—C10—H10A | 109.4 |
| C3—C2—H2 | 116.2 | C11—C10—H10A | 109.4 |
| C8—C3—C4 | 120.7 (2) | N1—C10—H10B | 109.4 |
| C8—C3—C2 | 117.5 (2) | C11—C10—H10B | 109.4 |
| C4—C3—C2 | 121.8 (2) | H10A—C10—H10B | 108 |
| O9—C4—C5 | 120.9 (2) | O12—C11—C10 | 112.3 (2) |
| O9—C4—C3 | 123.7 (2) | O12—C11—H11A | 109.1 |
| C5—C4—C3 | 115.4 (2) | C10—C11—H11A | 109.1 |
| C6—C5—C4 | 123.7 (2) | O12—C11—H11B | 109.1 |
| C6—C5—Br1 | 118.62 (19) | C10—C11—H11B | 109.1 |
| C4—C5—Br1 | 117.70 (18) | H11A—C11—H11B | 107.9 |
| C5—C6—C7 | 118.9 (2) | C11—O12—H12 | 107 (3) |
| | | |
| C10—N1—C2—C3 | 177.7 (2) | C5—C6—C7—C8 | 0.1 (4) |
| Cu1—N1—C2—C3 | −4.2 (4) | C5—C6—C7—Cl1 | −178.0 (2) |
| N1—C2—C3—C8 | 178.7 (2) | C6—C7—C8—C3 | 0.6 (4) |
| N1—C2—C3—C4 | 0.1 (4) | Cl1—C7—C8—C3 | 178.6 (2) |
| C8—C3—C4—O9 | 179.1 (2) | C4—C3—C8—C7 | −0.2 (4) |
| C2—C3—C4—O9 | −2.4 (4) | C2—C3—C8—C7 | −178.7 (2) |
| C8—C3—C4—C5 | −0.8 (3) | C5—C4—O9—Cu1 | −170.48 (17) |
| C2—C3—C4—C5 | 177.6 (2) | C3—C4—O9—Cu1 | 9.6 (4) |
| O9—C4—C5—C6 | −178.4 (2) | N1i—Cu1—O9—C4 | 169.6 (2) |
| C3—C4—C5—C6 | 1.5 (4) | N1—Cu1—O9—C4 | −10.4 (2) |
| O9—C4—C5—Br1 | 3.5 (3) | C2—N1—C10—C11 | −93.3 (3) |
| C3—C4—C5—Br1 | −176.59 (17) | Cu1—N1—C10—C11 | 88.6 (2) |
| C4—C5—C6—C7 | −1.2 (4) | N1—C10—C11—O12 | 72.5 (3) |
| Br1—C5—C6—C7 | 176.9 (2) | | |
| Symmetry code: (i) −x, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O12—H12···O12ii | 0.821 (19) | 1.98 (2) | 2.760 (2) | 158 (4) |
| Symmetry code: (ii) −x+1/2, y+1/2, −z+3/2. |

 journal menu
journal menu











