Buy article online - an online subscription or single-article purchase is required to access this article.
Molecules of the title compound (alternative name
p-methoxybenzaldehyde 2-pyridylhydrazone), C
13H
13N
3O, adopt an
E configuration about the azomethine C=N double bond. Molecules are almost planar, the dihedral angle between the pyridine and methoxyphenyl rings being only 6.19 (12)°. Pairwise N—H

N hydrogen bonds [
R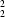
(8) in graph-set notation] link centrosymmetrically related molecules into discrete pairs.
Supporting information
CCDC reference: 211743
4-Methoxybenzaldehyde (4.10 g, 0.030 mol) was dissolved in hot methanol (22 ml), and 2-hydrazinopyridine (3.32 g, 0.030 mol) was dissolved in hot methanol (25 ml). The latter solution was added dropwise to the former at 298–303 K. The resulting yellow solid was filtered off, dried and recrystallized after refluxing in hot methanol for 4 h. The yellow crystals thus obtained were filtered off and dried in air.
H atoms were placed geometrically at distances of 0.86, 0.93 and 0.96 Å from their parent atoms for N—H, methyl C—H and sp2 C—H bonds, respectively. In refinement, a riding model was used for all H atoms with Uiso(H) equal to 1.3Ueq(C, N).
Data collection: CAD-4 EXPRESS (Enraf-Nonius, 1993); cell refinement: CAD-4 EXPRESS (Enraf-Nonius, 1993); data reduction: X-RED (Stoe & Cie, 1996); program(s) used to solve structure: SHELXS97 (Sheldrick, 1990); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: ORTEP-3 (Farrugia, 1997); software used to prepare material for publication: SHELXL97 (Sheldrick, 1997).
Crystal data top
| C13H13N3O | F(000) = 480 |
| Mr = 227.26 | Dx = 1.283 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 25 reflections |
| a = 5.5633 (16) Å | θ = 2.2–25.7° |
| b = 10.4599 (11) Å | µ = 0.09 mm−1 |
| c = 20.314 (4) Å | T = 293 K |
| β = 95.51 (2)° | Block, yellow |
| V = 1176.6 (4) Å3 | 0.30 × 0.27 × 0.21 mm |
| Z = 4 | |
Data collection top
Enraf-Nonius CAD-4
diffractometer | Rint = 0.021 |
| Radiation source: fine-focus sealed tube | θmax = 25.7°, θmin = 2.2° |
| Graphite monochromator | h = −6→0 |
| ω/2θ scans | k = 0→12 |
| 2461 measured reflections | l = −24→24 |
| 2223 independent reflections | 3 standard reflections every 120 min |
| 1177 reflections with I > 2σ(I) | intensity decay: 3% |
Refinement top
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.042 | H-atom parameters constrained |
| wR(F2) = 0.128 | w = 1/[σ2(Fo2) + (0.06P)2 + 0.057P],
where P = (Fo2 + 2Fc2)/3 |
| S = 0.99 | (Δ/σ)max < 0.001 |
| 2223 reflections | Δρmax = 0.11 e Å−3 |
| 154 parameters | Δρmin = −0.17 e Å−3 |
| Primary atom site location: structure-invariant direct methods | |
Crystal data top
| C13H13N3O | V = 1176.6 (4) Å3 |
| Mr = 227.26 | Z = 4 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 5.5633 (16) Å | µ = 0.09 mm−1 |
| b = 10.4599 (11) Å | T = 293 K |
| c = 20.314 (4) Å | 0.30 × 0.27 × 0.21 mm |
| β = 95.51 (2)° | |
Data collection top
Enraf-Nonius CAD-4
diffractometer | Rint = 0.021 |
| 2461 measured reflections | 3 standard reflections every 120 min |
| 2223 independent reflections | intensity decay: 3% |
| 1177 reflections with I > 2σ(I) | |
Refinement top
| R[F2 > 2σ(F2)] = 0.042 | 154 parameters |
| wR(F2) = 0.128 | H-atom parameters constrained |
| S = 0.99 | Δρmax = 0.11 e Å−3 |
| 2223 reflections | Δρmin = −0.17 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O | 0.0089 (3) | 0.97454 (15) | 0.34902 (7) | 0.0752 (5) | |
| N1 | 0.5950 (3) | 1.06807 (16) | 0.10031 (8) | 0.0585 (5) | |
| N2 | 0.7473 (5) | 1.06593 (16) | 0.05134 (8) | 0.0611 (5) | |
| N3 | 0.8670 (3) | 1.14892 (16) | −0.04462 (9) | 0.0595 (5) | |
| C1 | 0.4668 (3) | 0.98068 (18) | 0.19912 (9) | 0.0546 (5) | |
| C2 | 0.2753 (4) | 1.06599 (19) | 0.20170 (10) | 0.0597 (5) | |
| C3 | 0.1287 (4) | 1.0602 (2) | 0.25212 (10) | 0.0624 (6) | |
| C4 | 0.1656 (3) | 0.96965 (19) | 0.30124 (10) | 0.0561 (5) | |
| C5 | 0.3518 (4) | 0.8832 (2) | 0.29956 (10) | 0.0642 (6) | |
| C6 | 0.5000 (4) | 0.8903 (2) | 0.24861 (11) | 0.0657 (6) | |
| C7 | 0.0303 (5) | 0.8831 (2) | 0.40060 (12) | 0.0869 (8) | |
| C8 | 0.6241 (4) | 0.9844 (2) | 0.14568 (11) | 0.0597 (6) | |
| C9 | 0.7031 (3) | 1.14937 (19) | −0.00065 (10) | 0.0526 (5) | |
| C10 | 0.5007 (4) | 1.2284 (2) | −0.00650 (11) | 0.0633 (6) | |
| C11 | 0.4736 (4) | 1.3120 (2) | −0.05819 (12) | 0.0709 (6) | |
| C12 | 0.6429 (4) | 1.3167 (2) | −0.10354 (12) | 0.0718 (6) | |
| C13 | 0.8321 (4) | 1.2328 (2) | −0.09446 (11) | 0.0675 (6) | |
| H2A | 0.8673 | 1.0138 | 0.0530 | 0.073* | |
| H2 | 0.2468 | 1.1275 | 0.1688 | 0.072* | |
| H3 | 0.0025 | 1.1182 | 0.2532 | 0.075* | |
| H5 | 0.3776 | 0.8209 | 0.3322 | 0.077* | |
| H6 | 0.6262 | 0.8321 | 0.2478 | 0.079* | |
| H7A | −0.0910 | 0.8989 | 0.4302 | 0.130* | |
| H7B | 0.1875 | 0.8896 | 0.4244 | 0.130* | |
| H7C | 0.0089 | 0.7989 | 0.3822 | 0.130* | |
| H8 | 0.7475 | 0.9247 | 0.1448 | 0.072* | |
| H10 | 0.3869 | 1.2243 | 0.0241 | 0.076* | |
| H11 | 0.3404 | 1.3661 | −0.0629 | 0.085* | |
| H12 | 0.6291 | 1.3741 | −0.1386 | 0.086* | |
| H13 | 0.9453 | 1.2341 | −0.1253 | 0.081* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O | 0.067 (9) | 0.085 (1) | 0.076 (1) | 0.011 (9) | 0.0211 (8) | 0.009 (8) |
| N1 | 0.052 (1) | 0.059 (1) | 0.067 (1) | −0.005 (1) | 0.0118 (9) | −0.001 (9) |
| N2 | 0.050 (1) | 0.062 (1) | 0.073 (1) | 0.003 (1) | 0.0138 (9) | 0.009 (9) |
| N3 | 0.043 (9) | 0.060 (1) | 0.077 (1) | 0.007 (9) | 0.0101 (8) | −0.001 (8) |
| C1 | 0.051 (1) | 0.050 (1) | 0.063 (1) | −0.004 (1) | 0.0035 (1) | 0.001 (10) |
| C2 | 0.059 (1) | 0.055 (1) | 0.066 (1) | 0.003 (1) | 0.0057 (1) | 0.007 (11) |
| C3 | 0.053 (1) | 0.061 (1) | 0.074 (1) | 0.002 (1) | 0.0095 (1) | 0.010 (11) |
| C4 | 0.052 (1) | 0.056 (1) | 0.061 (1) | −0.003 (1) | 0.0053 (1) | −0.005 (11) |
| C5 | 0.066 (1) | 0.057 (1) | 0.069 (1) | 0.008 (1) | 0.0051 (1) | 0.006 (11) |
| C6 | 0.062 (1) | 0.059 (1) | 0.077 (1) | 0.004 (1) | 0.0086 (1) | 0.015 (11) |
| C7 | 0.097 (2) | 0.080 (2) | 0.087 (2) | 0.016 (2) | 0.0261 (1) | −0.008 (15) |
| C8 | 0.054 (1) | 0.056 (1) | 0.070 (1) | −0.005 (1) | 0.0092 (1) | 0.005 (10) |
| C9 | 0.041 (1) | 0.048 (1) | 0.068 (1) | −0.007 (1) | 0.0033 (1) | −0.003 (9) |
| C10 | 0.050 (1) | 0.062 (1) | 0.079 (1) | −0.002 (1) | 0.0106 (1) | 0.007 (11) |
| C11 | 0.051 (1) | 0.065 (1) | 0.096 (2) | 0.003 (1) | 0.0027 (1) | 0.010 (11) |
| C12 | 0.054 (1) | 0.065 (2) | 0.095 (2) | 0.018 (1) | 0.0008 (1) | 0.010 (11) |
| C13 | 0.047 (1) | 0.073 (2) | 0.084 (2) | 0.012 (1) | 0.0123 (1) | −0.004 (12) |
Geometric parameters (Å, º) top
| O—C4 | 1.367 (2) | C5—C6 | 1.386 (3) |
| O—C7 | 1.415 (2) | C5—H5 | 0.9300 |
| N1—C8 | 1.270 (2) | C6—H6 | 0.9300 |
| N1—N2 | 1.367 (2) | C7—H7A | 0.9600 |
| N2—C9 | 1.374 (2) | C7—H7B | 0.9600 |
| N2—H2A | 0.8600 | C7—H7C | 0.9600 |
| N3—C13 | 1.340 (2) | C8—H8 | 0.9300 |
| N3—C9 | 1.336 (2) | C9—C10 | 1.393 (3) |
| C1—C6 | 1.380 (3) | C10—C11 | 1.363 (3) |
| C1—C2 | 1.394 (3) | C10—H10 | 0.9300 |
| C1—C8 | 1.459 (3) | C11—C12 | 1.380 (3) |
| C2—C3 | 1.371 (3) | C11—H11 | 0.9300 |
| C2—H2 | 0.9300 | C12—C13 | 1.369 (3) |
| C3—C4 | 1.377 (3) | C12—H12 | 0.9300 |
| C3—H3 | 0.9300 | C13—H13 | 0.9300 |
| C4—C5 | 1.378 (3) | | |
| | | |
| C4—O—C7 | 119.11 (17) | O—C7—H7A | 109.5 |
| C8—N1—N2 | 118.18 (18) | O—C7—H7B | 109.5 |
| N1—N2—C9 | 118.12 (19) | H7A—C7—H7B | 109.5 |
| N1—N2—H2A | 120.87 | O—C7—H7C | 109.5 |
| C9—N2—H2A | 120.91 | H7A—C7—H7C | 109.5 |
| C9—N3—C13 | 116.38 (18) | H7B—C7—H7C | 109.5 |
| C6—C1—C2 | 117.42 (19) | N1—C8—C1 | 121.00 (19) |
| C6—C1—C8 | 120.84 (19) | N1—C8—H8 | 119.5 |
| C2—C1—C8 | 121.72 (19) | C1—C8—H8 | 119.5 |
| C3—C2—C1 | 120.8 (2) | N2—C9—C10 | 121.74 (18) |
| C3—C2—H2 | 119.9 | N3—C9—C10 | 122.9 (2) |
| C1—C2—H2 | 119.9 | N3—C9—N2 | 115.37 (17) |
| C2—C3—C4 | 120.9 (2) | C11—C10—C9 | 118.4 (2) |
| C2—C3—H3 | 119.6 | C11—C10—H10 | 121.3 |
| C4—C3—H3 | 119.6 | C9—C10—H10 | 121.3 |
| O—C4—C5 | 125.07 (19) | C10—C11—C12 | 120.3 (2) |
| O—C4—C3 | 115.33 (18) | C10—C11—H11 | 119.83 |
| C3—C4—C5 | 119.59 (19) | C12—C11—H11 | 119.85 |
| C4—C5—C6 | 119.08 (19) | C13—C12—C11 | 116.9 (2) |
| C4—C5—H5 | 120.5 | C13—C12—H12 | 121.8 |
| C6—C5—H5 | 120.5 | C11—C12—H12 | 121.8 |
| C1—C6—C5 | 122.2 (2) | N3—C13—C12 | 125.1 (2) |
| C1—C6—H6 | 119.0 | C12—C13—H13 | 117.48 |
| C5—C6—H6 | 119.0 | N3—C13—H13 | 117.40 |
| | | |
| C8—N1—N2—C9 | 174.6 (3) | C6—C1—C8—N1 | −179.4 (3) |
| C6—C1—C2—C3 | 0.7 (3) | C2—C1—C8—N1 | 1.5 (5) |
| C8—C1—C2—C3 | 179.5 (2) | N1—N2—C9—C10 | −4.3 (3) |
| C1—C2—C3—C4 | −0.4 (3) | N1—N2—C9—N3 | 175.66 (18) |
| C7—O—C4—C5 | −1.9 (5) | C13—N3—C9—C10 | 2.3 (3) |
| C7—O—C4—C3 | 178.56 (19) | C13—N3—C9—N2 | −177.69 (18) |
| C2—C3—C4—O | 179.5 (3) | N2—C9—C10—C11 | 177.7 (2) |
| C2—C3—C4—C5 | −0.3 (3) | N3—C9—C10—C11 | −2.3 (3) |
| O—C4—C5—C6 | −178.9 (3) | C9—C10—C11—C12 | 0.4 (3) |
| C3—C4—C5—C6 | 0.7 (3) | C10—C11—C12—C13 | 1.2 (3) |
| C2—C1—C6—C5 | −0.3 (3) | C11—C12—C13—N3 | −1.3 (3) |
| C8—C1—C6—C5 | −179.1 (2) | C9—N3—C13—C12 | −0.5 (3) |
| C4—C5—C6—C1 | −0.2 (6) | C11—C12—C13—N3 | −1.3 (3) |
| N2—N1—C8—C1 | −179.54 (19) | C9—N3—C13—C12 | −0.5 (3) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2A···N3i | 0.86 | 2.27 | 3.119 (4) | 169 |
| Symmetry code: (i) −x+2, −y+2, −z. |
Experimental details
| Crystal data |
| Chemical formula | C13H13N3O |
| Mr | 227.26 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 293 |
| a, b, c (Å) | 5.5633 (16), 10.4599 (11), 20.314 (4) |
| β (°) | 95.51 (2) |
| V (Å3) | 1176.6 (4) |
| Z | 4 |
| Radiation type | Mo Kα |
| µ (mm−1) | 0.09 |
| Crystal size (mm) | 0.30 × 0.27 × 0.21 |
| |
| Data collection |
| Diffractometer | Enraf-Nonius CAD-4
diffractometer |
| Absorption correction | – |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 2461, 2223, 1177 |
| Rint | 0.021 |
| (sin θ/λ)max (Å−1) | 0.609 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.042, 0.128, 0.99 |
| No. of reflections | 2223 |
| No. of parameters | 154 |
| No. of restraints | ? |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.11, −0.17 |
Selected geometric parameters (Å, º) top| O—C4 | 1.367 (2) | N2—C9 | 1.374 (2) |
| O—C7 | 1.415 (2) | N3—C13 | 1.340 (2) |
| N1—C8 | 1.270 (2) | N3—C9 | 1.336 (2) |
| N1—N2 | 1.367 (2) | C1—C8 | 1.459 (3) |
| | | |
| C4—O—C7 | 119.11 (17) | O—C4—C3 | 115.33 (18) |
| C8—N1—N2 | 118.18 (18) | C3—C4—C5 | 119.59 (19) |
| N1—N2—C9 | 118.12 (19) | N1—C8—C1 | 121.00 (19) |
| C9—N3—C13 | 116.38 (18) | N2—C9—C10 | 121.74 (18) |
| C2—C1—C8 | 121.72 (19) | N3—C9—N2 | 115.37 (17) |
| O—C4—C5 | 125.07 (19) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2A···N3i | 0.86 | 2.27 | 3.119 (4) | 169 |
| Symmetry code: (i) −x+2, −y+2, −z. |

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.
 N hydrogen bonds [R
N hydrogen bonds [R![[Figure 1]](http://journals.iucr.org/c/issues/2003/04/00/bm1525/bm1525fig1thm.gif)

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
 journal menu
journal menu









 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry



Hydrazones have been widely studied as chelating ligands for the spectrophotometric and fluorimetric determination of trace metal ions (Katyal & Dutt,1975; Galiano-Roth & Collum, 1988). Many of these compounds have found wide application in medicine, technology and analytical chemistry (Kitaev, 1977). In recent years there has been considerable interest in arlyhydrazone compounds containing a pyridyl group. Hydrazines condense with aldehydes and ketones to give hydrazones, which typically are crystalline compounds with sharp melting points that can therefore be used to identify the aldehydes and ketones from which the hydrazones have been formed (McMurry, 1999). Hydrazones are frequently more suitable than oximes for this purpose, since their greater molecular weight causes a lower solubility in most solvents and they can therefore often be more easily isolated and recrystallized. The hydrazones were obtained by the condensation reaction of aldehydes (or substituted aldehydes) with phenylhydrazine at pH 4–5 (McMurry, 1999).
??In the title compound, (I), ?? the configuration about the azomethine C═N double bond is E, with an N2—N1—C8—C1 torsion angle of −179.54 (19)°. Overall, the molecule is planar, the r.m.s. deviation of the non-H atoms from the least-squares mean plane being only 0.047 Å.
The N1—N2 [1.367 (2) Å] and N1—C8 [1.270 (2) Å] bond distances are comparable to those in the related compounds p-methoxbenzaldehyde isonicotinoyl hydrazone dihydrate (Fun et al., 1996) and p-methoxybenzaldehyde isonicotinoyl hydrazone monohydrate (Shanmuga Sundara Raj et al., 1999) and correspond to single and double bonds, respectively. There is only slight asymmetry of the exocyclic angles at C1 [C2—C1—C8 = 121.72 (19)° and C6—C1—C8 = 120.84 (19)°], while that at C4 is more pronounced [O—C4—C3 = 115.33 (18)° and O—C4—C5 = 125.07 (19)°]. This situation is typical of that found in anisoles and is caused by the tendency of the methoxy group to be coplanar with the phenyl through the conjugation of the O atom with the phenyl ring (Domiano et al., 1979). In (I) the methyl C atom shows no significant deviation from the plane of the phenyl ring. There is also asymmetry in the exocyclic angles at C9 [N2—C9—N3 = 115.37 (17)° and N2—C9—C10 = 121.74 (18)°, respectively]. The dihedral angle between the planes of the pyridine and methoxyphenyl rings is 6.19 (12)°, and these two planes make dihedral angles with the central hydrazone bridge (N2/N1/C8) of 7.59 (3)° and 1.41 (3)°, respectively.
Centrosymmetrically related molecules are linked into discrete pairs by pairwise N2—H2A···N3i hydrogen bonds [symmetry code: (i) 2 − x,2 − y,-z; graph-set notation R22(8)]. The NH···N and N···N distances of 2.27 and 3.119 (4) Å, respectively, are short compared with the relevant van der Waals radii (Bondi, 1964).