Buy article online - an online subscription or single-article purchase is required to access this article.
The title compound, bis(tetra-
n-butylammonium) tetrachlorotetra-μ
3-sulfidotetracopper(I)molybdenum(VI), (C
16H
36N)
2[MoS
4(CuCl)
4], has been obtained in a second polymorphic form. The anion, in which four of the six S

S edges of the central MoS
4 tetrahedron are bridged by CuCl neutral molecular units to give a planar pentanuclear MoCu
4 framework, is disordered on a crystallographic fourfold rotation axis, requiring equal occupancy of two sets of four positions for the S atoms. There is additional disorder as a result of 8.2 (2)% substitution of Cl by Br, arising from tetra-
n-butylammonium bromide used in the synthesis. Copper has distorted trigonal planar coordination, involving two μ
3-S atoms and a terminal halogen atom. The two independent cations lie on positions of symmetry
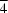
.
Supporting information
CCDC reference: 234818
Key indicators
- Single-crystal X-ray study
- T = 150 K
- Mean
![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.003 Å
(C-C) = 0.003 Å
- R factor = 0.024
- wR factor = 0.058
- Data-to-parameter ratio = 23.5
checkCIF/PLATON results
No syntax errors found
 Alert level B
ABSTM02_ALERT_3_B The ratio of expected to reported Tmax/Tmin(RR) is > 1.50
Tmin and Tmax reported: 0.701 0.876
Tmin and Tmax expected: 0.363 0.876
RR = 1.932
Please check that your absorption correction is appropriate.
PLAT060_ALERT_3_B Ratio Tmax/Tmin (Exp-to-Rep) (too) Large ....... 1.94
Alert level B
ABSTM02_ALERT_3_B The ratio of expected to reported Tmax/Tmin(RR) is > 1.50
Tmin and Tmax reported: 0.701 0.876
Tmin and Tmax expected: 0.363 0.876
RR = 1.932
Please check that your absorption correction is appropriate.
PLAT060_ALERT_3_B Ratio Tmax/Tmin (Exp-to-Rep) (too) Large ....... 1.94
 Alert level C
PLAT042_ALERT_1_C Calc. and Rep. MoietyFormula Strings Differ .... ?
PLAT068_ALERT_1_C Reported F000 Differs from Calcd (or Missing)... ?
PLAT077_ALERT_4_C Unitcell contains non-integer number of atoms .. ?
PLAT125_ALERT_4_C No _symmetry_space_group_name_Hall Given ....... ?
PLAT301_ALERT_3_C Main Residue Disorder ......................... 10.00 Perc.
Alert level C
PLAT042_ALERT_1_C Calc. and Rep. MoietyFormula Strings Differ .... ?
PLAT068_ALERT_1_C Reported F000 Differs from Calcd (or Missing)... ?
PLAT077_ALERT_4_C Unitcell contains non-integer number of atoms .. ?
PLAT125_ALERT_4_C No _symmetry_space_group_name_Hall Given ....... ?
PLAT301_ALERT_3_C Main Residue Disorder ......................... 10.00 Perc.
0 ALERT level A = In general: serious problem
2 ALERT level B = Potentially serious problem
5 ALERT level C = Check and explain
0 ALERT level G = General alerts; check
2 ALERT type 1 CIF construction/syntax error, inconsistent or missing data
0 ALERT type 2 Indicator that the structure model may be wrong or deficient
3 ALERT type 3 Indicator that the structure quality may be low
2 ALERT type 4 Improvement, methodology, query or suggestion
Data collection: COLLECT (Nonius, 1998); cell refinement: EVALCCD (Duisenberg et al., 2003); data reduction: EVALCCD; program(s) used to solve structure: SHELXTL (Sheldrick, 2001); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and local programs.
Crystal data top
| (C16H36N)2[MoS4(CuCl)4] | Dx = 1.532 Mg m−3 |
| Mr = 1119.51 | Mo Kα radiation, λ = 0.71073 Å |
| Tetragonal, P4/n | Cell parameters from 86 reflections |
| a = 13.3356 (6) Å | θ = 2.5–25.0° |
| c = 13.6495 (9) Å | µ = 2.64 mm−1 |
| V = 2427.4 (2) Å3 | T = 150 K |
| Z = 2 | Plate, purple |
| F(000) = 1148 | 0.40 × 0.40 × 0.05 mm |
Data collection top
Nonius KappaCCD
diffractometer | 2796 independent reflections |
| Radiation source: sealed tube | 2191 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.031 |
| φ and ω scans | θmax = 27.5°, θmin = 4.3° |
Absorption correction: multi-scan
(SADABS; Sheldrick, 2002) | h = −17→17 |
| Tmin = 0.701, Tmax = 0.876 | k = −15→17 |
| 19584 measured reflections | l = −17→17 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.024 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.058 | H-atom parameters constrained |
| S = 1.09 | w = 1/[σ2(Fo2) + (0.0191P)2 + 1.5691P]
where P = (Fo2 + 2Fc2)/3 |
| 2796 reflections | (Δ/σ)max = 0.001 |
| 119 parameters | Δρmax = 0.46 e Å−3 |
| 0 restraints | Δρmin = −0.36 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Mo | 0.2500 | 0.2500 | 0.26798 (2) | 0.02315 (9) | |
| Cu | 0.066050 (18) | 0.319184 (19) | 0.262395 (18) | 0.02955 (8) | |
| Cl | −0.08479 (3) | 0.37625 (4) | 0.25092 (3) | 0.0390 (2) | 0.918 (2) |
| Br | −0.08479 (3) | 0.37625 (4) | 0.25092 (3) | 0.0390 (2) | 0.082 (2) |
| S | 0.19314 (8) | 0.37513 (9) | 0.17358 (8) | 0.0326 (2) | 0.50 |
| S' | 0.19175 (8) | 0.37831 (8) | 0.35937 (7) | 0.0287 (2) | 0.50 |
| N1 | 0.7500 | 0.2500 | 0.0000 | 0.0221 (7) | |
| C1 | 0.83183 (14) | 0.28816 (14) | −0.06781 (13) | 0.0254 (4) | |
| H1A | 0.8485 | 0.2339 | −0.1145 | 0.030* | |
| H1B | 0.8040 | 0.3445 | −0.1064 | 0.030* | |
| C2 | 0.92831 (15) | 0.32343 (15) | −0.02096 (15) | 0.0294 (4) | |
| H2A | 0.9630 | 0.2664 | 0.0107 | 0.035* | |
| H2B | 0.9138 | 0.3745 | 0.0297 | 0.035* | |
| C3 | 0.99475 (17) | 0.36846 (18) | −0.10127 (17) | 0.0394 (5) | |
| H3A | 1.0003 | 0.3200 | −0.1559 | 0.047* | |
| H3B | 0.9625 | 0.4299 | −0.1270 | 0.047* | |
| C4 | 1.09892 (18) | 0.39429 (19) | −0.0651 (2) | 0.0546 (7) | |
| H4A | 1.0939 | 0.4405 | −0.0095 | 0.082* | |
| H4B | 1.1370 | 0.4262 | −0.1181 | 0.082* | |
| H4C | 1.1333 | 0.3329 | −0.0444 | 0.082* | |
| N2 | 0.7500 | 0.2500 | 0.5000 | 0.0243 (7) | |
| C5 | 0.82834 (15) | 0.29530 (15) | 0.56782 (14) | 0.0273 (4) | |
| H5A | 0.7941 | 0.3422 | 0.6130 | 0.033* | |
| H5B | 0.8573 | 0.2407 | 0.6080 | 0.033* | |
| C6 | 0.91377 (16) | 0.35107 (18) | 0.51841 (16) | 0.0383 (5) | |
| H6A | 0.8874 | 0.4116 | 0.4852 | 0.046* | |
| H6B | 0.9451 | 0.3075 | 0.4683 | 0.046* | |
| C7 | 0.99263 (18) | 0.38174 (17) | 0.59482 (18) | 0.0411 (6) | |
| H7A | 1.0366 | 0.4338 | 0.5660 | 0.049* | |
| H7B | 0.9580 | 0.4119 | 0.6518 | 0.049* | |
| C8 | 1.05661 (19) | 0.2959 (2) | 0.62988 (19) | 0.0497 (7) | |
| H8A | 1.0139 | 0.2449 | 0.6604 | 0.075* | |
| H8B | 1.1053 | 0.3206 | 0.6779 | 0.075* | |
| H8C | 1.0923 | 0.2664 | 0.5741 | 0.075* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Mo | 0.02494 (12) | 0.02494 (12) | 0.01956 (16) | 0.000 | 0.000 | 0.000 |
| Cu | 0.02496 (14) | 0.03581 (15) | 0.02788 (14) | 0.00148 (11) | −0.00074 (10) | 0.00147 (11) |
| Cl | 0.0252 (3) | 0.0633 (4) | 0.0286 (3) | 0.0060 (2) | −0.00074 (18) | 0.0018 (2) |
| Br | 0.0252 (3) | 0.0633 (4) | 0.0286 (3) | 0.0060 (2) | −0.00074 (18) | 0.0018 (2) |
| S | 0.0252 (5) | 0.0437 (6) | 0.0290 (5) | −0.0016 (5) | −0.0023 (4) | 0.0128 (5) |
| S' | 0.0285 (5) | 0.0318 (6) | 0.0259 (5) | −0.0023 (4) | 0.0028 (4) | −0.0072 (4) |
| N1 | 0.0235 (10) | 0.0235 (10) | 0.0192 (15) | 0.000 | 0.000 | 0.000 |
| C1 | 0.0296 (11) | 0.0239 (10) | 0.0225 (9) | 0.0005 (8) | 0.0054 (8) | 0.0008 (8) |
| C2 | 0.0299 (11) | 0.0280 (11) | 0.0305 (10) | −0.0010 (9) | 0.0065 (9) | −0.0008 (8) |
| C3 | 0.0387 (13) | 0.0354 (13) | 0.0442 (13) | −0.0028 (10) | 0.0155 (10) | 0.0022 (10) |
| C4 | 0.0377 (14) | 0.0411 (14) | 0.085 (2) | −0.0092 (11) | 0.0174 (14) | 0.0015 (14) |
| N2 | 0.0258 (11) | 0.0258 (11) | 0.0213 (16) | 0.000 | 0.000 | 0.000 |
| C5 | 0.0301 (11) | 0.0278 (11) | 0.0240 (9) | −0.0025 (8) | −0.0051 (8) | 0.0001 (8) |
| C6 | 0.0377 (13) | 0.0427 (13) | 0.0345 (11) | −0.0124 (10) | −0.0078 (10) | 0.0089 (10) |
| C7 | 0.0422 (14) | 0.0384 (13) | 0.0428 (13) | −0.0135 (11) | −0.0107 (11) | 0.0037 (10) |
| C8 | 0.0412 (14) | 0.0592 (17) | 0.0487 (15) | −0.0090 (12) | −0.0086 (11) | 0.0062 (13) |
Geometric parameters (Å, º) top
| Mo—Cu | 2.6220 (3) | C2—H2B | 0.990 |
| Mo—Cui | 2.6220 (3) | C2—C3 | 1.532 (3) |
| Mo—Cuii | 2.6220 (3) | C3—H3A | 0.990 |
| Mo—Cuiii | 2.6220 (3) | C3—H3B | 0.990 |
| Mo—S | 2.2406 (10) | C3—C4 | 1.514 (3) |
| Mo—Si | 2.2406 (10) | C4—H4A | 0.980 |
| Mo—Sii | 2.2406 (10) | C4—H4B | 0.980 |
| Mo—Siii | 2.2406 (10) | C4—H4C | 0.980 |
| Mo—S' | 2.2556 (10) | N2—C5 | 1.5210 (18) |
| Mo—S'i | 2.2556 (10) | N2—C5iv | 1.5210 (18) |
| Mo—S'iii | 2.2556 (10) | N2—C5vii | 1.5210 (18) |
| Mo—S'ii | 2.2556 (10) | N2—C5viii | 1.5210 (18) |
| Cu—Cl | 2.1563 (5) | C5—H5A | 0.990 |
| Cu—S | 2.2134 (10) | C5—H5B | 0.990 |
| Cu—Sii | 2.2159 (11) | C5—C6 | 1.519 (3) |
| Cu—S' | 2.2768 (10) | C6—H6A | 0.990 |
| Cu—S'ii | 2.2783 (10) | C6—H6B | 0.990 |
| N1—C1 | 1.5187 (18) | C6—C7 | 1.536 (3) |
| N1—C1iv | 1.5187 (18) | C7—H7A | 0.990 |
| N1—C1v | 1.5187 (18) | C7—H7B | 0.990 |
| N1—C1vi | 1.5187 (18) | C7—C8 | 1.505 (3) |
| C1—H1A | 0.990 | C8—H8A | 0.980 |
| C1—H1B | 0.990 | C8—H8B | 0.980 |
| C1—C2 | 1.512 (3) | C8—H8C | 0.980 |
| C2—H2A | 0.990 | | |
| | | |
| Sii—Mo—Siii | 109.78 (6) | C2—C3—H3A | 109.0 |
| S—Mo—Si | 109.78 (6) | C2—C3—H3B | 109.0 |
| Si—Mo—S'iii | 108.53 (4) | C2—C3—C4 | 112.7 (2) |
| S—Mo—S'iii | 108.56 (4) | H3A—C3—H3B | 107.8 |
| Sii—Mo—S' | 108.56 (4) | H3A—C3—C4 | 109.0 |
| Siii—Mo—S' | 108.53 (4) | H3B—C3—C4 | 109.0 |
| Si—Mo—S'ii | 108.56 (4) | C3—C4—H4A | 109.5 |
| S—Mo—S'ii | 108.53 (4) | C3—C4—H4B | 109.5 |
| Sii—Mo—S'i | 108.53 (4) | C3—C4—H4C | 109.5 |
| Siii—Mo—S'i | 108.56 (4) | H4A—C4—H4B | 109.5 |
| S'iii—Mo—S'ii | 112.85 (5) | H4A—C4—H4C | 109.5 |
| S'—Mo—S'i | 112.85 (5) | H4B—C4—H4C | 109.5 |
| Cl—Cu—S | 123.75 (3) | C5—N2—C5vii | 111.74 (8) |
| Cl—Cu—Sii | 123.93 (3) | C5—N2—C5viii | 111.74 (8) |
| Cl—Cu—S' | 127.36 (3) | C5vii—N2—C5viii | 105.02 (15) |
| Cl—Cu—S'ii | 127.53 (3) | C5—N2—C5iv | 105.02 (15) |
| Sii—Cu—S' | 108.67 (4) | C5vii—N2—C5iv | 111.74 (8) |
| S—Cu—S'ii | 108.68 (4) | C5viii—N2—C5iv | 111.74 (8) |
| Mo—S—Cu | 72.12 (3) | N2—C5—H5A | 108.3 |
| Mo—S—Cuiii | 72.08 (3) | N2—C5—H5B | 108.3 |
| Cu—S—Cuiii | 113.61 (5) | N2—C5—C6 | 116.08 (15) |
| Mo—S'—Cu | 70.69 (3) | H5A—C5—H5B | 107.4 |
| Mo—S'—Cuiii | 70.66 (3) | H5A—C5—C6 | 108.3 |
| Cu—S'—Cuiii | 108.91 (4) | H5B—C5—C6 | 108.3 |
| C1iv—N1—C1vi | 111.81 (8) | C5—C6—H6A | 109.7 |
| C1—N1—C1iv | 104.90 (14) | C5—C6—H6B | 109.7 |
| C1—N1—C1vi | 111.81 (8) | C5—C6—C7 | 110.02 (18) |
| C1iv—N1—C1v | 111.81 (8) | H6A—C6—H6B | 108.2 |
| C1vi—N1—C1v | 104.90 (14) | H6A—C6—C7 | 109.7 |
| C1—N1—C1v | 111.81 (8) | H6B—C6—C7 | 109.7 |
| N1—C1—H1A | 108.0 | C6—C7—H7A | 108.8 |
| N1—C1—H1B | 108.0 | C6—C7—H7B | 108.8 |
| N1—C1—C2 | 117.25 (14) | C6—C7—C8 | 113.7 (2) |
| H1A—C1—H1B | 107.2 | H7A—C7—H7B | 107.7 |
| H1A—C1—C2 | 108.0 | H7A—C7—C8 | 108.8 |
| H1B—C1—C2 | 108.0 | H7B—C7—C8 | 108.8 |
| C1—C2—H2A | 110.1 | C7—C8—H8A | 109.5 |
| C1—C2—H2B | 110.1 | C7—C8—H8B | 109.5 |
| C1—C2—C3 | 108.14 (17) | C7—C8—H8C | 109.5 |
| H2A—C2—H2B | 108.4 | H8A—C8—H8B | 109.5 |
| H2A—C2—C3 | 110.1 | H8A—C8—H8C | 109.5 |
| H2B—C2—C3 | 110.1 | H8B—C8—H8C | 109.5 |
| | | |
| C1iv—N1—C1—C2 | −175.0 (2) | C5vii—N2—C5—C6 | 61.8 (2) |
| C1vi—N1—C1—C2 | 63.7 (2) | C5viii—N2—C5—C6 | −55.6 (2) |
| C1v—N1—C1—C2 | −53.6 (2) | C5iv—N2—C5—C6 | −176.9 (2) |
| N1—C1—C2—C3 | −174.11 (15) | N2—C5—C6—C7 | −173.47 (16) |
| C1—C2—C3—C4 | −172.65 (19) | C5—C6—C7—C8 | 74.8 (3) |
| Symmetry codes: (i) −x+1/2, −y+1/2, z; (ii) −y+1/2, x, z; (iii) y, −x+1/2, z; (iv) −x+3/2, −y+1/2, z; (v) y+1/2, −x+1, −z; (vi) −y+1, x−1/2, −z; (vii) y+1/2, −x+1, −z+1; (viii) −y+1, x−1/2, −z+1. |

 Subscribe to Acta Crystallographica Section E: Crystallographic Communications
Subscribe to Acta Crystallographica Section E: Crystallographic Communications
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.
 S edges of the central MoS4 tetrahedron are bridged by CuCl neutral molecular units to give a planar pentanuclear MoCu4 framework, is disordered on a crystallographic fourfold rotation axis, requiring equal occupancy of two sets of four positions for the S atoms. There is additional disorder as a result of 8.2 (2)% substitution of Cl by Br, arising from tetra-n-butylammonium bromide used in the synthesis. Copper has distorted trigonal planar coordination, involving two μ3-S atoms and a terminal halogen atom. The two independent cations lie on positions of symmetry
S edges of the central MoS4 tetrahedron are bridged by CuCl neutral molecular units to give a planar pentanuclear MoCu4 framework, is disordered on a crystallographic fourfold rotation axis, requiring equal occupancy of two sets of four positions for the S atoms. There is additional disorder as a result of 8.2 (2)% substitution of Cl by Br, arising from tetra-n-butylammonium bromide used in the synthesis. Copper has distorted trigonal planar coordination, involving two μ3-S atoms and a terminal halogen atom. The two independent cations lie on positions of symmetry 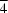 .
.  S edges of the central MoS4 tetrahedron are bridged by CuCl neutral molecular units to give a planar pentanuclear MoCu4 framework, is disordered on a crystallographic fourfold rotation axis, requiring equal occupancy of two sets of four positions for the S atoms. There is additional disorder as a result of 8.2 (2)% substitution of Cl by Br, arising from tetra-n-butylammonium bromide used in the synthesis. Copper has distorted trigonal planar coordination, involving two μ3-S atoms and a terminal halogen atom. The two independent cations lie on positions of symmetry
S edges of the central MoS4 tetrahedron are bridged by CuCl neutral molecular units to give a planar pentanuclear MoCu4 framework, is disordered on a crystallographic fourfold rotation axis, requiring equal occupancy of two sets of four positions for the S atoms. There is additional disorder as a result of 8.2 (2)% substitution of Cl by Br, arising from tetra-n-butylammonium bromide used in the synthesis. Copper has distorted trigonal planar coordination, involving two μ3-S atoms and a terminal halogen atom. The two independent cations lie on positions of symmetry 
 journal menu
journal menu











 Alert level B
Alert level B
 Alert level C
Alert level C



