Buy article online - an online subscription or single-article purchase is required to access this article.
We have isolated and crystallographically characterized the three homologous compounds
N,
N′-bis(2-methoxybenzylidene)ethane-1,2-diamine (MeSalen), C
18H
20N
2O
2,
N,
N′-bis(2-methoxybenzylidene)propane-1,3-diamine (MeSalpr), C
19H
22N
2O
2, and
N,
N′-bis(2-methoxybenzylidene)butane-1,4-diamine (MeSalbu), C
20H
24N
2O
2. In contrast with MeSalpr, the molecules of MeSalen and MeSalbu, which have an even number of methylene units, have crystallographic
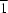
symmetry. Comparing these methoxy-substituted species with their hydroxy equivalents shows that the aryl rings rotate upon removal of the O—H

N hydrogen bonds. The packing of MeSalen and MeSalpr is controlled by C—H

π interactions, whereas that of MeSalbu has only van der Waals contacts.
Supporting information
CCDC references: 235331; 235332; 235333
MeSalen (Billman et al., 1963) and MeSalpr (Moneta et al., 1988) were prepared as previously reported and crystallized from chloroform: diethyl ether (Ratio?) by vapour diffusion. MeSalbu was prepared by refluxing 1,4-diaminobutane with two equivalents of ortho-anisaldehyde in methanol for 4 h. The solvent was removed, giving a yellow oil which was dissolved in a small amount of diethyl ether. Storing the solution in the freezer (258 K) overnight produced pale-yellow crystals which were recrystallized from chloroform:diethyl ether (Ratio?) by vapour diffussion. Analysis for MeSalbu: found: C 74.06, H 7.37, N 8.39%; expected for C20H24N2O2: C 74.05, H 7.46, N 8.64%; 1H NMR (270 MHz, CDCl3, δ, p.p.m.): 8.72 (s, 2H, C═N), 7.95 (d, 2H, arom), 7.39 (t, 2H, arom), 7.06 (t, 2H, arom), 6.91 (d, 2H, arom), 3.87 (s, 6H, OCH3), 3.67 (t, 4H, N—CH2–), 2.12 (p, 4H, –CH2—CH2–); 13C NMR (270 MHz, CDCl3, δ, p.p.m.): 159, 157, 132, 128, 125, 121 (arom), 111 (C═N), 62 (MeO–), 56 (N—CH2–), 29 (–CH2—CH2–); m.p. 332.5–334 K.
All H atoms were clearly revealed in difference maps and were then allowed for as riding atoms with idealized geometry in the final refinement cycles. C—H distances were in the range 0.93–0.99 Å, and Uiso(H) values were set to 1.2Ueq(C) for CH and CH2 groups, and to 1.5Ueq(C) for CH3 groups.
For all compounds, data collection: DENZO (Otwinowski & Minor, 1997) and COLLECT (Nonius, 1998); cell refinement: DENZO and COLLECT; data reduction: DENZO; program(s) used to solve structure: SHELXS97 (Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997) and WinGX (Farrugia, 1997); molecular graphics: ORTEPII (Johnson, 1976) and PLATON (Spek 2003); software used to prepare material for publication: SHELXL97.
(I)
N,
N-bis(2-methoxybenzylidene)ethane-1,2-diamine
top
Crystal data top
| C18H20N2O2 | F(000) = 316 |
| Mr = 296.36 | Dx = 1.229 Mg m−3 |
| Monoclinic, P21/n | Melting point: 115 K |
| Hall symbol: -P 2yn | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.8727 (2) Å | Cell parameters from 1815 reflections |
| b = 8.9846 (2) Å | θ = 3.5–27.5° |
| c = 11.3927 (3) Å | µ = 0.08 mm−1 |
| β = 96.345 (1)° | T = 123 K |
| V = 800.90 (3) Å3 | Prism, colourless |
| Z = 2 | 0.5 × 0.5 × 0.4 mm |
Data collection top
Nonius Kappa CCD area-detector
diffractometer | 1627 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.037 |
| Graphite monochromator | θmax = 27.5°, θmin = 3.5° |
| ϕ and ω scans | h = −10→10 |
| 9458 measured reflections | k = −11→10 |
| 1815 independent reflections | l = −14→14 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.036 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.098 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0465P)2 + 0.2339P]
where P = (Fo2 + 2Fc2)/3 |
| 1815 reflections | (Δ/σ)max = 0.001 |
| 101 parameters | Δρmax = 0.26 e Å−3 |
| 0 restraints | Δρmin = −0.17 e Å−3 |
Crystal data top
| C18H20N2O2 | V = 800.90 (3) Å3 |
| Mr = 296.36 | Z = 2 |
| Monoclinic, P21/n | Mo Kα radiation |
| a = 7.8727 (2) Å | µ = 0.08 mm−1 |
| b = 8.9846 (2) Å | T = 123 K |
| c = 11.3927 (3) Å | 0.5 × 0.5 × 0.4 mm |
| β = 96.345 (1)° | |
Data collection top
Nonius Kappa CCD area-detector
diffractometer | 1627 reflections with I > 2σ(I) |
| 9458 measured reflections | Rint = 0.037 |
| 1815 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.036 | 0 restraints |
| wR(F2) = 0.098 | H-atom parameters constrained |
| S = 1.02 | Δρmax = 0.26 e Å−3 |
| 1815 reflections | Δρmin = −0.17 e Å−3 |
| 101 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| O1 | 0.19421 (9) | 1.01402 (8) | 0.40164 (6) | 0.0244 (2) | |
| N1 | 0.54895 (11) | 0.68924 (9) | 0.44732 (7) | 0.0225 (2) | |
| C1 | 0.52413 (13) | 0.57243 (10) | 0.53260 (9) | 0.0239 (2) | |
| H1A | 0.4323 | 0.6017 | 0.5808 | 0.029* | |
| H1B | 0.6307 | 0.5581 | 0.5862 | 0.029* | |
| C11 | 0.46301 (12) | 0.92395 (10) | 0.36019 (8) | 0.0189 (2) | |
| C12 | 0.33329 (12) | 1.03201 (11) | 0.34116 (8) | 0.0189 (2) | |
| C13 | 0.34965 (13) | 1.14917 (11) | 0.26279 (8) | 0.0220 (2) | |
| H13 | 0.2627 | 1.2227 | 0.2508 | 0.026* | |
| C14 | 0.49296 (13) | 1.15812 (12) | 0.20244 (9) | 0.0256 (2) | |
| H14 | 0.5030 | 1.2377 | 0.1487 | 0.031* | |
| C15 | 0.62209 (13) | 1.05223 (12) | 0.21951 (9) | 0.0271 (2) | |
| H15 | 0.7199 | 1.0589 | 0.1778 | 0.033* | |
| C16 | 0.60611 (13) | 0.93660 (11) | 0.29828 (9) | 0.0234 (2) | |
| H16 | 0.6944 | 0.8644 | 0.3104 | 0.028* | |
| C17 | 0.44935 (12) | 0.80061 (10) | 0.44419 (8) | 0.0193 (2) | |
| H17 | 0.3642 | 0.8047 | 0.4970 | 0.023* | |
| C18 | 0.06398 (14) | 1.12653 (13) | 0.38476 (11) | 0.0319 (3) | |
| H18A | 0.1122 | 1.2228 | 0.4116 | 0.048* | |
| H18B | −0.0303 | 1.1007 | 0.4304 | 0.048* | |
| H18C | 0.0213 | 1.1327 | 0.3008 | 0.048* | |
| Cg1 | 0.47785 | 1.04201 | 0.28073 | 0.01* | 0.0 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0212 (4) | 0.0244 (4) | 0.0284 (4) | 0.0041 (3) | 0.0063 (3) | 0.0062 (3) |
| N1 | 0.0270 (4) | 0.0162 (4) | 0.0244 (4) | 0.0000 (3) | 0.0029 (3) | 0.0011 (3) |
| C1 | 0.0316 (5) | 0.0168 (5) | 0.0231 (5) | 0.0007 (4) | 0.0028 (4) | 0.0021 (4) |
| C11 | 0.0207 (5) | 0.0161 (5) | 0.0194 (4) | −0.0019 (3) | 0.0000 (3) | −0.0016 (3) |
| C12 | 0.0192 (4) | 0.0187 (5) | 0.0185 (4) | −0.0022 (3) | 0.0003 (3) | −0.0016 (3) |
| C13 | 0.0235 (5) | 0.0193 (5) | 0.0223 (5) | 0.0006 (4) | −0.0015 (4) | 0.0016 (4) |
| C14 | 0.0296 (5) | 0.0223 (5) | 0.0246 (5) | −0.0039 (4) | 0.0025 (4) | 0.0053 (4) |
| C15 | 0.0263 (5) | 0.0270 (5) | 0.0293 (5) | −0.0028 (4) | 0.0092 (4) | 0.0022 (4) |
| C16 | 0.0228 (5) | 0.0193 (5) | 0.0283 (5) | 0.0005 (4) | 0.0037 (4) | −0.0005 (4) |
| C17 | 0.0214 (4) | 0.0173 (4) | 0.0189 (4) | −0.0019 (4) | 0.0011 (3) | −0.0019 (3) |
| C18 | 0.0250 (5) | 0.0333 (6) | 0.0387 (6) | 0.0097 (4) | 0.0088 (4) | 0.0093 (5) |
Geometric parameters (Å, º) top
| O1—C12 | 1.3663 (11) | C13—C14 | 1.3869 (14) |
| O1—C18 | 1.4375 (12) | C13—H13 | 0.95 |
| N1—C17 | 1.2693 (13) | C14—C15 | 1.3903 (15) |
| N1—C1 | 1.4579 (12) | C14—H14 | 0.95 |
| C1—C1i | 1.5256 (19) | C15—C16 | 1.3876 (14) |
| C1—H1A | 0.99 | C15—H15 | 0.95 |
| C1—H1B | 0.99 | C16—H16 | 0.95 |
| C11—C16 | 1.3984 (14) | C17—H17 | 0.95 |
| C11—C12 | 1.4084 (13) | C18—H18A | 0.98 |
| C11—C17 | 1.4759 (13) | C18—H18B | 0.98 |
| C12—C13 | 1.3954 (13) | C18—H18C | 0.98 |
| | | |
| C12—O1—C18 | 116.67 (8) | C13—C14—H14 | 119.6 |
| C17—N1—C1 | 117.28 (8) | C15—C14—H14 | 119.6 |
| N1—C1—C1i | 109.52 (10) | C16—C15—C14 | 119.06 (9) |
| N1—C1—H1A | 109.8 | C16—C15—H15 | 120.5 |
| C1i—C1—H1A | 109.8 | C14—C15—H15 | 120.5 |
| N1—C1—H1B | 109.8 | C15—C16—C11 | 121.52 (9) |
| C1i—C1—H1B | 109.8 | C15—C16—H16 | 119.2 |
| H1A—C1—H1B | 108.2 | C11—C16—H16 | 119.2 |
| C16—C11—C12 | 118.50 (9) | N1—C17—C11 | 121.40 (9) |
| C16—C11—C17 | 120.57 (9) | N1—C17—H17 | 119.3 |
| C12—C11—C17 | 120.93 (8) | C11—C17—H17 | 119.3 |
| O1—C12—C13 | 123.21 (9) | O1—C18—H18A | 109.5 |
| O1—C12—C11 | 116.66 (8) | O1—C18—H18B | 109.5 |
| C13—C12—C11 | 120.13 (9) | H18A—C18—H18B | 109.5 |
| C14—C13—C12 | 119.91 (9) | O1—C18—H18C | 109.5 |
| C14—C13—H13 | 120.0 | H18A—C18—H18C | 109.5 |
| C12—C13—H13 | 120.0 | H18B—C18—H18C | 109.5 |
| C13—C14—C15 | 120.88 (9) | | |
| | | |
| C17—N1—C1—C1i | 117.42 (11) | C12—C13—C14—C15 | −0.50 (15) |
| C18—O1—C12—C13 | −2.37 (14) | C13—C14—C15—C16 | −0.07 (16) |
| C18—O1—C12—C11 | 178.32 (9) | C14—C15—C16—C11 | 0.33 (16) |
| C16—C11—C12—O1 | 178.79 (8) | C12—C11—C16—C15 | −0.02 (15) |
| C17—C11—C12—O1 | −1.68 (13) | C17—C11—C16—C15 | −179.55 (9) |
| C16—C11—C12—C13 | −0.55 (14) | C1—N1—C17—C11 | −179.39 (8) |
| C17—C11—C12—C13 | 178.98 (8) | C16—C11—C17—N1 | −12.87 (14) |
| O1—C12—C13—C14 | −178.48 (9) | C12—C11—C17—N1 | 167.61 (9) |
| C11—C12—C13—C14 | 0.81 (14) | | |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C17—H17···Cg1ii | 0.95 | 3.02 | 3.428 | 107 |
| Symmetry code: (ii) −x+1, −y+2, −z+1. |
(II)
N,
N-bis(2-methoxybenzylidene)propane-1,3-diamine
top
Crystal data top
| C19H22N2O2 | Z = 2 |
| Mr = 310.39 | F(000) = 332 |
| Triclinic, P1 | Dx = 1.242 Mg m−3 |
| Hall symbol: -P 1 | Melting point: 75 K |
| a = 8.8669 (2) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 9.9139 (3) Å | Cell parameters from 3737 reflections |
| c = 10.2724 (4) Å | θ = 2.0–27.5° |
| α = 101.140 (2)° | µ = 0.08 mm−1 |
| β = 92.341 (2)° | T = 123 K |
| γ = 109.442 (2)° | Prism, yellow |
| V = 830.05 (5) Å3 | 0.4 × 0.3 × 0.2 mm |
Data collection top
Nonius Kappa CCD area-detector
diffractometer | 2279 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.063 |
| Graphite monochromator | θmax = 27.6°, θmin = 2.0° |
| ϕ and ω scans | h = −11→11 |
| 12958 measured reflections | k = −12→12 |
| 3737 independent reflections | l = −12→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.047 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.122 | H-atom parameters constrained |
| S = 0.99 | w = 1/[σ2(Fo2) + (0.0622P)2]
where P = (Fo2 + 2Fc2)/3 |
| 3737 reflections | (Δ/σ)max < 0.001 |
| 210 parameters | Δρmax = 0.24 e Å−3 |
| 0 restraints | Δρmin = −0.22 e Å−3 |
Crystal data top
| C19H22N2O2 | γ = 109.442 (2)° |
| Mr = 310.39 | V = 830.05 (5) Å3 |
| Triclinic, P1 | Z = 2 |
| a = 8.8669 (2) Å | Mo Kα radiation |
| b = 9.9139 (3) Å | µ = 0.08 mm−1 |
| c = 10.2724 (4) Å | T = 123 K |
| α = 101.140 (2)° | 0.4 × 0.3 × 0.2 mm |
| β = 92.341 (2)° | |
Data collection top
Nonius Kappa CCD area-detector
diffractometer | 2279 reflections with I > 2σ(I) |
| 12958 measured reflections | Rint = 0.063 |
| 3737 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.047 | 0 restraints |
| wR(F2) = 0.122 | H-atom parameters constrained |
| S = 0.99 | Δρmax = 0.24 e Å−3 |
| 3737 reflections | Δρmin = −0.22 e Å−3 |
| 210 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| O1 | 0.77317 (12) | 0.14374 (12) | 1.07537 (10) | 0.0282 (3) | |
| O2 | 1.05404 (13) | 0.82837 (12) | 0.48211 (10) | 0.0335 (3) | |
| N1 | 0.50559 (15) | 0.15505 (14) | 0.75501 (12) | 0.0241 (3) | |
| N2 | 0.72851 (15) | 0.43107 (14) | 0.49220 (12) | 0.0269 (3) | |
| C1 | 0.46774 (19) | 0.28801 (17) | 0.75720 (15) | 0.0250 (4) | |
| H1A | 0.5282 | 0.3644 | 0.8364 | 0.030* | |
| H1B | 0.3513 | 0.2668 | 0.7636 | 0.030* | |
| C2 | 0.51246 (18) | 0.34378 (17) | 0.63030 (15) | 0.0241 (4) | |
| H2A | 0.4581 | 0.2639 | 0.5515 | 0.029* | |
| H2B | 0.4726 | 0.4255 | 0.6274 | 0.029* | |
| C3 | 0.69278 (18) | 0.39654 (18) | 0.62193 (15) | 0.0269 (4) | |
| H3A | 0.7465 | 0.4850 | 0.6935 | 0.032* | |
| H3B | 0.7357 | 0.3195 | 0.6358 | 0.032* | |
| C11 | 0.62244 (17) | 0.01624 (17) | 0.86792 (14) | 0.0219 (4) | |
| C12 | 0.72085 (17) | 0.01728 (17) | 0.97853 (14) | 0.0228 (4) | |
| C13 | 0.76131 (19) | −0.10518 (18) | 0.98533 (15) | 0.0280 (4) | |
| H13 | 0.8282 | −0.1042 | 1.0604 | 0.034* | |
| C14 | 0.7039 (2) | −0.22879 (19) | 0.88232 (16) | 0.0316 (4) | |
| H14 | 0.7313 | −0.3126 | 0.8876 | 0.038* | |
| C15 | 0.60735 (19) | −0.23176 (18) | 0.77203 (16) | 0.0293 (4) | |
| H15 | 0.5694 | −0.3165 | 0.7014 | 0.035* | |
| C16 | 0.56651 (18) | −0.10966 (17) | 0.76566 (15) | 0.0253 (4) | |
| H16 | 0.4993 | −0.1118 | 0.6903 | 0.030* | |
| C17 | 0.57997 (17) | 0.14626 (17) | 0.85944 (15) | 0.0226 (4) | |
| H17 | 0.6093 | 0.2261 | 0.9347 | 0.027* | |
| C18 | 0.8520 (2) | 0.1415 (2) | 1.19883 (15) | 0.0340 (4) | |
| H18A | 0.7847 | 0.0593 | 1.2341 | 0.051* | |
| H18B | 0.8696 | 0.2337 | 1.2632 | 0.051* | |
| H18C | 0.9559 | 0.1300 | 1.1837 | 0.051* | |
| C21 | 0.87439 (17) | 0.59806 (17) | 0.36295 (14) | 0.0217 (4) | |
| C22 | 0.98652 (17) | 0.73724 (17) | 0.36087 (15) | 0.0235 (4) | |
| C23 | 1.02586 (18) | 0.77502 (18) | 0.24003 (15) | 0.0256 (4) | |
| H23 | 1.0991 | 0.8698 | 0.2392 | 0.031* | |
| C24 | 0.95873 (19) | 0.67495 (18) | 0.12104 (15) | 0.0289 (4) | |
| H24 | 0.9878 | 0.7007 | 0.0388 | 0.035* | |
| C25 | 0.8495 (2) | 0.53756 (19) | 0.12092 (16) | 0.0317 (4) | |
| H25 | 0.8039 | 0.4688 | 0.0391 | 0.038* | |
| C26 | 0.80752 (18) | 0.50115 (18) | 0.24110 (15) | 0.0267 (4) | |
| H26 | 0.7309 | 0.4074 | 0.2404 | 0.032* | |
| C27 | 0.83081 (17) | 0.55534 (18) | 0.48955 (15) | 0.0240 (4) | |
| H27 | 0.8805 | 0.6225 | 0.5714 | 0.029* | |
| C28 | 1.1828 (2) | 0.9626 (2) | 0.48021 (17) | 0.0428 (5) | |
| H28A | 1.2697 | 0.9397 | 0.4357 | 0.064* | |
| H28B | 1.2239 | 1.0176 | 0.5720 | 0.064* | |
| H28C | 1.1423 | 1.0218 | 0.4317 | 0.064* | |
| Cg1 | 0.66373 | −0.10698 | 0.87530 (6) | 0.01* | 0.0 |
| Cg2 | 0.91709 | 0.63733 | 0.24115 (6) | 0.01* | 0.0 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0310 (7) | 0.0254 (7) | 0.0246 (6) | 0.0049 (5) | −0.0008 (5) | 0.0068 (5) |
| O2 | 0.0337 (7) | 0.0264 (7) | 0.0281 (6) | −0.0067 (5) | 0.0009 (5) | 0.0091 (5) |
| N1 | 0.0235 (7) | 0.0216 (8) | 0.0267 (7) | 0.0039 (6) | 0.0050 (6) | 0.0108 (6) |
| N2 | 0.0262 (8) | 0.0248 (8) | 0.0313 (8) | 0.0066 (6) | 0.0068 (6) | 0.0129 (6) |
| C1 | 0.0220 (9) | 0.0224 (9) | 0.0319 (9) | 0.0071 (7) | 0.0044 (7) | 0.0099 (7) |
| C2 | 0.0228 (9) | 0.0207 (9) | 0.0287 (8) | 0.0048 (7) | 0.0024 (6) | 0.0103 (7) |
| C3 | 0.0239 (9) | 0.0258 (9) | 0.0325 (9) | 0.0054 (7) | 0.0048 (7) | 0.0156 (7) |
| C11 | 0.0190 (8) | 0.0207 (9) | 0.0257 (8) | 0.0030 (7) | 0.0076 (6) | 0.0101 (7) |
| C12 | 0.0207 (8) | 0.0234 (9) | 0.0223 (8) | 0.0024 (7) | 0.0066 (6) | 0.0086 (7) |
| C13 | 0.0263 (9) | 0.0314 (10) | 0.0285 (9) | 0.0092 (8) | 0.0044 (7) | 0.0133 (8) |
| C14 | 0.0342 (10) | 0.0259 (10) | 0.0395 (10) | 0.0135 (8) | 0.0089 (8) | 0.0118 (8) |
| C15 | 0.0315 (10) | 0.0227 (9) | 0.0307 (9) | 0.0065 (8) | 0.0062 (7) | 0.0037 (7) |
| C16 | 0.0229 (9) | 0.0250 (9) | 0.0258 (8) | 0.0034 (7) | 0.0046 (7) | 0.0085 (7) |
| C17 | 0.0197 (8) | 0.0199 (9) | 0.0242 (8) | 0.0013 (7) | 0.0051 (6) | 0.0047 (7) |
| C18 | 0.0341 (10) | 0.0366 (11) | 0.0264 (9) | 0.0053 (8) | −0.0045 (7) | 0.0097 (8) |
| C21 | 0.0164 (8) | 0.0227 (9) | 0.0283 (9) | 0.0072 (7) | 0.0049 (6) | 0.0097 (7) |
| C22 | 0.0176 (8) | 0.0241 (9) | 0.0284 (9) | 0.0053 (7) | 0.0013 (6) | 0.0085 (7) |
| C23 | 0.0225 (9) | 0.0253 (9) | 0.0308 (9) | 0.0058 (7) | 0.0070 (7) | 0.0139 (7) |
| C24 | 0.0273 (9) | 0.0343 (11) | 0.0287 (9) | 0.0110 (8) | 0.0083 (7) | 0.0139 (8) |
| C25 | 0.0318 (10) | 0.0290 (10) | 0.0298 (9) | 0.0060 (8) | 0.0048 (7) | 0.0040 (8) |
| C26 | 0.0217 (9) | 0.0238 (9) | 0.0345 (9) | 0.0056 (7) | 0.0064 (7) | 0.0096 (8) |
| C27 | 0.0187 (8) | 0.0241 (9) | 0.0282 (8) | 0.0045 (7) | 0.0004 (7) | 0.0089 (7) |
| C28 | 0.0450 (11) | 0.0297 (11) | 0.0351 (10) | −0.0123 (9) | −0.0004 (8) | 0.0104 (8) |
Geometric parameters (Å, º) top
| O1—C12 | 1.3674 (17) | C14—H14 | 0.95 |
| O1—C18 | 1.4307 (17) | C15—C16 | 1.386 (2) |
| O2—C22 | 1.3674 (17) | C15—H15 | 0.95 |
| O2—C28 | 1.4404 (19) | C16—H16 | 0.95 |
| N1—C17 | 1.2688 (18) | C17—H17 | 0.95 |
| N1—C1 | 1.4609 (19) | C18—H18A | 0.98 |
| N2—C27 | 1.270 (2) | C18—H18B | 0.98 |
| N2—C3 | 1.4600 (18) | C18—H18C | 0.98 |
| C1—C2 | 1.529 (2) | C21—C26 | 1.392 (2) |
| C1—H1A | 0.99 | C21—C22 | 1.412 (2) |
| C1—H1B | 0.99 | C21—C27 | 1.471 (2) |
| C2—C3 | 1.520 (2) | C22—C23 | 1.390 (2) |
| C2—H2A | 0.99 | C23—C24 | 1.382 (2) |
| C2—H2B | 0.99 | C23—H23 | 0.95 |
| C3—H3A | 0.99 | C24—C25 | 1.383 (2) |
| C3—H3B | 0.99 | C24—H24 | 0.95 |
| C11—C16 | 1.395 (2) | C25—C26 | 1.383 (2) |
| C11—C12 | 1.400 (2) | C25—H25 | 0.95 |
| C11—C17 | 1.475 (2) | C26—H26 | 0.95 |
| C12—C13 | 1.389 (2) | C27—H27 | 0.95 |
| C13—C14 | 1.386 (2) | C28—H28A | 0.98 |
| C13—H13 | 0.95 | C28—H28B | 0.98 |
| C14—C15 | 1.382 (2) | C28—H28C | 0.98 |
| | | |
| C12—O1—C18 | 117.86 (13) | C15—C16—H16 | 119.4 |
| C22—O2—C28 | 116.49 (12) | C11—C16—H16 | 119.4 |
| C17—N1—C1 | 117.99 (13) | N1—C17—C11 | 122.23 (14) |
| C27—N2—C3 | 118.37 (13) | N1—C17—H17 | 118.9 |
| N1—C1—C2 | 110.05 (12) | C11—C17—H17 | 118.9 |
| N1—C1—H1A | 109.7 | O1—C18—H18A | 109.5 |
| C2—C1—H1A | 109.7 | O1—C18—H18B | 109.5 |
| N1—C1—H1B | 109.7 | H18A—C18—H18B | 109.5 |
| C2—C1—H1B | 109.7 | O1—C18—H18C | 109.5 |
| H1A—C1—H1B | 108.2 | H18A—C18—H18C | 109.5 |
| C3—C2—C1 | 112.78 (12) | H18B—C18—H18C | 109.5 |
| C3—C2—H2A | 109.0 | C26—C21—C22 | 117.86 (14) |
| C1—C2—H2A | 109.0 | C26—C21—C27 | 120.79 (14) |
| C3—C2—H2B | 109.0 | C22—C21—C27 | 121.34 (14) |
| C1—C2—H2B | 109.0 | O2—C22—C23 | 123.15 (14) |
| H2A—C2—H2B | 107.8 | O2—C22—C21 | 116.56 (13) |
| N2—C3—C2 | 110.90 (12) | C23—C22—C21 | 120.28 (14) |
| N2—C3—H3A | 109.5 | C24—C23—C22 | 120.16 (15) |
| C2—C3—H3A | 109.5 | C24—C23—H23 | 119.9 |
| N2—C3—H3B | 109.5 | C22—C23—H23 | 119.9 |
| C2—C3—H3B | 109.5 | C23—C24—C25 | 120.45 (15) |
| H3A—C3—H3B | 108.0 | C23—C24—H24 | 119.8 |
| C16—C11—C12 | 118.70 (14) | C25—C24—H24 | 119.8 |
| C16—C11—C17 | 120.75 (14) | C26—C25—C24 | 119.39 (15) |
| C12—C11—C17 | 120.55 (14) | C26—C25—H25 | 120.3 |
| O1—C12—C13 | 123.50 (14) | C24—C25—H25 | 120.3 |
| O1—C12—C11 | 116.29 (14) | C25—C26—C21 | 121.82 (15) |
| C13—C12—C11 | 120.22 (14) | C25—C26—H26 | 119.1 |
| C14—C13—C12 | 119.84 (15) | C21—C26—H26 | 119.1 |
| C14—C13—H13 | 120.1 | N2—C27—C21 | 121.70 (14) |
| C12—C13—H13 | 120.1 | N2—C27—H27 | 119.1 |
| C15—C14—C13 | 120.84 (16) | C21—C27—H27 | 119.1 |
| C15—C14—H14 | 119.6 | O2—C28—H28A | 109.5 |
| C13—C14—H14 | 119.6 | O2—C28—H28B | 109.5 |
| C14—C15—C16 | 119.25 (16) | H28A—C28—H28B | 109.5 |
| C14—C15—H15 | 120.4 | O2—C28—H28C | 109.5 |
| C16—C15—H15 | 120.4 | H28A—C28—H28C | 109.5 |
| C15—C16—C11 | 121.15 (15) | H28B—C28—H28C | 109.5 |
| | | |
| C17—N1—C1—C2 | 133.21 (14) | C16—C11—C17—N1 | −7.1 (2) |
| N1—C1—C2—C3 | −66.40 (17) | C12—C11—C17—N1 | 172.39 (13) |
| C27—N2—C3—C2 | 128.37 (15) | C28—O2—C22—C23 | −6.3 (2) |
| C1—C2—C3—N2 | 172.76 (13) | C28—O2—C22—C21 | 172.46 (15) |
| C18—O1—C12—C13 | −10.2 (2) | C26—C21—C22—O2 | −177.99 (13) |
| C18—O1—C12—C11 | 170.25 (13) | C27—C21—C22—O2 | 1.2 (2) |
| C16—C11—C12—O1 | 179.57 (13) | C26—C21—C22—C23 | 0.8 (2) |
| C17—C11—C12—O1 | 0.0 (2) | C27—C21—C22—C23 | 179.94 (13) |
| C16—C11—C12—C13 | 0.0 (2) | O2—C22—C23—C24 | 176.90 (14) |
| C17—C11—C12—C13 | −179.52 (13) | C21—C22—C23—C24 | −1.8 (2) |
| O1—C12—C13—C14 | −179.58 (14) | C22—C23—C24—C25 | 1.3 (2) |
| C11—C12—C13—C14 | −0.1 (2) | C23—C24—C25—C26 | 0.2 (2) |
| C12—C13—C14—C15 | 0.4 (2) | C24—C25—C26—C21 | −1.2 (2) |
| C13—C14—C15—C16 | −0.7 (2) | C22—C21—C26—C25 | 0.7 (2) |
| C14—C15—C16—C11 | 0.6 (2) | C27—C21—C26—C25 | −178.42 (14) |
| C12—C11—C16—C15 | −0.3 (2) | C3—N2—C27—C21 | 179.51 (13) |
| C17—C11—C16—C15 | 179.23 (13) | C26—C21—C27—N2 | −1.8 (2) |
| C1—N1—C17—C11 | 179.40 (13) | C22—C21—C27—N2 | 179.08 (14) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1B···Cg2i | 0.99 | 2.84 | 3.725 | 149 |
| C23—H23···Cg1ii | 0.95 | 3.05 | 3.944 | 157 |
| C17—H17···Cg1iii | 0.95 | 3.23 | 3.546 | 102 |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+2, −y+1, −z+1; (iii) −x+1, −y, −z+2. |
(III)
N,
N-bis(2-methoxybenzylidene)butane-1,4-diamine
top
Crystal data top
| C20H24N2O2 | F(000) = 348 |
| Mr = 324.41 | Dx = 1.221 Mg m−3 |
| Monoclinic, P21/n | Melting point: 60 K |
| Hall symbol: -P 2yn | Mo Kα radiation, λ = 0.71073 Å |
| a = 4.6903 (2) Å | Cell parameters from 2004 reflections |
| b = 14.2387 (6) Å | θ = 2.1–27.5° |
| c = 13.2170 (5) Å | µ = 0.08 mm−1 |
| β = 91.002 (2)° | T = 123 K |
| V = 882.55 (6) Å3 | Needle, pale yellow |
| Z = 2 | 0.40 × 0.15 × 0.10 mm |
Data collection top
Nonius Kappa CCD area-detector
diffractometer | 1303 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.046 |
| Graphite monochromator | θmax = 27.5°, θmin = 2.1° |
| ϕ and ω scans | h = −6→6 |
| 10154 measured reflections | k = −18→18 |
| 2004 independent reflections | l = −17→17 |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.042 | H-atom parameters constrained |
| wR(F2) = 0.103 | w = 1/[σ2(Fo2) + (0.0411P)2 + 0.144P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max < 0.001 |
| 2004 reflections | Δρmax = 0.19 e Å−3 |
| 111 parameters | Δρmin = −0.19 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 1997), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.021 (4) |
Crystal data top
| C20H24N2O2 | V = 882.55 (6) Å3 |
| Mr = 324.41 | Z = 2 |
| Monoclinic, P21/n | Mo Kα radiation |
| a = 4.6903 (2) Å | µ = 0.08 mm−1 |
| b = 14.2387 (6) Å | T = 123 K |
| c = 13.2170 (5) Å | 0.40 × 0.15 × 0.10 mm |
| β = 91.002 (2)° | |
Data collection top
Nonius Kappa CCD area-detector
diffractometer | 1303 reflections with I > 2σ(I) |
| 10154 measured reflections | Rint = 0.046 |
| 2004 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.042 | 0 restraints |
| wR(F2) = 0.103 | H-atom parameters constrained |
| S = 1.03 | Δρmax = 0.19 e Å−3 |
| 2004 reflections | Δρmin = −0.19 e Å−3 |
| 111 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N1 | 0.6705 (2) | 0.39640 (8) | 0.30317 (8) | 0.0288 (3) | |
| O1 | 1.0906 (2) | 0.15552 (7) | 0.33652 (7) | 0.0420 (3) | |
| C1 | 0.4697 (3) | 0.41660 (10) | 0.38382 (10) | 0.0284 (3) | |
| H1A | 0.4272 | 0.3582 | 0.4213 | 0.034* | |
| H1B | 0.2889 | 0.4407 | 0.3541 | 0.034* | |
| C2 | 0.5965 (3) | 0.48897 (9) | 0.45585 (10) | 0.0268 (3) | |
| H2A | 0.6335 | 0.5476 | 0.4180 | 0.032* | |
| H2B | 0.7815 | 0.4654 | 0.4826 | 0.032* | |
| C11 | 0.9857 (3) | 0.28276 (10) | 0.22940 (10) | 0.0268 (3) | |
| C12 | 1.1410 (3) | 0.20003 (10) | 0.24681 (10) | 0.0308 (4) | |
| C13 | 1.3370 (3) | 0.16890 (11) | 0.17602 (11) | 0.0356 (4) | |
| H13 | 1.4399 | 0.1123 | 0.1872 | 0.043* | |
| C14 | 1.3799 (3) | 0.22110 (11) | 0.08954 (12) | 0.0385 (4) | |
| H14 | 1.5146 | 0.2002 | 0.0416 | 0.046* | |
| C15 | 1.2308 (3) | 0.30294 (11) | 0.07162 (11) | 0.0380 (4) | |
| H15 | 1.2629 | 0.3383 | 0.0119 | 0.046* | |
| C16 | 1.0337 (3) | 0.33327 (10) | 0.14122 (10) | 0.0318 (4) | |
| H16 | 0.9297 | 0.3894 | 0.1286 | 0.038* | |
| C17 | 0.7745 (3) | 0.31425 (10) | 0.30313 (10) | 0.0267 (3) | |
| H17 | 0.7132 | 0.2710 | 0.3530 | 0.032* | |
| C18 | 1.2349 (4) | 0.06902 (12) | 0.35660 (13) | 0.0512 (5) | |
| H18A | 1.1717 | 0.0217 | 0.3073 | 0.077* | |
| H18B | 1.1908 | 0.0476 | 0.4250 | 0.077* | |
| H18C | 1.4410 | 0.0784 | 0.3513 | 0.077* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N1 | 0.0274 (7) | 0.0315 (7) | 0.0276 (7) | −0.0003 (5) | 0.0046 (5) | −0.0048 (5) |
| O1 | 0.0522 (7) | 0.0381 (6) | 0.0359 (6) | 0.0136 (5) | 0.0082 (5) | 0.0020 (5) |
| C1 | 0.0234 (7) | 0.0311 (8) | 0.0308 (8) | −0.0003 (6) | 0.0048 (6) | −0.0029 (6) |
| C2 | 0.0220 (7) | 0.0276 (8) | 0.0310 (8) | −0.0005 (6) | 0.0042 (6) | −0.0015 (6) |
| C11 | 0.0233 (7) | 0.0291 (8) | 0.0280 (8) | −0.0046 (6) | 0.0013 (6) | −0.0081 (6) |
| C12 | 0.0280 (8) | 0.0334 (9) | 0.0310 (8) | −0.0027 (6) | 0.0009 (6) | −0.0088 (6) |
| C13 | 0.0290 (8) | 0.0365 (9) | 0.0414 (9) | −0.0005 (7) | 0.0028 (7) | −0.0142 (7) |
| C14 | 0.0305 (8) | 0.0457 (10) | 0.0398 (9) | −0.0083 (7) | 0.0124 (7) | −0.0195 (7) |
| C15 | 0.0415 (9) | 0.0426 (9) | 0.0303 (9) | −0.0116 (8) | 0.0106 (7) | −0.0075 (7) |
| C16 | 0.0314 (8) | 0.0330 (8) | 0.0310 (8) | −0.0051 (7) | 0.0029 (6) | −0.0058 (6) |
| C17 | 0.0251 (7) | 0.0293 (8) | 0.0257 (8) | −0.0033 (6) | 0.0020 (6) | −0.0034 (6) |
| C18 | 0.0665 (12) | 0.0401 (10) | 0.0469 (10) | 0.0193 (9) | −0.0004 (9) | −0.0002 (8) |
Geometric parameters (Å, º) top
| N1—C17 | 1.2673 (17) | C12—C13 | 1.3954 (19) |
| N1—C1 | 1.4634 (17) | C13—C14 | 1.381 (2) |
| O1—C12 | 1.3686 (17) | C13—H13 | 0.95 |
| O1—C18 | 1.4279 (18) | C14—C15 | 1.378 (2) |
| C1—C2 | 1.5172 (18) | C14—H14 | 0.95 |
| C1—H1A | 0.99 | C15—C16 | 1.384 (2) |
| C1—H1B | 0.99 | C15—H15 | 0.95 |
| C2—C2i | 1.522 (3) | C16—H16 | 0.95 |
| C2—H2A | 0.99 | C17—H17 | 0.95 |
| C2—H2B | 0.99 | C18—H18A | 0.98 |
| C11—C16 | 1.391 (2) | C18—H18B | 0.98 |
| C11—C12 | 1.402 (2) | C18—H18C | 0.98 |
| C11—C17 | 1.4719 (18) | | |
| | | |
| C17—N1—C1 | 115.73 (12) | C14—C13—H13 | 120.3 |
| C12—O1—C18 | 118.18 (11) | C12—C13—H13 | 120.3 |
| N1—C1—C2 | 109.90 (11) | C15—C14—C13 | 121.10 (14) |
| N1—C1—H1A | 109.7 | C15—C14—H14 | 119.4 |
| C2—C1—H1A | 109.7 | C13—C14—H14 | 119.4 |
| N1—C1—H1B | 109.7 | C14—C15—C16 | 119.51 (14) |
| C2—C1—H1B | 109.7 | C14—C15—H15 | 120.2 |
| H1A—C1—H1B | 108.2 | C16—C15—H15 | 120.2 |
| C1—C2—C2i | 112.93 (14) | C15—C16—C11 | 121.00 (15) |
| C1—C2—H2A | 109.0 | C15—C16—H16 | 119.5 |
| C2i—C2—H2A | 109.0 | C11—C16—H16 | 119.5 |
| C1—C2—H2B | 109.0 | N1—C17—C11 | 123.01 (13) |
| C2i—C2—H2B | 109.0 | N1—C17—H17 | 118.5 |
| H2A—C2—H2B | 107.8 | C11—C17—H17 | 118.5 |
| C16—C11—C12 | 118.74 (13) | O1—C18—H18A | 109.5 |
| C16—C11—C17 | 121.19 (13) | O1—C18—H18B | 109.5 |
| C12—C11—C17 | 120.07 (13) | H18A—C18—H18B | 109.5 |
| O1—C12—C13 | 124.01 (13) | O1—C18—H18C | 109.5 |
| O1—C12—C11 | 115.75 (12) | H18A—C18—H18C | 109.5 |
| C13—C12—C11 | 120.22 (13) | H18B—C18—H18C | 109.5 |
| C14—C13—C12 | 119.43 (15) | | |
| | | |
| C17—N1—C1—C2 | 113.14 (13) | C11—C12—C13—C14 | 1.1 (2) |
| N1—C1—C2—C2i | −177.94 (14) | C12—C13—C14—C15 | −0.6 (2) |
| C18—O1—C12—C13 | −3.8 (2) | C13—C14—C15—C16 | −0.2 (2) |
| C18—O1—C12—C11 | 177.69 (14) | C14—C15—C16—C11 | 0.5 (2) |
| C16—C11—C12—O1 | 177.84 (12) | C12—C11—C16—C15 | 0.0 (2) |
| C17—C11—C12—O1 | −2.81 (19) | C17—C11—C16—C15 | −179.39 (12) |
| C16—C11—C12—C13 | −0.7 (2) | C1—N1—C17—C11 | −178.18 (12) |
| C17—C11—C12—C13 | 178.61 (12) | C16—C11—C17—N1 | −15.5 (2) |
| O1—C12—C13—C14 | −177.41 (13) | C12—C11—C17—N1 | 165.12 (13) |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Experimental details
| | (I) | (II) | (III) |
| Crystal data |
| Chemical formula | C18H20N2O2 | C19H22N2O2 | C20H24N2O2 |
| Mr | 296.36 | 310.39 | 324.41 |
| Crystal system, space group | Monoclinic, P21/n | Triclinic, P1 | Monoclinic, P21/n |
| Temperature (K) | 123 | 123 | 123 |
| a, b, c (Å) | 7.8727 (2), 8.9846 (2), 11.3927 (3) | 8.8669 (2), 9.9139 (3), 10.2724 (4) | 4.6903 (2), 14.2387 (6), 13.2170 (5) |
| α, β, γ (°) | 90, 96.345 (1), 90 | 101.140 (2), 92.341 (2), 109.442 (2) | 90, 91.002 (2), 90 |
| V (Å3) | 800.90 (3) | 830.05 (5) | 882.55 (6) |
| Z | 2 | 2 | 2 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα |
| µ (mm−1) | 0.08 | 0.08 | 0.08 |
| Crystal size (mm) | 0.5 × 0.5 × 0.4 | 0.4 × 0.3 × 0.2 | 0.40 × 0.15 × 0.10 |
| |
| Data collection |
| Diffractometer | Nonius Kappa CCD area-detector
diffractometer | Nonius Kappa CCD area-detector
diffractometer | Nonius Kappa CCD area-detector
diffractometer |
| Absorption correction | – | – | – |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 9458, 1815, 1627 | 12958, 3737, 2279 | 10154, 2004, 1303 |
| Rint | 0.037 | 0.063 | 0.046 |
| (sin θ/λ)max (Å−1) | 0.649 | 0.651 | 0.649 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.036, 0.098, 1.02 | 0.047, 0.122, 0.99 | 0.042, 0.103, 1.03 |
| No. of reflections | 1815 | 3737 | 2004 |
| No. of parameters | 101 | 210 | 111 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.26, −0.17 | 0.24, −0.22 | 0.19, −0.19 |
Selected geometric parameters (Å, º) for (I) top| O1—C12 | 1.3663 (11) | N1—C1 | 1.4579 (12) |
| O1—C18 | 1.4375 (12) | C11—C17 | 1.4759 (13) |
| N1—C17 | 1.2693 (13) | | |
| | | |
| C12—O1—C18 | 116.67 (8) | O1—C12—C13 | 123.21 (9) |
| C17—N1—C1 | 117.28 (8) | O1—C12—C11 | 116.66 (8) |
| C16—C11—C17 | 120.57 (9) | N1—C17—C11 | 121.40 (9) |
| C12—C11—C17 | 120.93 (8) | | |
Hydrogen-bond geometry (Å, º) for (I) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C17—H17···Cg1i | 0.95 | 3.02 | 3.428 | 107 |
| Symmetry code: (i) −x+1, −y+2, −z+1. |
Selected geometric parameters (Å, º) for (II) top| O1—C12 | 1.3674 (17) | N1—C1 | 1.4609 (19) |
| O1—C18 | 1.4307 (17) | N2—C27 | 1.270 (2) |
| O2—C22 | 1.3674 (17) | N2—C3 | 1.4600 (18) |
| O2—C28 | 1.4404 (19) | C11—C17 | 1.475 (2) |
| N1—C17 | 1.2688 (18) | C21—C27 | 1.471 (2) |
| | | |
| C12—O1—C18 | 117.86 (13) | O1—C12—C11 | 116.29 (14) |
| C22—O2—C28 | 116.49 (12) | N1—C17—C11 | 122.23 (14) |
| C17—N1—C1 | 117.99 (13) | C26—C21—C27 | 120.79 (14) |
| C27—N2—C3 | 118.37 (13) | C22—C21—C27 | 121.34 (14) |
| C16—C11—C17 | 120.75 (14) | O2—C22—C23 | 123.15 (14) |
| C12—C11—C17 | 120.55 (14) | O2—C22—C21 | 116.56 (13) |
| O1—C12—C13 | 123.50 (14) | N2—C27—C21 | 121.70 (14) |
Hydrogen-bond geometry (Å, º) for (II) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1B···Cg2i | 0.99 | 2.84 | 3.725 | 149 |
| C23—H23···Cg1ii | 0.95 | 3.05 | 3.944 | 157 |
| C17—H17···Cg1iii | 0.95 | 3.23 | 3.546 | 102 |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+2, −y+1, −z+1; (iii) −x+1, −y, −z+2. |
Selected geometric parameters (Å, º) for (III) top| N1—C17 | 1.2673 (17) | O1—C18 | 1.4279 (18) |
| N1—C1 | 1.4634 (17) | C11—C17 | 1.4719 (18) |
| O1—C12 | 1.3686 (17) | | |
| | | |
| C17—N1—C1 | 115.73 (12) | O1—C12—C13 | 124.01 (13) |
| C12—O1—C18 | 118.18 (11) | O1—C12—C11 | 115.75 (12) |
| C16—C11—C17 | 121.19 (13) | N1—C17—C11 | 123.01 (13) |
| C12—C11—C17 | 120.07 (13) | | |

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.
 N hydrogen bonds. The packing of MeSalen and MeSalpr is controlled by C—H
N hydrogen bonds. The packing of MeSalen and MeSalpr is controlled by C—H π interactions, whereas that of MeSalbu has only van der Waals contacts.
π interactions, whereas that of MeSalbu has only van der Waals contacts.
 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2004/03/00/fg1727/fg1727fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2004/03/00/fg1727/fg1727fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2004/03/00/fg1727/fg1727fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/c/issues/2004/03/00/fg1727/fg1727fig4thm.gif)

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry



We have been investigating the structures adopted by the complexes of the late transition metals Ni, Cu and Zn (Reglinski et al., 2002a,b) with symmetric salicylidene Schiff base ligands, as a function of the number of methylene units (n = 2, 3 or 4) between the two imine groups. As part of our on-going study, we recently turned our attention to the preparation of the homologous compounds derived from o-anisaldehyde. As part of the characterization process, we have isolated and crystallographically characterized the three title compounds, namely N,N-bis(2-methoxybenzylidene)ethane-1,2-diamine (MeSalen), (I) (Billman & Dorman, 1963), N,N-bis(2-methoxybenzylidene)propane-1,3-diamine (MeSalpr), (II) (Moneta et al., 1988), and N,N-bis(2-methoxybenzylidene)butane-1,4-diamine (MeSalbu), (III). A search of the December 2003 release of the Cambridge Structural Database (Allen, 2002) shows that no higher homologues have been reported to date. The availability of the structures of these three compounds has allowed us to make some simple but instructive observations regarding the effects of methylene chain lengths and the effects of hydrogen bonding in these systems, compared with the parent salicylidene complexes, N,N-bis(2-hydroxybenzylidene)ethane-1,2-diamine (salen; Pahor et al., 1978), N,N-bis(2-hydroxybenzylidene)propane-1,3-diamine (Salpr; Elderman et al., 1991) and N,N-bis(2-hydroxybenzylidene)butane-1,4-diamine (Salbu; Kennedy & Reglinski, 2001), respectively. \sch
The three title molecules adopt extended conformations, as shown in Figs. 1a, 1 b and 1c. In salen, (I), and Salbu, (III), where there are an even number of C atoms in the chain, the molecules lie about inversion centres and adopt an essentially planar extended conformation, with the methoxy moieties, of necessity, in a trans arrangement (Figs. 1a and 1c). However, for MeSalpr, (II), which has an odd number of C atoms in the linking chains, no symmetry is imposed by the P1 space group and a markedly non-planar conformation is adopted, with the methoxy moieties in a cis arrangement (Fig. 1 b).
In all three cases, replacement of the salicylidene moiety (–OH) with the anisaldehyde (–OMe) moiety has removed the stabilizing influence of intramolecular hydrogen bonding exhibited in the parent compounds (Pahor et al., 1978; Elderman et al., 1991; Kennedy & Reglinski, 2001), thus allowing a rotation of the aromatic ring such that the methoxy group is remote from the influence of the imine N atom. The lack of intramolecular hydrogen bonding does not alter the bond lengths or angles of the molecules. The largest systematic difference occurs in the angles to the OR group, where O1—C12—C11 is significantly smaller for the methoxy compounds than for their hydroxy equivalents [116.66 (8), 116.29 (14) and 115.75 (12)° for (I), (II) and (III), respectively, and 121.23, 121.68 and 121.37° for the equivalent literature species). This difference is simply explained by the steric effect of the Me group, which lies in the aryl ring plane.
Geometry calculations with PLATON (Spek, 2003) show that the packing of the three compounds is quite different. In MeSalen (Fig. 2), the molecules are linked by pairs of inversion-related C—H···π interactions to generate infinite chains extending in the b direction (details in Table 2). In MeSalpr, chains of inversion-related molecules are linked by two independent C—H···π interactions, as shown in Fig. 3a (details in Table 4). These chains are then linked to form sheets via pairs of inversion-related C17—H17···Cg1iii interactions similar to those observed in MeSalen [Fig. 2; symmetry code: (iii) 1 − x, −y, 2 − z]. In this way, the three independent C—H···π interactions generate sheets of molecules in the [011] plane, as shown in the stereo pair (Fig. 3 b). By contrast, in MeSalbu (Fig. 4), with a short [4.6903 (2) Å] a axis, there are no signficant intermolecular interactions and contacts are all of the van der Waals variety.