
Determination of the crystal structures of the homologous (1
R*,2
R*)-
trans-2-hydroxy-1-cyclopentanecarboxylic acid (5T), (1
R*,2
S*)-
cis-2-hydroxy-1-cyclohexanecarboxylic acid (6C) and (1
R*,2
S*)-
cis-2-hydroxy-1-cycloheptanecarboxylic acid (7C) proved a predicted pattern of supramolecular close packing. The prediction was based on the common features observed in the crystal structures of six related 2-hydroxy-1-cyclopentanecarboxylic acids and analogous carboxamides [Kálmán
et al. (2001).
Acta Cryst. B
57, 539–550]. This pattern is characterized by tetrameric
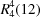
rings of
C2 symmetry formed from dimeric
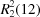
rings. The
C2 symmetry of such tetramers is not common in the literature, usually they have
Ci symmetry. Both types of tetramers are formed from dimers with similar or opposite orientation. The
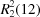
dimers differ in their hydrogen bonds. In 5T the monomers are joined by a pair of O1—H

O2=C bonds, whereas in 7C they are joined by a pair of O3—H

O1-H bonds. In 6C 60% of the disordered
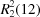
dimers are similar to those in 7C, while 40% resemble those in 5T. Apart from these hydrogen-bonding differences and the ring-size differences, the three crystals exhibit isostructurality.
Supporting information
CCDC references: 188096; 188097; 188098
For all compounds, data collection: CAD-4 EXPRESS; cell refinement: CAD-4 EXPRESS; data reduction: XCAD4 (Harms 1996); program(s) used to solve structure: SHELXS97 (Sheldrick 1997); program(s) used to refine structure: SHELXL97 (Sheldrick 1997); molecular graphics: PLATON (Spek 1998).
(5ts)
trans-2-hydroxy-1-cyclopentane carboxylic acid
top
Crystal data top
| C6H10O3 | F(000) = 560 |
| Mr = 130.14 | Dx = 1.325 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71070 Å |
| a = 17.383 (2) Å | Cell parameters from 25 reflections |
| b = 6.188 (1) Å | θ = 13.1–14.3° |
| c = 12.361 (1) Å | µ = 0.11 mm−1 |
| β = 101.16 (1)° | T = 293 K |
| V = 1304.5 (3) Å3 | Block, colourless |
| Z = 8 | 0.40 × 0.25 × 0.15 mm |
Data collection top
Enraf-Nonius CAD4
diffractometer | 838 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.022 |
| Graphite monochromator | θmax = 28.0°, θmin = 3.4° |
| ω–2θ scans | h = −22→22 |
Absorption correction: ψ scan
North A.C., Philips, D.C. & Mathews, F. (1968) Acta Cryst. A24 350-359 | k = −8→8 |
| Tmin = 0.959, Tmax = 0.984 | l = −16→16 |
| 3396 measured reflections | 3 standard reflections every 60 min |
| 1575 independent reflections | intensity decay: 1% |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.043 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.142 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.82 | w = 1/[σ2(Fo2) + (0.1P)2]
where P = (Fo2 + 2Fc2)/3 |
| 1575 reflections | (Δ/σ)max < 0.001 |
| 84 parameters | Δρmax = 0.18 e Å−3 |
| 21 restraints | Δρmin = −0.16 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.08144 (9) | 0.4250 (3) | 0.21183 (10) | 0.0579 (5) | |
| H1 | 0.0490 | 0.5042 | 0.1743 | 0.075* | |
| O2 | 0.02571 (8) | 0.3088 (2) | −0.07966 (11) | 0.0546 (4) | |
| O3 | 0.14061 (9) | 0.4528 (3) | −0.08640 (12) | 0.0707 (5) | |
| H3 | 0.1164 | 0.4997 | −0.1452 | 0.092* | |
| C1 | 0.13442 (10) | 0.2386 (3) | 0.06747 (13) | 0.0377 (4) | |
| H1A | 0.1823 | 0.3203 | 0.0960 | 0.049* | |
| C2 | 0.08541 (11) | 0.2263 (3) | 0.15561 (13) | 0.0419 (5) | |
| H2 | 0.0323 | 0.1789 | 0.1225 | 0.054* | |
| C3 | 0.12508 (14) | 0.0535 (4) | 0.23511 (17) | 0.0576 (6) | |
| H3A | 0.1566 | 0.1198 | 0.2999 | 0.075* | |
| H3B | 0.0861 | −0.0387 | 0.2584 | 0.075* | |
| C4 | 0.17620 (14) | −0.0766 (4) | 0.17334 (17) | 0.0585 (6) | |
| H4A | 0.1652 | −0.2298 | 0.1776 | 0.076* | |
| H4B | 0.2312 | −0.0520 | 0.2039 | 0.076* | |
| C5 | 0.15589 (13) | 0.0017 (4) | 0.05415 (16) | 0.0501 (5) | |
| H5A | 0.1120 | −0.0783 | 0.0124 | 0.065* | |
| H5B | 0.2005 | −0.0114 | 0.0179 | 0.065* | |
| C6 | 0.09401 (11) | 0.3382 (3) | −0.03940 (13) | 0.0386 (4) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0760 (10) | 0.0679 (10) | 0.0260 (6) | 0.0230 (7) | 0.0003 (7) | −0.0076 (6) |
| O2 | 0.0520 (8) | 0.0710 (11) | 0.0384 (7) | 0.0009 (7) | 0.0026 (6) | 0.0138 (7) |
| O3 | 0.0698 (10) | 0.0993 (13) | 0.0376 (8) | −0.0248 (9) | −0.0029 (7) | 0.0256 (8) |
| C1 | 0.0428 (9) | 0.0445 (11) | 0.0246 (8) | −0.0027 (8) | 0.0035 (7) | 0.0014 (7) |
| C2 | 0.0521 (10) | 0.0490 (11) | 0.0240 (7) | −0.0008 (8) | 0.0057 (7) | 0.0011 (7) |
| C3 | 0.0799 (15) | 0.0599 (14) | 0.0326 (9) | 0.0006 (11) | 0.0096 (10) | 0.0146 (9) |
| C4 | 0.0706 (14) | 0.0508 (13) | 0.0504 (12) | 0.0069 (10) | 0.0024 (10) | 0.0145 (10) |
| C5 | 0.0598 (11) | 0.0514 (11) | 0.0379 (10) | 0.0103 (10) | 0.0064 (9) | −0.0010 (9) |
| C6 | 0.0507 (10) | 0.0411 (10) | 0.0232 (7) | 0.0010 (8) | 0.0051 (7) | −0.0015 (7) |
Geometric parameters (Å, º) top
| O1—C2 | 1.421 (2) | C2—H2 | 0.9800 |
| O1—H1 | 0.8200 | C3—C4 | 1.511 (3) |
| O2—C6 | 1.209 (2) | C3—H3A | 0.9700 |
| O3—C6 | 1.296 (2) | C3—H3B | 0.9700 |
| O3—H3 | 0.8200 | C4—C5 | 1.526 (3) |
| C1—C6 | 1.503 (2) | C4—H4A | 0.9700 |
| C1—C2 | 1.509 (3) | C4—H4B | 0.9700 |
| C1—C5 | 1.530 (3) | C5—H5A | 0.9700 |
| C1—H1A | 0.9800 | C5—H5B | 0.9700 |
| C2—C3 | 1.524 (3) | | |
| | | |
| C2—O1—H1 | 109.5 | C2—C3—H3B | 110.4 |
| C6—O3—H3 | 109.5 | H3A—C3—H3B | 108.6 |
| C6—C1—C2 | 114.96 (15) | C3—C4—C5 | 105.66 (16) |
| C6—C1—C5 | 112.37 (15) | C3—C4—H4A | 110.6 |
| C2—C1—C5 | 102.29 (16) | C5—C4—H4A | 110.6 |
| C6—C1—H1A | 109.0 | C3—C4—H4B | 110.6 |
| C2—C1—H1A | 109.0 | C5—C4—H4B | 110.6 |
| C5—C1—H1A | 109.0 | H4A—C4—H4B | 108.7 |
| O1—C2—C1 | 113.39 (16) | C4—C5—C1 | 102.48 (16) |
| O1—C2—C3 | 110.53 (15) | C4—C5—H5A | 111.3 |
| C1—C2—C3 | 104.64 (16) | C1—C5—H5A | 111.3 |
| O1—C2—H2 | 109.4 | C4—C5—H5B | 111.3 |
| C1—C2—H2 | 109.4 | C1—C5—H5B | 111.3 |
| C3—C2—H2 | 109.4 | H5A—C5—H5B | 109.2 |
| C4—C3—C2 | 106.56 (16) | O2—C6—O3 | 123.17 (16) |
| C4—C3—H3A | 110.4 | O2—C6—C1 | 123.71 (17) |
| C2—C3—H3A | 110.4 | O3—C6—C1 | 113.08 (16) |
| C4—C3—H3B | 110.4 | | |
| | | |
| C6—C1—C2—O1 | −79.5 (2) | C3—C4—C5—C1 | 31.3 (2) |
| C5—C1—C2—O1 | 158.39 (15) | C6—C1—C5—C4 | −166.49 (16) |
| C6—C1—C2—C3 | 159.94 (17) | C2—C1—C5—C4 | −42.66 (19) |
| C5—C1—C2—C3 | 37.87 (18) | C2—C1—C6—O2 | −38.4 (3) |
| O1—C2—C3—C4 | −140.97 (18) | C5—C1—C6—O2 | 78.1 (2) |
| C1—C2—C3—C4 | −18.6 (2) | C2—C1—C6—O3 | 144.04 (18) |
| C2—C3—C4—C5 | −8.2 (2) | C5—C1—C6—O3 | −99.5 (2) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O2i | 0.82 | 1.95 | 2.7701 (19) | 178 |
| O3—H3···O1ii | 0.82 | 1.81 | 2.6187 (19) | 167 |
| Symmetry codes: (i) −x, −y+1, −z; (ii) x, −y+1, z−1/2. |
(6cs) (1
R*,2
S*)-2-hydroxy-1-cyclohexanecarboxylic acid
top
Crystal data top
| C7H12O3 | F(000) = 624 |
| Mr = 144.17 | Dx = 1.248 Mg m−3 |
| Monoclinic, C2/c | Cu Kα radiation, λ = 1.54180 Å |
| a = 21.436 (8) Å | Cell parameters from 25 reflections |
| b = 5.974 (1) Å | θ = 30.1–34.5° |
| c = 12.095 (3) Å | µ = 0.81 mm−1 |
| β = 97.70 (3)° | T = 293 K |
| V = 1534.9 (7) Å3 | Block, colourless |
| Z = 8 | 0.40 × 0.25 × 0.03 mm |
Data collection top
Enraf-Nonius CAD4
diffractometer | 1287 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.010 |
| Graphite monochromator | θmax = 75.6°, θmin = 4.2° |
| ω–2θ scans | h = −26→26 |
Absorption correction: ψ scan
North A.C., Philips, D.C. & Mathews, F. (1968) Acta Cryst. A24, 350-359 | k = 0→7 |
| Tmin = 0.738, Tmax = 0.976 | l = 0→15 |
| 1794 measured reflections | 3 standard reflections every 60 min |
| 1597 independent reflections | intensity decay: 16% |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.036 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.136 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.13 | w = 1/[σ2(Fo2) + (0.1P)2]
where P = (Fo2 + 2Fc2)/3 |
| 1597 reflections | (Δ/σ)max = 0.001 |
| 122 parameters | Δρmax = 0.19 e Å−3 |
| 40 restraints | Δρmin = −0.16 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| O1 | 0.06910 (4) | 0.07106 (14) | 0.18797 (6) | 0.0511 (3) | |
| H1X | 0.0753 | 0.0656 | 0.2563 | 0.066* | 0.604 (12) |
| H1Y | 0.0385 | 0.0066 | 0.1544 | 0.066* | 0.396 (12) |
| O2A | 0.0897 (3) | 0.0166 (8) | −0.0891 (4) | 0.0667 (9) | 0.604 (12) |
| O3A | 0.0056 (3) | 0.2159 (15) | −0.0642 (6) | 0.0504 (12) | 0.604 (12) |
| H3X | −0.0115 | 0.1223 | −0.1077 | 0.066* | 0.604 (12) |
| O2B | 0.0174 (4) | 0.202 (3) | −0.0774 (11) | 0.0556 (17) | 0.396 (12) |
| O3B | 0.1107 (4) | 0.0603 (13) | −0.0999 (5) | 0.0624 (15) | 0.396 (12) |
| H3Y | 0.0915 | 0.0148 | −0.1588 | 0.081* | 0.396 (12) |
| C1 | 0.10260 (5) | 0.31317 (18) | 0.04841 (8) | 0.0395 (3) | |
| H1A | 0.1005 | 0.4708 | 0.0258 | 0.051* | 0.604 (12) |
| H1B | 0.0980 | 0.4653 | 0.0180 | 0.051* | 0.396 (12) |
| C2 | 0.06923 (5) | 0.30032 (16) | 0.15243 (8) | 0.0375 (3) | |
| H2 | 0.0257 | 0.3518 | 0.1336 | 0.049* | |
| C3 | 0.10238 (6) | 0.4462 (2) | 0.24469 (10) | 0.0508 (3) | |
| H3A | 0.0965 | 0.6021 | 0.2233 | 0.066* | |
| H3B | 0.0831 | 0.4232 | 0.3119 | 0.066* | |
| C4 | 0.17203 (6) | 0.3984 (2) | 0.27000 (11) | 0.0580 (4) | |
| H4A | 0.1782 | 0.2491 | 0.3011 | 0.075* | |
| H4B | 0.1912 | 0.5043 | 0.3251 | 0.075* | |
| C5 | 0.20362 (6) | 0.4158 (3) | 0.16539 (13) | 0.0670 (4) | |
| H5A | 0.2478 | 0.3770 | 0.1829 | 0.087* | |
| H5B | 0.2009 | 0.5688 | 0.1383 | 0.087* | |
| C6 | 0.17235 (6) | 0.2601 (3) | 0.07521 (11) | 0.0593 (4) | |
| H6A | 0.1776 | 0.1062 | 0.1003 | 0.077* | |
| H6B | 0.1924 | 0.2764 | 0.0084 | 0.077* | |
| C7A | 0.0700 (3) | 0.1837 (13) | −0.0512 (6) | 0.0381 (12) | 0.604 (12) |
| C7B | 0.0683 (5) | 0.154 (2) | −0.0350 (8) | 0.040 (2) | 0.396 (12) |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0646 (5) | 0.0517 (5) | 0.0351 (4) | −0.0160 (4) | −0.0003 (3) | 0.0076 (3) |
| O2A | 0.076 (2) | 0.0853 (19) | 0.0387 (11) | 0.0076 (15) | 0.0088 (13) | −0.0234 (12) |
| O3A | 0.056 (2) | 0.0437 (11) | 0.046 (2) | −0.0028 (16) | −0.0124 (16) | 0.0014 (14) |
| O2B | 0.056 (3) | 0.063 (4) | 0.046 (2) | −0.015 (2) | −0.003 (2) | −0.001 (2) |
| O3B | 0.078 (3) | 0.076 (3) | 0.0316 (16) | 0.018 (2) | −0.0002 (19) | −0.0143 (16) |
| C1 | 0.0488 (6) | 0.0422 (6) | 0.0274 (5) | −0.0026 (4) | 0.0046 (4) | 0.0018 (4) |
| C2 | 0.0401 (5) | 0.0415 (6) | 0.0308 (5) | 0.0024 (4) | 0.0042 (4) | −0.0028 (4) |
| C3 | 0.0625 (7) | 0.0504 (6) | 0.0392 (6) | 0.0000 (5) | 0.0052 (5) | −0.0136 (5) |
| C4 | 0.0602 (7) | 0.0631 (8) | 0.0460 (7) | −0.0109 (6) | −0.0103 (5) | −0.0082 (6) |
| C5 | 0.0453 (6) | 0.0846 (10) | 0.0697 (9) | −0.0165 (6) | 0.0023 (6) | −0.0025 (7) |
| C6 | 0.0464 (7) | 0.0844 (9) | 0.0493 (7) | −0.0023 (6) | 0.0152 (5) | −0.0089 (7) |
| C7A | 0.0610 (18) | 0.0359 (17) | 0.0153 (19) | −0.0005 (11) | −0.0020 (14) | 0.0092 (19) |
| C7B | 0.063 (3) | 0.043 (4) | 0.015 (3) | 0.005 (2) | 0.0080 (16) | 0.015 (2) |
Geometric parameters (Å, º) top
| O1—C2 | 1.4356 (13) | C1—H1B | 0.9800 |
| O1—H1X | 0.8200 | C2—C3 | 1.5158 (15) |
| O1—H1Y | 0.8200 | C2—H2 | 0.9800 |
| O2A—C7A | 1.198 (8) | C3—C4 | 1.5105 (19) |
| O3A—C7A | 1.380 (10) | C3—H3A | 0.9700 |
| O3A—H3X | 0.8200 | C3—H3B | 0.9700 |
| O2B—C7B | 1.178 (12) | C4—C5 | 1.516 (2) |
| O3B—C7B | 1.393 (11) | C4—H4A | 0.9700 |
| O3B—H3Y | 0.8200 | C4—H4B | 0.9700 |
| C1—C7B | 1.505 (6) | C5—C6 | 1.519 (2) |
| C1—C6 | 1.5204 (18) | C5—H5A | 0.9700 |
| C1—C2 | 1.5302 (14) | C5—H5B | 0.9700 |
| C1—C7A | 1.521 (4) | C6—H6A | 0.9700 |
| C1—H1A | 0.9800 | C6—H6B | 0.9700 |
| | | |
| C2—O1—H1X | 109.5 | H3A—C3—H3B | 107.8 |
| C2—O1—H1Y | 109.5 | C3—C4—C5 | 110.95 (10) |
| C7B—O3B—H3Y | 109.5 | C3—C4—H4A | 109.4 |
| C7B—C1—C6 | 112.9 (5) | C5—C4—H4A | 109.4 |
| C7B—C1—C2 | 106.2 (5) | C3—C4—H4B | 109.4 |
| C6—C1—C2 | 111.64 (9) | C5—C4—H4B | 109.4 |
| C6—C1—C7A | 113.4 (3) | H4A—C4—H4B | 108.0 |
| C2—C1—C7A | 114.2 (4) | C4—C5—C6 | 110.72 (11) |
| C6—C1—H1A | 105.6 | C4—C5—H5A | 109.5 |
| C2—C1—H1A | 105.6 | C6—C5—H5A | 109.5 |
| C7A—C1—H1A | 105.6 | C4—C5—H5B | 109.5 |
| C7B—C1—H1B | 108.7 | C6—C5—H5B | 109.5 |
| C6—C1—H1B | 108.7 | H5A—C5—H5B | 108.1 |
| C2—C1—H1B | 108.7 | C5—C6—C1 | 110.60 (11) |
| O1—C2—C3 | 110.35 (9) | C5—C6—H6A | 109.5 |
| O1—C2—C1 | 108.29 (8) | C1—C6—H6A | 109.5 |
| C3—C2—C1 | 110.60 (9) | C5—C6—H6B | 109.5 |
| O1—C2—H2 | 109.2 | C1—C6—H6B | 109.5 |
| C3—C2—H2 | 109.2 | H6A—C6—H6B | 108.1 |
| C1—C2—H2 | 109.2 | O2A—C7A—O3A | 118.1 (6) |
| C4—C3—C2 | 113.18 (10) | O2A—C7A—C1 | 125.3 (6) |
| C4—C3—H3A | 108.9 | O3A—C7A—C1 | 111.4 (6) |
| C2—C3—H3A | 108.9 | O2B—C7B—O3B | 119.2 (9) |
| C4—C3—H3B | 108.9 | O2B—C7B—C1 | 118.6 (10) |
| C2—C3—H3B | 108.9 | O3B—C7B—C1 | 109.6 (7) |
| | | |
| C7B—C1—C2—O1 | 55.9 (5) | C7B—C1—C6—C5 | −176.1 (6) |
| C6—C1—C2—O1 | −67.52 (12) | C2—C1—C6—C5 | −56.56 (14) |
| C7A—C1—C2—O1 | 62.8 (4) | C7A—C1—C6—C5 | 172.7 (4) |
| C7B—C1—C2—C3 | 176.9 (5) | C6—C1—C7A—O2A | 18.7 (12) |
| C6—C1—C2—C3 | 53.51 (13) | C2—C1—C7A—O2A | −110.7 (10) |
| C7A—C1—C2—C3 | −176.2 (4) | C6—C1—C7A—O3A | 172.6 (6) |
| O1—C2—C3—C4 | 67.02 (13) | C2—C1—C7A—O3A | 43.2 (8) |
| C1—C2—C3—C4 | −52.78 (13) | C6—C1—C7B—O2B | −166.1 (12) |
| C2—C3—C4—C5 | 54.55 (16) | C2—C1—C7B—O2B | 71.2 (14) |
| C3—C4—C5—C6 | −56.27 (17) | C6—C1—C7B—O3B | −24.6 (10) |
| C4—C5—C6—C1 | 57.65 (16) | C2—C1—C7B—O3B | −147.2 (7) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1X···O2Ai | 0.82 | 1.92 | 2.724 (4) | 167 |
| O1—H1Y···O2Bii | 0.82 | 1.89 | 2.687 (14) | 166 |
| O3A—H3X···O1ii | 0.82 | 1.87 | 2.665 (9) | 165 |
| O3B—H3Y···O1iii | 0.82 | 1.92 | 2.717 (6) | 163 |
| Symmetry codes: (i) x, −y, z+1/2; (ii) −x, −y, −z; (iii) x, −y, z−1/2. |
(7cs) (1
R*,2
S*)-2-hydroxy-1-cycloheptane carboxylic acid
top
Crystal data top
| C8H14O3 | F(000) = 688 |
| Mr = 158.19 | Dx = 1.257 Mg m−3 |
| Monoclinic, I2/c | Cu Kα radiation, λ = 1.54180 Å |
| a = 22.876 (5) Å | Cell parameters from 25 reflections |
| b = 6.224 (1) Å | θ = 37.1–39.9° |
| c = 11.793 (2) Å | µ = 0.79 mm−1 |
| β = 95.56 (3)° | T = 293 K |
| V = 1671.2 (5) Å3 | Block, colourless |
| Z = 8 | 0.35 × 0.25 × 0.20 mm |
Data collection top
Enraf-Nonius CAD4
diffractometer | 1574 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.019 |
| Graphite monochromator | θmax = 76.0°, θmin = 3.9° |
| ω–2θ scans | h = −28→28 |
Absorption correction: ψ scan
North A.C., Philips, D.C. & Mathews, F. (1968) Acta Cryst. A24, 350-359 | k = −7→7 |
| Tmin = 0.771, Tmax = 0.859 | l = −14→14 |
| 6926 measured reflections | 3 standard reflections every 60 min |
| 1720 independent reflections | intensity decay: none |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.038 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.148 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.38 | w = 1/[σ2(Fo2) + (0.1P)2]
where P = (Fo2 + 2Fc2)/3 |
| 1720 reflections | (Δ/σ)max < 0.001 |
| 148 parameters | Δρmax = 0.15 e Å−3 |
| 245 restraints | Δρmin = −0.15 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| O1 | 0.05831 (4) | −0.11228 (13) | 0.17608 (5) | 0.0565 (3) | |
| H1 | 0.0642 | −0.1176 | 0.2457 | 0.073* | |
| O2 | 0.08762 (4) | 0.03752 (15) | −0.09191 (7) | 0.0663 (3) | |
| O3 | 0.01314 (3) | −0.18594 (12) | −0.08259 (6) | 0.0536 (3) | |
| H3 | −0.0054 | −0.0868 | −0.1145 | 0.070* | |
| C1 | 0.10206 (4) | −0.28259 (14) | 0.02337 (7) | 0.0420 (3) | |
| H1A | 0.1028 | −0.4202 | −0.0168 | 0.055* | 0.851 (5) |
| H1B | 0.0957 | −0.4189 | −0.0175 | 0.055* | 0.149 (5) |
| C2 | 0.06954 (4) | −0.31900 (15) | 0.12895 (7) | 0.0409 (3) | |
| H2A | 0.0319 | −0.3887 | 0.1056 | 0.053* | 0.851 (5) |
| H2B | 0.0307 | −0.3720 | 0.0990 | 0.053* | 0.149 (5) |
| C3A | 0.10321 (11) | −0.4563 (5) | 0.22091 (19) | 0.0577 (6) | 0.851 (5) |
| H3A | 0.0748 | −0.5242 | 0.2653 | 0.075* | 0.851 (5) |
| H3B | 0.1268 | −0.3609 | 0.2717 | 0.075* | 0.851 (5) |
| C3B | 0.0935 (5) | −0.491 (3) | 0.2101 (14) | 0.076 (5) | 0.149 (5) |
| H3C | 0.0733 | −0.4825 | 0.2785 | 0.099* | 0.149 (5) |
| H3D | 0.0844 | −0.6300 | 0.1754 | 0.099* | 0.149 (5) |
| C4A | 0.14359 (7) | −0.6321 (2) | 0.18140 (13) | 0.0642 (5) | 0.851 (5) |
| H4A | 0.1479 | −0.7445 | 0.2386 | 0.084* | 0.851 (5) |
| H4B | 0.1255 | −0.6951 | 0.1114 | 0.084* | 0.851 (5) |
| C4B | 0.1583 (3) | −0.4808 (19) | 0.2436 (7) | 0.076 (3) | 0.149 (5) |
| H4C | 0.1661 | −0.5505 | 0.3171 | 0.099* | 0.149 (5) |
| H4D | 0.1691 | −0.3309 | 0.2538 | 0.099* | 0.149 (5) |
| C5A | 0.20469 (12) | −0.5463 (6) | 0.1610 (3) | 0.0757 (10) | 0.851 (5) |
| H5A | 0.2205 | −0.4718 | 0.2294 | 0.098* | 0.851 (5) |
| H5B | 0.2300 | −0.6688 | 0.1513 | 0.098* | 0.851 (5) |
| C5B | 0.1984 (8) | −0.581 (3) | 0.1626 (13) | 0.072 (4) | 0.149 (5) |
| H5C | 0.2380 | −0.5846 | 0.2006 | 0.094* | 0.149 (5) |
| H5D | 0.1861 | −0.7282 | 0.1484 | 0.094* | 0.149 (5) |
| C6A | 0.20894 (12) | −0.3976 (5) | 0.0615 (3) | 0.0712 (8) | 0.851 (5) |
| H6A | 0.2032 | −0.4819 | −0.0079 | 0.093* | 0.851 (5) |
| H6B | 0.2485 | −0.3403 | 0.0663 | 0.093* | 0.851 (5) |
| C6B | 0.2005 (6) | −0.470 (2) | 0.0491 (12) | 0.064 (4) | 0.149 (5) |
| H6C | 0.1856 | −0.5689 | −0.0105 | 0.083* | 0.149 (5) |
| H6D | 0.2413 | −0.4408 | 0.0384 | 0.083* | 0.149 (5) |
| C7A | 0.16627 (8) | −0.2101 (5) | 0.0507 (3) | 0.0570 (7) | 0.851 (5) |
| H7A | 0.1768 | −0.1148 | −0.0091 | 0.074* | 0.851 (5) |
| H7B | 0.1697 | −0.1297 | 0.1216 | 0.074* | 0.851 (5) |
| C7B | 0.1666 (4) | −0.264 (2) | 0.0341 (17) | 0.058 (4) | 0.149 (5) |
| H7C | 0.1783 | −0.1716 | 0.0987 | 0.076* | 0.149 (5) |
| H7D | 0.1780 | −0.1921 | −0.0334 | 0.076* | 0.149 (5) |
| C8 | 0.06781 (4) | −0.12552 (15) | −0.05584 (7) | 0.0442 (3) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0779 (6) | 0.0603 (5) | 0.0308 (4) | 0.0234 (4) | 0.0021 (3) | −0.0044 (3) |
| O2 | 0.0744 (6) | 0.0700 (6) | 0.0534 (5) | −0.0034 (4) | 0.0012 (4) | 0.0278 (4) |
| O3 | 0.0593 (5) | 0.0507 (5) | 0.0491 (5) | 0.0084 (3) | −0.0032 (3) | 0.0002 (3) |
| C1 | 0.0528 (5) | 0.0411 (5) | 0.0331 (5) | 0.0070 (3) | 0.0094 (4) | 0.0029 (3) |
| C2 | 0.0467 (5) | 0.0436 (5) | 0.0330 (5) | −0.0023 (3) | 0.0062 (3) | 0.0027 (3) |
| C3A | 0.0777 (10) | 0.0584 (12) | 0.0386 (7) | 0.0136 (10) | 0.0132 (8) | 0.0161 (7) |
| C3B | 0.090 (6) | 0.055 (6) | 0.082 (10) | −0.027 (4) | −0.008 (6) | 0.031 (6) |
| C4A | 0.0905 (11) | 0.0408 (7) | 0.0606 (8) | 0.0106 (6) | 0.0034 (7) | 0.0105 (6) |
| C4B | 0.063 (4) | 0.099 (8) | 0.064 (5) | 0.011 (4) | −0.001 (4) | 0.034 (5) |
| C5A | 0.0693 (11) | 0.073 (2) | 0.0847 (19) | 0.0286 (12) | 0.0052 (10) | 0.0195 (13) |
| C5B | 0.127 (11) | 0.033 (4) | 0.057 (7) | 0.005 (5) | 0.009 (6) | 0.004 (4) |
| C6A | 0.0563 (9) | 0.0718 (17) | 0.0878 (14) | 0.0193 (10) | 0.0182 (9) | 0.0182 (12) |
| C6B | 0.061 (6) | 0.049 (7) | 0.083 (7) | 0.030 (5) | 0.015 (5) | 0.008 (5) |
| C7A | 0.0448 (8) | 0.0537 (13) | 0.0739 (14) | 0.0065 (6) | 0.0132 (7) | 0.0181 (11) |
| C7B | 0.068 (5) | 0.047 (7) | 0.063 (6) | 0.010 (4) | 0.027 (4) | 0.016 (6) |
| C8 | 0.0571 (5) | 0.0483 (5) | 0.0280 (4) | 0.0081 (4) | 0.0084 (3) | 0.0011 (3) |
Geometric parameters (Å, º) top
| O1—C2 | 1.4346 (11) | C4A—H4A | 0.9700 |
| O1—H1 | 0.8200 | C4A—H4B | 0.9700 |
| O2—C8 | 1.2055 (13) | C4B—C5B | 1.520 (10) |
| O3—C8 | 1.3147 (13) | C4B—H4C | 0.9700 |
| O3—H3 | 0.8200 | C4B—H4D | 0.9700 |
| C1—C7B | 1.474 (10) | C5A—C6A | 1.505 (3) |
| C1—C8 | 1.5167 (13) | C5A—H5A | 0.9700 |
| C1—C2 | 1.5278 (11) | C5A—H5B | 0.9700 |
| C1—C7A | 1.541 (2) | C5B—C6B | 1.510 (11) |
| C1—H1A | 0.9800 | C5B—H5C | 0.9700 |
| C1—H1B | 0.9800 | C5B—H5D | 0.9700 |
| C2—C3B | 1.504 (9) | C6A—C7A | 1.519 (3) |
| C2—C3A | 1.5289 (19) | C6A—H6A | 0.9700 |
| C2—H2A | 0.9800 | C6A—H6B | 0.9700 |
| C2—H2B | 0.9800 | C6B—C7B | 1.504 (10) |
| C3A—C4A | 1.533 (3) | C6B—H6C | 0.9700 |
| C3A—H3A | 0.9700 | C6B—H6D | 0.9700 |
| C3A—H3B | 0.9700 | C7A—H7A | 0.9700 |
| C3B—C4B | 1.498 (10) | C7A—H7B | 0.9700 |
| C3B—H3C | 0.9700 | C7B—H7C | 0.9700 |
| C3B—H3D | 0.9700 | C7B—H7D | 0.9700 |
| C4A—C5A | 1.537 (4) | | |
| | | |
| C2—O1—H1 | 109.5 | C5B—C4B—H4C | 108.0 |
| C8—O3—H3 | 109.5 | C3B—C4B—H4D | 108.0 |
| C7B—C1—C8 | 117.0 (6) | C5B—C4B—H4D | 108.0 |
| C7B—C1—C2 | 120.3 (7) | H4C—C4B—H4D | 107.2 |
| C8—C1—C2 | 109.53 (7) | C6A—C5A—C4A | 117.7 (2) |
| C8—C1—C7A | 111.43 (13) | C6A—C5A—H5A | 107.9 |
| C2—C1—C7A | 113.72 (16) | C4A—C5A—H5A | 107.9 |
| C8—C1—H1A | 107.3 | C6A—C5A—H5B | 107.9 |
| C2—C1—H1A | 107.3 | C4A—C5A—H5B | 107.9 |
| C7A—C1—H1A | 107.3 | H5A—C5A—H5B | 107.2 |
| C7B—C1—H1B | 102.2 | C6B—C5B—C4B | 116.3 (9) |
| C8—C1—H1B | 102.2 | C6B—C5B—H5C | 108.2 |
| C2—C1—H1B | 102.2 | C4B—C5B—H5C | 108.2 |
| O1—C2—C3B | 117.4 (8) | C6B—C5B—H5D | 108.2 |
| O1—C2—C3A | 108.86 (13) | C4B—C5B—H5D | 108.2 |
| O1—C2—C1 | 107.60 (7) | H5C—C5B—H5D | 107.4 |
| C3B—C2—C1 | 116.6 (6) | C5A—C6A—C7A | 116.6 (2) |
| C3A—C2—C1 | 114.16 (9) | C5A—C6A—H6A | 108.1 |
| O1—C2—H2A | 108.7 | C7A—C6A—H6A | 108.1 |
| C3A—C2—H2A | 108.7 | C5A—C6A—H6B | 108.1 |
| C1—C2—H2A | 108.7 | C7A—C6A—H6B | 108.1 |
| O1—C2—H2B | 104.6 | H6A—C6A—H6B | 107.3 |
| C3B—C2—H2B | 104.6 | C5B—C6B—C7B | 115.7 (10) |
| C1—C2—H2B | 104.6 | C5B—C6B—H6C | 108.4 |
| C2—C3A—C4A | 117.40 (16) | C7B—C6B—H6C | 108.4 |
| C2—C3A—H3A | 108.0 | C5B—C6B—H6D | 108.4 |
| C4A—C3A—H3A | 108.0 | C7B—C6B—H6D | 108.4 |
| C2—C3A—H3B | 108.0 | H6C—C6B—H6D | 107.4 |
| C4A—C3A—H3B | 108.0 | C6A—C7A—C1 | 112.6 (2) |
| H3A—C3A—H3B | 107.2 | C6A—C7A—H7A | 109.1 |
| C4B—C3B—C2 | 115.3 (8) | C1—C7A—H7A | 109.1 |
| C4B—C3B—H3C | 108.4 | C6A—C7A—H7B | 109.1 |
| C2—C3B—H3C | 108.4 | C1—C7A—H7B | 109.1 |
| C4B—C3B—H3D | 108.4 | H7A—C7A—H7B | 107.8 |
| C2—C3B—H3D | 108.4 | C1—C7B—C6B | 116.3 (11) |
| H3C—C3B—H3D | 107.5 | C1—C7B—H7C | 108.2 |
| C5A—C4A—C3A | 112.6 (2) | C6B—C7B—H7C | 108.2 |
| C5A—C4A—H4A | 109.1 | C1—C7B—H7D | 108.2 |
| C3A—C4A—H4A | 109.1 | C6B—C7B—H7D | 108.2 |
| C5A—C4A—H4B | 109.1 | H7C—C7B—H7D | 107.4 |
| C3A—C4A—H4B | 109.1 | O2—C8—O3 | 122.52 (9) |
| H4A—C4A—H4B | 107.8 | O2—C8—C1 | 124.78 (9) |
| C3B—C4B—C5B | 117.3 (11) | O3—C8—C1 | 112.71 (8) |
| C3B—C4B—H4C | 108.0 | | |
| | | |
| C7B—C1—C2—O1 | 87.1 (7) | C4A—C5A—C6A—C7A | 49.4 (4) |
| C8—C1—C2—O1 | −52.91 (10) | C4B—C5B—C6B—C7B | 5 (2) |
| C7A—C1—C2—O1 | 72.49 (15) | C5A—C6A—C7A—C1 | −67.1 (4) |
| C7B—C1—C2—C3B | −47.3 (10) | C8—C1—C7A—C6A | −144.1 (2) |
| C8—C1—C2—C3B | 172.7 (8) | C2—C1—C7A—C6A | 91.6 (2) |
| C8—C1—C2—C3A | −173.84 (16) | C8—C1—C7B—C6B | −144.8 (10) |
| C7A—C1—C2—C3A | −48.4 (2) | C2—C1—C7B—C6B | 78.1 (15) |
| O1—C2—C3A—C4A | −152.4 (2) | C5B—C6B—C7B—C1 | −70 (2) |
| C1—C2—C3A—C4A | −32.2 (3) | C7B—C1—C8—O2 | −15.8 (9) |
| O1—C2—C3B—C4B | −80.8 (14) | C2—C1—C8—O2 | 125.63 (10) |
| C1—C2—C3B—C4B | 49.0 (17) | C7A—C1—C8—O2 | −1.08 (19) |
| C2—C3A—C4A—C5A | 84.5 (3) | C7B—C1—C8—O3 | 163.9 (8) |
| C2—C3B—C4B—C5B | −82.9 (17) | C2—C1—C8—O3 | −54.66 (10) |
| C3A—C4A—C5A—C6A | −68.9 (3) | C7A—C1—C8—O3 | 178.62 (16) |
| C3B—C4B—C5B—C6B | 67.7 (19) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O2i | 0.82 | 2.00 | 2.7923 (12) | 162 |
| O3—H3···O1ii | 0.82 | 1.83 | 2.6425 (11) | 170 |
| Symmetry codes: (i) x, −y, z+1/2; (ii) −x, −y, −z. |
 O2=C bonds, whereas in 7C they are joined by a pair of O3—H
O2=C bonds, whereas in 7C they are joined by a pair of O3—H O1-H bonds. In 6C 60% of the disordered
O1-H bonds. In 6C 60% of the disordered 
 journal menu
journal menu











