Buy article online - an online subscription or single-article purchase is required to access this article.
In the crystal structure of the title compound, C
14H
20ClNO
3·C
2H
2O
4·H
2O, there are infinitive flat ribbons, parallel to the [0
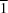
1] direction. Molecules in the ribbons are linked together by an extensive network of hydrogen bonds.
Supporting information
CCDC reference: 209991
Key indicators
- Single-crystal X-ray study
- T = 120 K
- Mean
![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.002 Å
(C-C) = 0.002 Å
- R factor = 0.033
- wR factor = 0.085
- Data-to-parameter ratio = 14.4
checkCIF results
No syntax errors found
ADDSYM reports no extra symmetry
 Alert Level A:
Alert Level A:
PLAT_353 Alert A Long N-H Bond (0.87A) N1 - H7O = 1.34 Ang.
 Alert Level C:
Alert Level C:
PLAT_369 Alert C Long C(sp2)-C(sp2) Bond C15 - C16 = 1.55 Ang.
1 Alert Level A = Potentially serious problem
0 Alert Level B = Potential problem
1 Alert Level C = Please check
Crystals of (III) were obtained by slow evaporation from a solution in Et2O at 277 K.
Data collection: SMART (Bruker, 1997); cell refinement: SMART; data reduction: SHELXTL (Bruker, 1997); program(s) used to solve structure: SHELXS97 (Sheldrick, 1990); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Methyl (±)-(1
α,2
β,8
α,9
α,10
β)-2-chloro-3-oxa- 4-azatetracyclo[8.4.0
2,90
4,8]tetradecane-9-carboxylate oxalic acid monohydrate
top
Crystal data top
| C14H20ClNO3·C2H2O4·H2O | Z = 2 |
| Mr = 393.81 | F(000) = 416 |
| Triclinic, P1 | Dx = 1.428 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 6.6133 (2) Å | Cell parameters from 999 reflections |
| b = 8.0737 (2) Å | θ = 10.3–20.8° |
| c = 17.8848 (5) Å | µ = 0.25 mm−1 |
| α = 77.746 (1)° | T = 120 K |
| β = 83.211 (1)° | Block, colourless |
| γ = 80.169 (1)° | 0.42 × 0.35 × 0.17 mm |
| V = 916.14 (4) Å3 | |
Data collection top
Bruker SMART-CCD 1K
diffractometer | 3835 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.026 |
| Graphite monochromator | θmax = 29.0°, θmin = 2.3° |
| ω scans | h = −8→9 |
| 11202 measured reflections | k = −10→10 |
| 4763 independent reflections | l = −24→23 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.033 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.085 | All H-atom parameters refined |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.04P)2 + 0.3P]
where P = (Fo2 + 2Fc2)/3 |
| 4763 reflections | (Δ/σ)max < 0.001 |
| 331 parameters | Δρmax = 0.39 e Å−3 |
| 0 restraints | Δρmin = −0.28 e Å−3 |
Crystal data top
| C14H20ClNO3·C2H2O4·H2O | γ = 80.169 (1)° |
| Mr = 393.81 | V = 916.14 (4) Å3 |
| Triclinic, P1 | Z = 2 |
| a = 6.6133 (2) Å | Mo Kα radiation |
| b = 8.0737 (2) Å | µ = 0.25 mm−1 |
| c = 17.8848 (5) Å | T = 120 K |
| α = 77.746 (1)° | 0.42 × 0.35 × 0.17 mm |
| β = 83.211 (1)° | |
Data collection top
Bruker SMART-CCD 1K
diffractometer | 3835 reflections with I > 2σ(I) |
| 11202 measured reflections | Rint = 0.026 |
| 4763 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.033 | 0 restraints |
| wR(F2) = 0.085 | All H-atom parameters refined |
| S = 1.02 | Δρmax = 0.39 e Å−3 |
| 4763 reflections | Δρmin = −0.28 e Å−3 |
| 331 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Cl1 | −0.09069 (5) | 0.02471 (4) | 0.163575 (19) | 0.02746 (9) | |
| O1 | −0.45991 (14) | −0.02751 (11) | 0.22730 (5) | 0.02126 (19) | |
| O2 | −0.06398 (14) | −0.55704 (11) | 0.24933 (5) | 0.0231 (2) | |
| O3 | 0.08735 (14) | −0.35793 (12) | 0.16422 (6) | 0.0268 (2) | |
| O4 | 0.05205 (15) | 0.17594 (14) | 0.43769 (7) | 0.0320 (2) | |
| O5 | −0.23363 (15) | 0.34249 (12) | 0.47483 (6) | 0.0282 (2) | |
| O6 | −0.45876 (16) | 0.16682 (17) | 0.41118 (9) | 0.0484 (3) | |
| O7 | −0.16835 (14) | 0.03342 (12) | 0.35997 (5) | 0.02272 (19) | |
| O1W | 0.26688 (16) | 0.31727 (14) | 0.50902 (6) | 0.0253 (2) | |
| N1 | −0.41902 (16) | −0.11384 (13) | 0.30664 (6) | 0.0183 (2) | |
| C1 | −0.62385 (19) | −0.10868 (17) | 0.35159 (8) | 0.0214 (2) | |
| C2 | −0.71691 (19) | −0.25532 (18) | 0.33415 (8) | 0.0227 (3) | |
| C3 | −0.52807 (19) | −0.39647 (16) | 0.33350 (8) | 0.0204 (2) | |
| C4 | −0.34408 (18) | −0.29981 (15) | 0.30182 (7) | 0.0167 (2) | |
| C5 | −0.26763 (17) | −0.29163 (15) | 0.21625 (7) | 0.0160 (2) | |
| C6 | −0.42686 (18) | −0.30992 (16) | 0.15951 (7) | 0.0176 (2) | |
| C7 | −0.4024 (2) | −0.46438 (17) | 0.12020 (8) | 0.0215 (3) | |
| C8 | −0.2486 (2) | −0.45580 (18) | 0.04882 (8) | 0.0245 (3) | |
| C9 | −0.2821 (2) | −0.27937 (19) | −0.00365 (8) | 0.0274 (3) | |
| C10 | −0.2398 (2) | −0.14239 (18) | 0.03772 (8) | 0.0244 (3) | |
| C11 | −0.3909 (2) | −0.13198 (17) | 0.10907 (7) | 0.0212 (2) | |
| C12 | −0.29973 (19) | −0.09861 (16) | 0.17885 (7) | 0.0195 (2) | |
| C13 | −0.05962 (18) | −0.40003 (16) | 0.20571 (7) | 0.0179 (2) | |
| C14 | 0.1218 (2) | −0.68069 (18) | 0.24252 (9) | 0.0253 (3) | |
| C15 | −0.14498 (19) | 0.22915 (16) | 0.44224 (7) | 0.0206 (2) | |
| C16 | −0.2716 (2) | 0.13487 (17) | 0.40170 (8) | 0.0232 (3) | |
| H011 | −0.593 (2) | −0.134 (2) | 0.4052 (10) | 0.023 (4)* | |
| H012 | −0.704 (3) | 0.003 (2) | 0.3391 (9) | 0.026 (4)* | |
| H21 | −0.773 (2) | −0.2169 (19) | 0.2834 (9) | 0.021 (4)* | |
| H22 | −0.825 (3) | −0.289 (2) | 0.3741 (11) | 0.037 (5)* | |
| H31 | −0.508 (2) | −0.458 (2) | 0.3860 (10) | 0.024 (4)* | |
| H32 | −0.540 (2) | −0.483 (2) | 0.3035 (10) | 0.026 (4)* | |
| H4 | −0.231 (2) | −0.341 (2) | 0.3330 (9) | 0.021 (4)* | |
| H6 | −0.562 (2) | −0.2980 (19) | 0.1868 (9) | 0.018 (4)* | |
| H71 | −0.535 (3) | −0.466 (2) | 0.1042 (10) | 0.029 (4)* | |
| H72 | −0.370 (2) | −0.572 (2) | 0.1588 (9) | 0.023 (4)* | |
| H81 | −0.108 (2) | −0.4761 (19) | 0.0627 (9) | 0.020 (4)* | |
| H82 | −0.264 (2) | −0.547 (2) | 0.0234 (10) | 0.027 (4)* | |
| H91 | −0.194 (3) | −0.277 (2) | −0.0507 (11) | 0.035 (5)* | |
| H92 | −0.422 (3) | −0.253 (2) | −0.0200 (10) | 0.031 (4)* | |
| H101 | −0.103 (2) | −0.1732 (19) | 0.0534 (9) | 0.021 (4)* | |
| H102 | −0.248 (3) | −0.027 (2) | 0.0034 (11) | 0.038 (5)* | |
| H11 | −0.514 (3) | −0.054 (2) | 0.0970 (9) | 0.026 (4)* | |
| H141 | 0.167 (3) | −0.678 (2) | 0.1889 (11) | 0.038 (5)* | |
| H142 | 0.223 (3) | −0.657 (2) | 0.2676 (10) | 0.033 (4)* | |
| H143 | 0.081 (3) | −0.793 (2) | 0.2680 (11) | 0.037 (5)* | |
| H1W | 0.243 (3) | 0.426 (3) | 0.5090 (12) | 0.051 (6)* | |
| H2W | 0.381 (4) | 0.294 (3) | 0.4852 (13) | 0.050 (6)* | |
| H4O | 0.116 (4) | 0.236 (3) | 0.4653 (14) | 0.069 (7)* | |
| H7O | −0.287 (4) | −0.037 (3) | 0.3342 (14) | 0.071 (7)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cl1 | 0.03382 (18) | 0.02492 (16) | 0.02669 (17) | −0.01474 (13) | 0.00010 (12) | −0.00510 (12) |
| O1 | 0.0257 (5) | 0.0189 (4) | 0.0179 (4) | 0.0015 (3) | −0.0037 (3) | −0.0036 (3) |
| O2 | 0.0191 (4) | 0.0190 (4) | 0.0278 (5) | 0.0010 (3) | 0.0028 (3) | −0.0026 (4) |
| O3 | 0.0175 (4) | 0.0281 (5) | 0.0319 (5) | −0.0036 (4) | 0.0024 (4) | −0.0018 (4) |
| O4 | 0.0212 (5) | 0.0402 (6) | 0.0435 (6) | −0.0067 (4) | −0.0001 (4) | −0.0278 (5) |
| O5 | 0.0317 (5) | 0.0255 (5) | 0.0307 (5) | −0.0002 (4) | −0.0036 (4) | −0.0157 (4) |
| O6 | 0.0203 (5) | 0.0540 (7) | 0.0859 (10) | −0.0025 (5) | 0.0006 (5) | −0.0524 (7) |
| O7 | 0.0219 (4) | 0.0241 (5) | 0.0259 (5) | −0.0050 (3) | 0.0004 (4) | −0.0133 (4) |
| O1W | 0.0250 (5) | 0.0259 (5) | 0.0283 (5) | −0.0052 (4) | −0.0010 (4) | −0.0121 (4) |
| N1 | 0.0203 (5) | 0.0185 (5) | 0.0167 (5) | −0.0019 (4) | −0.0026 (4) | −0.0050 (4) |
| C1 | 0.0191 (6) | 0.0230 (6) | 0.0219 (6) | 0.0004 (5) | −0.0010 (5) | −0.0075 (5) |
| C2 | 0.0173 (6) | 0.0301 (7) | 0.0222 (6) | −0.0043 (5) | −0.0008 (5) | −0.0084 (5) |
| C3 | 0.0214 (6) | 0.0204 (6) | 0.0201 (6) | −0.0056 (5) | −0.0002 (5) | −0.0048 (5) |
| C4 | 0.0167 (5) | 0.0163 (5) | 0.0179 (6) | −0.0010 (4) | −0.0032 (4) | −0.0052 (4) |
| C5 | 0.0145 (5) | 0.0167 (5) | 0.0173 (5) | −0.0030 (4) | −0.0019 (4) | −0.0040 (4) |
| C6 | 0.0149 (5) | 0.0220 (6) | 0.0173 (6) | −0.0031 (4) | −0.0022 (4) | −0.0061 (5) |
| C7 | 0.0200 (6) | 0.0256 (6) | 0.0220 (6) | −0.0063 (5) | −0.0016 (5) | −0.0094 (5) |
| C8 | 0.0229 (6) | 0.0298 (7) | 0.0233 (6) | −0.0040 (5) | 0.0005 (5) | −0.0122 (5) |
| C9 | 0.0293 (7) | 0.0361 (8) | 0.0186 (6) | −0.0058 (6) | −0.0015 (5) | −0.0089 (6) |
| C10 | 0.0281 (7) | 0.0281 (7) | 0.0174 (6) | −0.0060 (5) | −0.0027 (5) | −0.0030 (5) |
| C11 | 0.0229 (6) | 0.0219 (6) | 0.0187 (6) | −0.0008 (5) | −0.0053 (5) | −0.0039 (5) |
| C12 | 0.0214 (6) | 0.0183 (6) | 0.0193 (6) | −0.0045 (4) | −0.0017 (4) | −0.0037 (5) |
| C13 | 0.0165 (5) | 0.0208 (6) | 0.0182 (6) | −0.0032 (4) | −0.0038 (4) | −0.0059 (5) |
| C14 | 0.0206 (6) | 0.0223 (7) | 0.0317 (7) | 0.0018 (5) | −0.0012 (5) | −0.0069 (5) |
| C15 | 0.0231 (6) | 0.0203 (6) | 0.0196 (6) | −0.0047 (5) | 0.0010 (5) | −0.0066 (5) |
| C16 | 0.0215 (6) | 0.0215 (6) | 0.0292 (7) | −0.0031 (5) | −0.0018 (5) | −0.0110 (5) |
Geometric parameters (Å, º) top
| Cl1—C12 | 1.801 (1) | C4—C5 | 1.545 (2) |
| O1—C12 | 1.410 (2) | C4—H4 | 0.96 (2) |
| O1—N1 | 1.476 (1) | C5—C13 | 1.514 (2) |
| O2—C13 | 1.343 (1) | C5—C12 | 1.549 (2) |
| O2—C14 | 1.456 (2) | C5—C6 | 1.590 (2) |
| O3—C13 | 1.203 (1) | C6—C7 | 1.534 (2) |
| O4—C15 | 1.299 (2) | C6—C11 | 1.565 (2) |
| O4—H4O | 0.94 (3) | C6—H6 | 0.96 (2) |
| O5—C15 | 1.218 (2) | C7—C8 | 1.533 (2) |
| O6—C16 | 1.219 (2) | C7—H71 | 0.96 (2) |
| O7—C16 | 1.282 (2) | C7—H72 | 1.00 (2) |
| O7—H7O | 1.22 (2) | C8—C9 | 1.527 (2) |
| O1W—H1W | 0.86 (2) | C8—H81 | 0.97 (2) |
| O1W—H2W | 0.83 (2) | C8—H82 | 0.97 (2) |
| N1—C1 | 1.490 (2) | C9—C10 | 1.533 (2) |
| N1—C4 | 1.516 (2) | C9—H91 | 0.96 (2) |
| N1—H7O | 1.34 (2) | C9—H92 | 0.98 (2) |
| C1—C2 | 1.526 (2) | C10—C11 | 1.533 (2) |
| C1—H011 | 0.98 (2) | C10—H101 | 0.95 (2) |
| C1—H012 | 0.96 (2) | C10—H102 | 1.00 (2) |
| C2—C3 | 1.541 (2) | C11—C12 | 1.540 (2) |
| C2—H21 | 0.99 (2) | C11—H11 | 0.96 (2) |
| C2—H22 | 0.98 (2) | C14—H141 | 0.97 (2) |
| C3—C4 | 1.544 (2) | C14—H142 | 0.92 (2) |
| C3—H31 | 0.98 (2) | C14—H143 | 0.99 (2) |
| C3—H32 | 0.98 (2) | C15—C16 | 1.550 (2) |
| | | |
| C12—O1—N1 | 106.47 (8) | C6—C7—H71 | 106.0 (10) |
| C13—O2—C14 | 115.94 (10) | C8—C7—H72 | 112.1 (9) |
| C15—O4—H4O | 109.2 (14) | C6—C7—H72 | 109.4 (9) |
| C16—O7—H7O | 109.4 (11) | H71—C7—H72 | 106.6 (13) |
| H1W—O1W—H2W | 109 (2) | C9—C8—C7 | 110.94 (11) |
| O1—N1—C1 | 105.85 (9) | C9—C8—H81 | 107.4 (9) |
| O1—N1—C4 | 104.78 (8) | C7—C8—H81 | 111.3 (9) |
| C1—N1—C4 | 106.68 (9) | C9—C8—H82 | 112.5 (10) |
| O1—N1—H7O | 110.8 (10) | C7—C8—H82 | 107.6 (10) |
| C1—N1—H7O | 113.1 (10) | H81—C8—H82 | 107.1 (13) |
| C4—N1—H7O | 114.9 (10) | C8—C9—C10 | 109.93 (11) |
| N1—C1—C2 | 105.03 (10) | C8—C9—H91 | 111.0 (10) |
| N1—C1—H011 | 104.9 (9) | C10—C9—H91 | 109.8 (11) |
| C2—C1—H011 | 109.9 (9) | C8—C9—H92 | 110.7 (10) |
| N1—C1—H012 | 110.4 (10) | C10—C9—H92 | 110.6 (10) |
| C2—C1—H012 | 115.3 (10) | H91—C9—H92 | 104.6 (15) |
| H011—C1—H012 | 110.6 (13) | C11—C10—C9 | 110.96 (11) |
| C1—C2—C3 | 101.90 (10) | C11—C10—H101 | 108.7 (9) |
| C1—C2—H21 | 108.3 (9) | C9—C10—H101 | 108.3 (9) |
| C3—C2—H21 | 112.4 (9) | C11—C10—H102 | 109.3 (10) |
| C1—C2—H22 | 110.4 (11) | C9—C10—H102 | 112.1 (11) |
| C3—C2—H22 | 112.5 (10) | H101—C10—H102 | 107.3 (14) |
| H21—C2—H22 | 110.8 (14) | C10—C11—C12 | 115.46 (11) |
| C2—C3—C4 | 105.01 (10) | C10—C11—C6 | 114.49 (11) |
| C2—C3—H31 | 109.5 (9) | C12—C11—C6 | 86.74 (9) |
| C4—C3—H31 | 109.3 (9) | C10—C11—H11 | 111.7 (10) |
| C2—C3—H32 | 114.2 (9) | C12—C11—H11 | 111.6 (10) |
| C4—C3—H32 | 112.0 (9) | C6—C11—H11 | 114.8 (10) |
| H31—C3—H32 | 106.8 (13) | O1—C12—C11 | 109.23 (10) |
| N1—C4—C3 | 106.00 (9) | O1—C12—C5 | 103.35 (9) |
| N1—C4—C5 | 103.60 (9) | C11—C12—C5 | 92.32 (9) |
| C3—C4—C5 | 117.43 (10) | O1—C12—Cl1 | 109.52 (8) |
| N1—C4—H4 | 108.3 (9) | C11—C12—Cl1 | 119.37 (9) |
| C3—C4—H4 | 110.9 (9) | C5—C12—Cl1 | 121.05 (9) |
| C5—C4—H4 | 109.9 (9) | O3—C13—O2 | 123.94 (11) |
| C13—C5—C4 | 112.30 (10) | O3—C13—C5 | 127.02 (11) |
| C13—C5—C12 | 120.05 (10) | O2—C13—C5 | 109.02 (10) |
| C4—C5—C12 | 105.36 (9) | O2—C14—H141 | 109.6 (11) |
| C13—C5—C6 | 113.31 (10) | O2—C14—H142 | 110.3 (11) |
| C4—C5—C6 | 117.89 (9) | H141—C14—H142 | 110.4 (15) |
| C12—C5—C6 | 85.55 (9) | O2—C14—H143 | 104.8 (10) |
| C7—C6—C11 | 118.41 (10) | H141—C14—H143 | 111.8 (15) |
| C7—C6—C5 | 122.38 (10) | H142—C14—H143 | 109.8 (15) |
| C11—C6—C5 | 89.80 (9) | O5—C15—O4 | 126.07 (12) |
| C7—C6—H6 | 108.7 (9) | O5—C15—C16 | 119.35 (11) |
| C11—C6—H6 | 108.9 (9) | O4—C15—C16 | 114.58 (11) |
| C5—C6—H6 | 106.9 (9) | O6—C16—O7 | 126.40 (12) |
| C8—C7—C6 | 114.62 (11) | O6—C16—C15 | 117.18 (12) |
| C8—C7—H71 | 107.7 (10) | O7—C16—C15 | 116.39 (11) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O7—H7O···N1 | 1.22 (2) | 1.34 (2) | 2.5690 (14) | 179 (2) |
| O4—H4O···O1W | 0.94 (3) | 1.63 (3) | 2.5589 (14) | 169 (2) |
| O1W—H1W···O5i | 0.86 (2) | 1.94 (2) | 2.7917 (14) | 168 (2) |
| O1W—H2W···O6ii | 0.83 (2) | 1.95 (2) | 2.6886 (15) | 147 (2) |
| Symmetry codes: (i) −x, −y+1, −z+1; (ii) x+1, y, z. |
Experimental details
| Crystal data |
| Chemical formula | C14H20ClNO3·C2H2O4·H2O |
| Mr | 393.81 |
| Crystal system, space group | Triclinic, P1 |
| Temperature (K) | 120 |
| a, b, c (Å) | 6.6133 (2), 8.0737 (2), 17.8848 (5) |
| α, β, γ (°) | 77.746 (1), 83.211 (1), 80.169 (1) |
| V (Å3) | 916.14 (4) |
| Z | 2 |
| Radiation type | Mo Kα |
| µ (mm−1) | 0.25 |
| Crystal size (mm) | 0.42 × 0.35 × 0.17 |
| |
| Data collection |
| Diffractometer | Bruker SMART-CCD 1K
diffractometer |
| Absorption correction | – |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 11202, 4763, 3835 |
| Rint | 0.026 |
| (sin θ/λ)max (Å−1) | 0.682 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.033, 0.085, 1.02 |
| No. of reflections | 4763 |
| No. of parameters | 331 |
| H-atom treatment | All H-atom parameters refined |
| Δρmax, Δρmin (e Å−3) | 0.39, −0.28 |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O7—H7O···N1 | 1.22 (2) | 1.34 (2) | 2.5690 (14) | 179 (2) |
| O4—H4O···O1W | 0.94 (3) | 1.63 (3) | 2.5589 (14) | 169 (2) |
| O1W—H1W···O5i | 0.86 (2) | 1.94 (2) | 2.7917 (14) | 168 (2) |
| O1W—H2W···O6ii | 0.83 (2) | 1.95 (2) | 2.6886 (15) | 147 (2) |
| Symmetry codes: (i) −x, −y+1, −z+1; (ii) x+1, y, z. |

 Subscribe to Acta Crystallographica Section E: Crystallographic Communications
Subscribe to Acta Crystallographica Section E: Crystallographic Communications
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.
![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.002 Å
(C-C) = 0.002 ÅAlert Level A:
Alert Level C:

 Subscribe to Acta Crystallographica Section E: Crystallographic Communications
Subscribe to Acta Crystallographica Section E: Crystallographic Communications
 journal menu
journal menu











![[Figure 1]](http://journals.iucr.org/e/issues/2003/04/00/cv6174/cv6174fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/e/issues/2003/04/00/cv6174/cv6174fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/e/issues/2003/04/00/cv6174/cv6174fig3thm.gif)




The tetracyclic title compound, (III), was obtained upon an attempted 1,3-dipolar cycloaddition of 3,4-dihydro-2H-pyrrole 1-oxide, (I), to methyl 2-chloro-2-(bicyclo[4.1.0]hept-7-ylidene)acetate, (II) (Brandi et al., 2003), during systematic studies of 1,3-dipolar cycloadditions to substituted methylenecyclopropanes (Goti et al., 1996; de Meijere, Kozhushkov & Khlebnikov, 2000; de Meijere & Kozhushkov, 2000). The relative configuration of (III) was established by X-ray crystal structure analysis of its solvate with oxalic acid and water (1:1:1) (IV).
Chlorine and methoxycarbonyl substitutents at the two bridgehead atoms of the tetracyclic system are cis-oriented with respect to each other (Fig. 1).
All four cycles are substantially non-planar. The four-membered ring is folded around the C6···C12 line by 14.7(?)°. The cyclohexane ring has a slightly flattened chair conformation, while the five-membered heterocycles adopt conformations of O1-envelope and twisted half-chair. The solvent molecule of oxalic acid is also not entirely plana, with a torsion angle O5—C15—C16—O6 of −7.5(?)°.
The most remarkable feature of the studied structure is the packing of the molecules in the crystal. The molecles of oxalic acid and water form flat ribbons; the tetracyclic molecules are attached to both edges of the ribbons by very strong hydrogen bonds between oxalic acid and the tetradecane N atom (Fig. 2). The molecules of oxalic acid and water are linked together by a network of O—H···O hydrogen bonds. Tetracyclic molecules in the same ribbon are also connected by C6—H6···O3 [C···O = 3.290 (1) Å] interactions. In the crystal structure, these ribbons are parallel to the [011] direction and show a number of weak C—H···O interactions.