Buy article online - an online subscription or single-article purchase is required to access this article.

The crystal structures of the title compounds have been determined in the temperature range 140–290 K for the zinc complex, and 190–270 K for the copper complex. The two structures are isostructural in the trigonal space group
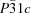
with the sulfate anion severely disordered on a site with 32 (
D3) symmetry. This sulfate disorder leads to a disordered three-dimensional hydrogen-bond network, with the N—H atoms acting as donors and the sulfate O atoms as acceptors. The displacement parameters of the N and C atoms in both compounds contain disorder contributions in the out-of-ligand plane direction owing to ring puckering and/or disorder in hydrogen bonding. In the Zn compound the vibrational amplitudes in the bond directions are closely similar. Their differences show no significant deviations from rigid-bond behaviour. In the Cu compound, a (presumably) dynamic Jahn–Teller effect is identified from a temperature-independent contribution to the displacement ellipsoids of the N atom along the N—Cu bond. These conclusions derive from analyses of the atomic displacement parameters with the Hirshfeld test, with rigid-body models at different temperatures, and with a normal coordinate analysis. This analysis considers the atomic displacement parameters (ADPs) from all different temperatures simultaneously and provides a detailed description of both the thermal motion and the disorder in the cation. The Jahn–Teller radii of the Cu compound derived on the basis of the ADP analysis and from the bond distances in the statically distorted low-temperature phase [Lutz (2010).
Acta Cryst. C
66, m330–m335] are found to be the same.
Supporting information
 | Crystallographic Information File (CIF) https://doi.org/10.1107/S0108768110054571/bk5101sup1.cif
Contains datablocks 1a, 1b, 1c, 1d, 1e, 1f, 2a, 2b, 2c, 2d, 2e, global |
 | Structure factor file (CIF format) https://doi.org/10.1107/S0108768110054571/bk51011asup2.hkl
Contains datablock 1a |
 | Structure factor file (CIF format) https://doi.org/10.1107/S0108768110054571/bk51011bsup3.hkl
Contains datablock 1b |
 | Structure factor file (CIF format) https://doi.org/10.1107/S0108768110054571/bk51011csup4.hkl
Contains datablock 1c |
 | Structure factor file (CIF format) https://doi.org/10.1107/S0108768110054571/bk51011dsup5.hkl
Contains datablock 1d |
 | Structure factor file (CIF format) https://doi.org/10.1107/S0108768110054571/bk51011esup6.hkl
Contains datablock 1e |
 | Structure factor file (CIF format) https://doi.org/10.1107/S0108768110054571/bk51011fsup7.hkl
Contains datablock 1f |
 | Structure factor file (CIF format) https://doi.org/10.1107/S0108768110054571/bk51012asup8.hkl
Contains datablock 2a |
 | Structure factor file (CIF format) https://doi.org/10.1107/S0108768110054571/bk51012bsup9.hkl
Contains datablock 2b |
 | Structure factor file (CIF format) https://doi.org/10.1107/S0108768110054571/bk51012csup10.hkl
Contains datablock 2c |
 | Structure factor file (CIF format) https://doi.org/10.1107/S0108768110054571/bk51012dsup11.hkl
Contains datablock 2d |
 | Structure factor file (CIF format) https://doi.org/10.1107/S0108768110054571/bk51012esup12.hkl
Contains datablock 2e |
 | Portable Document Format (PDF) file https://doi.org/10.1107/S0108768110054571/bk5101sup13.pdf
Extra figures and tables |
CCDC references: 814500; 814501; 814502; 814503; 814504; 814505; 814506; 814507; 814508; 814509; 814510
For all structures, data collection: COLLECT (Nonius, 1999); cell refinement: PEAKREF (Schreurs, 2005); data reduction: EVAL15 (Schreurs et al., 2010), SADABS2008/1 (Sheldrick, 2008b); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008a); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008a).
Crystal data top
| C6H24N6Zn·O4S | Dx = 1.708 Mg m−3 |
| Mr = 341.74 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, P31c | Cell parameters from 11284 reflections |
| Hall symbol: -P 3 2c | θ = 2.1–35.0° |
| a = 8.9594 (1) Å | µ = 2.02 mm−1 |
| c = 9.5583 (1) Å | T = 140 K |
| V = 664.46 (2) Å3 | Needle, colourless |
| Z = 2 | 0.30 × 0.09 × 0.09 mm |
| F(000) = 360 | |
Data collection top
Nonius KappaCCD
diffractometer | 683 independent reflections |
| Radiation source: rotating anode | 649 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.027 |
| φ and ω scans | θmax = 30.5°, θmin = 2.6° |
Absorption correction: analytical
SADABS-2008/1 (Sheldrick, 2008b) | h = −12→12 |
| Tmin = 0.619, Tmax = 0.878 | k = −12→12 |
| 10729 measured reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.020 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.048 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.10 | w = 1/[σ2(Fo2) + (0.0206P)2 + 0.4531P]
where P = (Fo2 + 2Fc2)/3 |
| 683 reflections | (Δ/σ)max < 0.001 |
| 49 parameters | Δρmax = 0.56 e Å−3 |
| 16 restraints | Δρmin = −0.25 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Zn1 | 0.6667 | 0.3333 | 0.2500 | 0.01092 (9) | |
| N1 | 0.45179 (14) | 0.30945 (15) | 0.12505 (12) | 0.0197 (2) | |
| H1N | 0.436 (3) | 0.393 (3) | 0.136 (2) | 0.033 (5)* | |
| H2N | 0.470 (3) | 0.304 (3) | 0.040 (2) | 0.035 (5)* | |
| C1 | 0.29463 (16) | 0.15271 (17) | 0.17058 (13) | 0.0199 (2) | |
| H1A | 0.2912 | 0.0497 | 0.1292 | 0.024* | |
| H1B | 0.1915 | 0.1565 | 0.1386 | 0.024* | |
| S1 | 0.3333 | 0.6667 | 0.2500 | 0.01097 (13) | |
| O1 | 0.3589 (9) | 0.5639 (7) | 0.1489 (3) | 0.0623 (18) | 0.33333 |
| O2 | 0.2850 (10) | 0.7776 (9) | 0.1764 (4) | 0.093 (3) | 0.33333 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Zn1 | 0.01117 (11) | 0.01117 (11) | 0.01043 (15) | 0.00559 (6) | 0.000 | 0.000 |
| N1 | 0.0162 (5) | 0.0244 (5) | 0.0171 (5) | 0.0090 (4) | −0.0021 (4) | 0.0045 (4) |
| C1 | 0.0133 (5) | 0.0228 (6) | 0.0200 (5) | 0.0063 (4) | −0.0032 (4) | −0.0023 (4) |
| S1 | 0.01203 (18) | 0.01203 (18) | 0.0089 (3) | 0.00601 (9) | 0.000 | 0.000 |
| O1 | 0.156 (6) | 0.075 (3) | 0.0164 (16) | 0.103 (4) | 0.004 (2) | −0.0032 (18) |
| O2 | 0.212 (8) | 0.136 (6) | 0.026 (2) | 0.158 (6) | 0.020 (3) | 0.024 (3) |
Geometric parameters (Å, º) top
| Zn1—N1i | 2.1832 (11) | S1—O1vi | 1.431 (3) |
| Zn1—N1ii | 2.1832 (11) | S1—O1vii | 1.431 (3) |
| Zn1—N1 | 2.1832 (11) | S1—O1 | 1.431 (3) |
| Zn1—N1iii | 2.1833 (11) | S1—O1viii | 1.431 (3) |
| Zn1—N1iv | 2.1833 (11) | S1—O1ix | 1.431 (3) |
| Zn1—N1v | 2.1833 (11) | S1—O1v | 1.431 (3) |
| N1—C1 | 1.4720 (17) | S1—O2viii | 1.449 (4) |
| N1—H1N | 0.84 (2) | S1—O2ix | 1.450 (4) |
| N1—H2N | 0.84 (2) | S1—O2vi | 1.450 (4) |
| C1—C1ii | 1.521 (3) | S1—O2 | 1.450 (4) |
| C1—H1A | 0.9900 | S1—O2vii | 1.450 (4) |
| C1—H1B | 0.9900 | S1—O2v | 1.450 (4) |
| | | |
| N1i—Zn1—N1ii | 94.50 (7) | Zn1—N1—H1N | 112.5 (14) |
| N1i—Zn1—N1 | 92.93 (4) | C1—N1—H2N | 110.8 (14) |
| N1ii—Zn1—N1 | 80.43 (6) | Zn1—N1—H2N | 110.2 (15) |
| N1i—Zn1—N1iii | 92.93 (4) | H1N—N1—H2N | 107 (2) |
| N1ii—Zn1—N1iii | 170.26 (6) | N1—C1—C1ii | 108.98 (9) |
| N1—Zn1—N1iii | 92.93 (4) | N1—C1—H1A | 109.9 |
| N1i—Zn1—N1iv | 80.43 (6) | C1ii—C1—H1A | 109.9 |
| N1ii—Zn1—N1iv | 92.93 (4) | N1—C1—H1B | 109.9 |
| N1—Zn1—N1iv | 170.26 (6) | C1ii—C1—H1B | 109.9 |
| N1iii—Zn1—N1iv | 94.50 (7) | H1A—C1—H1B | 108.3 |
| N1i—Zn1—N1v | 170.26 (6) | O1vii—S1—O1 | 112.2 (5) |
| N1ii—Zn1—N1v | 92.93 (4) | O1vii—S1—O2 | 110.5 (4) |
| N1—Zn1—N1v | 94.50 (7) | O1—S1—O2 | 108.2 (2) |
| N1iii—Zn1—N1v | 80.43 (6) | O1vii—S1—O2vii | 108.2 (2) |
| N1iv—Zn1—N1v | 92.93 (4) | O1—S1—O2vii | 110.5 (4) |
| C1—N1—Zn1 | 107.92 (8) | O2—S1—O2vii | 107.1 (6) |
| C1—N1—H1N | 108.0 (14) | | |
| | | |
| N1i—Zn1—N1—C1 | 79.09 (11) | N1v—Zn1—N1—C1 | −107.21 (9) |
| N1ii—Zn1—N1—C1 | −14.99 (6) | Zn1—N1—C1—C1ii | 42.08 (14) |
| N1iii—Zn1—N1—C1 | 172.18 (8) | | |
| Symmetry codes: (i) −y+1, x−y, z; (ii) x, x−y, −z+1/2; (iii) −x+y+1, −x+1, z; (iv) −x+y+1, y, −z+1/2; (v) −y+1, −x+1, −z+1/2; (vi) −x+y, −x+1, z; (vii) −x+y, y, −z+1/2; (viii) −y+1, x−y+1, z; (ix) x, x−y+1, −z+1/2. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1 | 0.84 (2) | 1.97 (2) | 2.799 (3) | 170 (2) |
| N1—H2N···O2x | 0.84 (2) | 2.12 (2) | 2.951 (4) | 178 (2) |
| Symmetry code: (x) x−y+1, x, −z. |
Crystal data top
| C6H24N6Zn·O4S | Dx = 1.704 Mg m−3 |
| Mr = 341.74 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, P31c | Cell parameters from 11070 reflections |
| Hall symbol: -P 3 2c | θ = 2.1–35.0° |
| a = 8.9649 (1) Å | µ = 2.02 mm−1 |
| c = 9.5685 (1) Å | T = 170 K |
| V = 665.99 (2) Å3 | Needle, colourless |
| Z = 2 | 0.30 × 0.09 × 0.09 mm |
| F(000) = 360 | |
Data collection top
Nonius KappaCCD
diffractometer | 689 independent reflections |
| Radiation source: rotating anode | 650 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.026 |
| φ and ω scans | θmax = 30.5°, θmin = 2.6° |
Absorption correction: analytical
SADABS-2008/1 (Sheldrick, 2008b) | h = −12→12 |
| Tmin = 0.625, Tmax = 0.876 | k = −12→12 |
| 10818 measured reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.021 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.051 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0226P)2 + 0.4729P]
where P = (Fo2 + 2Fc2)/3 |
| 689 reflections | (Δ/σ)max < 0.001 |
| 49 parameters | Δρmax = 0.53 e Å−3 |
| 16 restraints | Δρmin = −0.26 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Zn1 | 0.6667 | 0.3333 | 0.2500 | 0.01325 (10) | |
| N1 | 0.45189 (15) | 0.30936 (16) | 0.12505 (12) | 0.0222 (2) | |
| H1N | 0.437 (3) | 0.393 (3) | 0.136 (2) | 0.031 (5)* | |
| H2N | 0.472 (3) | 0.305 (3) | 0.038 (2) | 0.038 (6)* | |
| C1 | 0.29493 (17) | 0.15272 (18) | 0.17071 (14) | 0.0231 (3) | |
| H1A | 0.2916 | 0.0498 | 0.1296 | 0.028* | |
| H1B | 0.1918 | 0.1563 | 0.1387 | 0.028* | |
| S1 | 0.3333 | 0.6667 | 0.2500 | 0.01318 (14) | |
| O1 | 0.3574 (10) | 0.5636 (7) | 0.1483 (4) | 0.071 (2) | 0.33333 |
| O2 | 0.2849 (10) | 0.7773 (9) | 0.1777 (5) | 0.100 (3) | 0.33333 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Zn1 | 0.01350 (12) | 0.01350 (12) | 0.01276 (16) | 0.00675 (6) | 0.000 | 0.000 |
| N1 | 0.0190 (5) | 0.0270 (6) | 0.0198 (5) | 0.0109 (4) | −0.0024 (4) | 0.0040 (4) |
| C1 | 0.0155 (5) | 0.0262 (6) | 0.0235 (6) | 0.0074 (5) | −0.0040 (4) | −0.0035 (5) |
| S1 | 0.01415 (19) | 0.01415 (19) | 0.0112 (3) | 0.00707 (9) | 0.000 | 0.000 |
| O1 | 0.171 (7) | 0.086 (4) | 0.0215 (18) | 0.114 (5) | 0.005 (3) | −0.004 (2) |
| O2 | 0.222 (9) | 0.145 (6) | 0.032 (2) | 0.166 (7) | 0.020 (4) | 0.027 (3) |
Geometric parameters (Å, º) top
| Zn1—N1i | 2.1838 (11) | S1—O1vi | 1.430 (3) |
| Zn1—N1 | 2.1838 (11) | S1—O1vii | 1.430 (3) |
| Zn1—N1ii | 2.1838 (11) | S1—O1 | 1.430 (3) |
| Zn1—N1iii | 2.1838 (11) | S1—O1viii | 1.430 (3) |
| Zn1—N1iv | 2.1838 (11) | S1—O1ix | 1.430 (3) |
| Zn1—N1v | 2.1839 (11) | S1—O1iii | 1.430 (3) |
| N1—C1 | 1.4720 (18) | S1—O2 | 1.443 (4) |
| N1—H1N | 0.83 (2) | S1—O2vii | 1.443 (4) |
| N1—H2N | 0.85 (2) | S1—O2iii | 1.443 (4) |
| C1—C1ii | 1.520 (3) | S1—O2viii | 1.443 (4) |
| C1—H1A | 0.9900 | S1—O2vi | 1.443 (4) |
| C1—H1B | 0.9900 | S1—O2ix | 1.443 (4) |
| | | |
| N1i—Zn1—N1 | 92.89 (4) | Zn1—N1—H1N | 112.1 (14) |
| N1i—Zn1—N1ii | 170.22 (7) | C1—N1—H2N | 111.4 (15) |
| N1—Zn1—N1ii | 80.44 (6) | Zn1—N1—H2N | 110.0 (15) |
| N1i—Zn1—N1iii | 80.44 (6) | H1N—N1—H2N | 107 (2) |
| N1—Zn1—N1iii | 94.58 (7) | N1—C1—C1ii | 109.01 (9) |
| N1ii—Zn1—N1iii | 92.89 (4) | N1—C1—H1A | 109.9 |
| N1i—Zn1—N1iv | 92.89 (4) | C1ii—C1—H1A | 109.9 |
| N1—Zn1—N1iv | 92.89 (4) | N1—C1—H1B | 109.9 |
| N1ii—Zn1—N1iv | 94.58 (7) | C1ii—C1—H1B | 109.9 |
| N1iii—Zn1—N1iv | 170.22 (7) | H1A—C1—H1B | 108.3 |
| N1i—Zn1—N1v | 94.57 (7) | O1vii—S1—O1 | 112.0 (5) |
| N1—Zn1—N1v | 170.22 (7) | O1vii—S1—O2 | 110.7 (4) |
| N1ii—Zn1—N1v | 92.89 (4) | O1—S1—O2 | 108.2 (2) |
| N1iii—Zn1—N1v | 92.89 (4) | O1vii—S1—O2vii | 108.2 (2) |
| N1iv—Zn1—N1v | 80.44 (6) | O1—S1—O2vii | 110.7 (4) |
| C1—N1—Zn1 | 107.86 (8) | O2—S1—O2vii | 106.9 (6) |
| C1—N1—H1N | 108.5 (14) | | |
| | | |
| N1i—Zn1—N1—C1 | 172.21 (9) | N1iv—Zn1—N1—C1 | 79.17 (12) |
| N1ii—Zn1—N1—C1 | −14.99 (6) | Zn1—N1—C1—C1ii | 42.12 (15) |
| N1iii—Zn1—N1—C1 | −107.16 (10) | | |
| Symmetry codes: (i) −x+y+1, −x+1, z; (ii) x, x−y, −z+1/2; (iii) −y+1, −x+1, −z+1/2; (iv) −y+1, x−y, z; (v) −x+y+1, y, −z+1/2; (vi) −x+y, −x+1, z; (vii) −x+y, y, −z+1/2; (viii) −y+1, x−y+1, z; (ix) x, x−y+1, −z+1/2. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1 | 0.83 (2) | 1.99 (2) | 2.810 (4) | 169 (2) |
| N1—H2N···O2x | 0.85 (2) | 2.11 (2) | 2.966 (5) | 177 (2) |
| Symmetry code: (x) x−y+1, x, −z. |
Crystal data top
| C6H24N6Zn·O4S | Dx = 1.700 Mg m−3 |
| Mr = 341.74 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, P31c | Cell parameters from 10704 reflections |
| Hall symbol: -P 3 2c | θ = 2.1–35.0° |
| a = 8.9696 (2) Å | µ = 2.01 mm−1 |
| c = 9.5842 (1) Å | T = 200 K |
| V = 667.78 (3) Å3 | Needle, colourless |
| Z = 2 | 0.30 × 0.09 × 0.09 mm |
| F(000) = 360 | |
Data collection top
Nonius KappaCCD
diffractometer | 690 independent reflections |
| Radiation source: rotating anode | 652 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.027 |
| φ and ω scans | θmax = 30.5°, θmin = 2.6° |
Absorption correction: analytical
SADABS-2008/1 (Sheldrick, 2008b) | h = −12→12 |
| Tmin = 0.622, Tmax = 0.882 | k = −12→12 |
| 10832 measured reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.022 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.058 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0316P)2 + 0.3881P]
where P = (Fo2 + 2Fc2)/3 |
| 690 reflections | (Δ/σ)max = 0.001 |
| 49 parameters | Δρmax = 0.56 e Å−3 |
| 16 restraints | Δρmin = −0.25 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Zn1 | 0.6667 | 0.3333 | 0.2500 | 0.01591 (11) | |
| N1 | 0.45178 (16) | 0.30936 (17) | 0.12531 (13) | 0.0251 (3) | |
| H1N | 0.434 (3) | 0.394 (3) | 0.137 (2) | 0.040 (6)* | |
| H2N | 0.472 (3) | 0.305 (3) | 0.039 (2) | 0.036 (5)* | |
| C1 | 0.29525 (18) | 0.1529 (2) | 0.17090 (16) | 0.0278 (3) | |
| H1A | 0.2921 | 0.0501 | 0.1298 | 0.033* | |
| H1B | 0.1920 | 0.1562 | 0.1389 | 0.033* | |
| S1 | 0.3333 | 0.6667 | 0.2500 | 0.01605 (15) | |
| O1 | 0.3538 (11) | 0.5626 (8) | 0.1466 (4) | 0.082 (3) | 0.33333 |
| O2 | 0.2837 (11) | 0.7780 (10) | 0.1814 (5) | 0.105 (3) | 0.33333 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Zn1 | 0.01605 (13) | 0.01605 (13) | 0.01564 (17) | 0.00802 (7) | 0.000 | 0.000 |
| N1 | 0.0225 (6) | 0.0308 (6) | 0.0222 (5) | 0.0136 (5) | −0.0030 (4) | 0.0028 (4) |
| C1 | 0.0187 (6) | 0.0320 (7) | 0.0287 (7) | 0.0096 (5) | −0.0048 (5) | −0.0044 (5) |
| S1 | 0.0165 (2) | 0.0165 (2) | 0.0152 (3) | 0.00825 (10) | 0.000 | 0.000 |
| O1 | 0.190 (8) | 0.099 (5) | 0.028 (2) | 0.125 (6) | 0.010 (3) | −0.003 (3) |
| O2 | 0.223 (10) | 0.152 (7) | 0.042 (3) | 0.170 (8) | 0.018 (4) | 0.027 (4) |
Geometric parameters (Å, º) top
| Zn1—N1i | 2.1852 (12) | S1—O1vi | 1.435 (4) |
| Zn1—N1ii | 2.1852 (12) | S1—O1vii | 1.435 (4) |
| Zn1—N1 | 2.1852 (12) | S1—O1viii | 1.435 (4) |
| Zn1—N1iii | 2.1852 (12) | S1—O1ix | 1.435 (4) |
| Zn1—N1iv | 2.1852 (12) | S1—O1 | 1.435 (4) |
| Zn1—N1v | 2.1852 (12) | S1—O1ii | 1.435 (4) |
| N1—C1 | 1.470 (2) | S1—O2ii | 1.439 (4) |
| N1—H1N | 0.86 (3) | S1—O2 | 1.439 (4) |
| N1—H2N | 0.86 (2) | S1—O2ix | 1.439 (4) |
| C1—C1iii | 1.519 (3) | S1—O2vii | 1.439 (4) |
| C1—H1A | 0.9900 | S1—O2vi | 1.439 (4) |
| C1—H1B | 0.9900 | S1—O2viii | 1.439 (4) |
| | | |
| N1i—Zn1—N1ii | 80.38 (7) | Zn1—N1—H1N | 113.1 (16) |
| N1i—Zn1—N1 | 92.95 (5) | C1—N1—H2N | 111.5 (15) |
| N1ii—Zn1—N1 | 94.52 (7) | Zn1—N1—H2N | 110.0 (15) |
| N1i—Zn1—N1iii | 170.22 (7) | H1N—N1—H2N | 107 (2) |
| N1ii—Zn1—N1iii | 92.94 (5) | N1—C1—C1iii | 109.03 (10) |
| N1—Zn1—N1iii | 80.38 (7) | N1—C1—H1A | 109.9 |
| N1i—Zn1—N1iv | 92.94 (5) | C1iii—C1—H1A | 109.9 |
| N1ii—Zn1—N1iv | 170.22 (7) | N1—C1—H1B | 109.9 |
| N1—Zn1—N1iv | 92.95 (5) | C1iii—C1—H1B | 109.9 |
| N1iii—Zn1—N1iv | 94.52 (7) | H1A—C1—H1B | 108.3 |
| N1i—Zn1—N1v | 94.52 (7) | O1ix—S1—O1 | 111.4 (6) |
| N1ii—Zn1—N1v | 92.94 (5) | O1ix—S1—O2 | 110.9 (4) |
| N1—Zn1—N1v | 170.22 (7) | O1—S1—O2 | 108.7 (3) |
| N1iii—Zn1—N1v | 92.94 (5) | O1ix—S1—O2ix | 108.7 (3) |
| N1iv—Zn1—N1v | 80.38 (7) | O1—S1—O2ix | 110.9 (4) |
| C1—N1—Zn1 | 107.80 (9) | O2—S1—O2ix | 106.2 (7) |
| C1—N1—H1N | 107.0 (16) | | |
| | | |
| N1i—Zn1—N1—C1 | 172.18 (10) | N1iv—Zn1—N1—C1 | 79.08 (12) |
| N1ii—Zn1—N1—C1 | −107.24 (10) | Zn1—N1—C1—C1iii | 42.20 (17) |
| N1iii—Zn1—N1—C1 | −15.02 (7) | | |
| Symmetry codes: (i) −x+y+1, −x+1, z; (ii) −y+1, −x+1, −z+1/2; (iii) x, x−y, −z+1/2; (iv) −y+1, x−y, z; (v) −x+y+1, y, −z+1/2; (vi) −x+y, −x+1, z; (vii) −y+1, x−y+1, z; (viii) x, x−y+1, −z+1/2; (ix) −x+y, y, −z+1/2. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1 | 0.86 (3) | 1.97 (3) | 2.823 (5) | 170 (2) |
| N1—H2N···O2x | 0.86 (2) | 2.15 (2) | 3.006 (5) | 177 (2) |
| Symmetry code: (x) x−y+1, x, −z. |
Crystal data top
| C6H24N6Zn·O4S | Dx = 1.694 Mg m−3 |
| Mr = 341.74 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, P31c | Cell parameters from 10489 reflections |
| Hall symbol: -P 3 2c | θ = 2.1–35.0° |
| a = 8.9790 (1) Å | µ = 2.01 mm−1 |
| c = 9.5973 (2) Å | T = 230 K |
| V = 670.09 (2) Å3 | Needle, colourless |
| Z = 2 | 0.30 × 0.09 × 0.09 mm |
| F(000) = 360 | |
Data collection top
Nonius KappaCCD
diffractometer | 694 independent reflections |
| Radiation source: rotating anode | 649 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.027 |
| φ and ω scans | θmax = 30.5°, θmin = 2.6° |
Absorption correction: analytical
SADABS-2008/1 (Sheldrick, 2008b) | h = −12→12 |
| Tmin = 0.625, Tmax = 0.878 | k = −12→12 |
| 10896 measured reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.023 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.061 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.08 | w = 1/[σ2(Fo2) + (0.0331P)2 + 0.3399P]
where P = (Fo2 + 2Fc2)/3 |
| 694 reflections | (Δ/σ)max < 0.001 |
| 49 parameters | Δρmax = 0.59 e Å−3 |
| 16 restraints | Δρmin = −0.24 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Zn1 | 0.6667 | 0.3333 | 0.2500 | 0.01825 (12) | |
| N1 | 0.45172 (17) | 0.30921 (18) | 0.12541 (14) | 0.0278 (3) | |
| H1N | 0.434 (3) | 0.395 (3) | 0.138 (3) | 0.043 (6)* | |
| H2N | 0.471 (3) | 0.305 (3) | 0.040 (2) | 0.046 (6)* | |
| C1 | 0.29538 (19) | 0.1530 (2) | 0.17114 (17) | 0.0312 (3) | |
| H1A | 0.2921 | 0.0513 | 0.1305 | 0.037* | |
| H1B | 0.1933 | 0.1564 | 0.1396 | 0.037* | |
| S1 | 0.3333 | 0.6667 | 0.2500 | 0.01843 (16) | |
| O1 | 0.3535 (11) | 0.5634 (9) | 0.1464 (5) | 0.089 (3) | 0.33333 |
| O2 | 0.2836 (11) | 0.7783 (10) | 0.1823 (6) | 0.110 (4) | 0.33333 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Zn1 | 0.01837 (14) | 0.01837 (14) | 0.01802 (18) | 0.00918 (7) | 0.000 | 0.000 |
| N1 | 0.0254 (6) | 0.0340 (7) | 0.0248 (6) | 0.0153 (5) | −0.0031 (5) | 0.0026 (5) |
| C1 | 0.0212 (6) | 0.0354 (8) | 0.0328 (7) | 0.0109 (6) | −0.0055 (5) | −0.0055 (6) |
| S1 | 0.0188 (2) | 0.0188 (2) | 0.0177 (3) | 0.00939 (11) | 0.000 | 0.000 |
| O1 | 0.196 (9) | 0.106 (5) | 0.035 (2) | 0.127 (7) | 0.014 (4) | −0.004 (3) |
| O2 | 0.228 (10) | 0.156 (8) | 0.049 (3) | 0.173 (8) | 0.019 (5) | 0.028 (4) |
Geometric parameters (Å, º) top
| Zn1—N1i | 2.1871 (13) | S1—O1vi | 1.432 (4) |
| Zn1—N1ii | 2.1871 (13) | S1—O1vii | 1.432 (4) |
| Zn1—N1iii | 2.1871 (13) | S1—O1viii | 1.432 (4) |
| Zn1—N1iv | 2.1871 (13) | S1—O1 | 1.432 (4) |
| Zn1—N1v | 2.1871 (13) | S1—O1ix | 1.432 (4) |
| Zn1—N1 | 2.1871 (13) | S1—O1i | 1.432 (4) |
| N1—C1 | 1.470 (2) | S1—O2i | 1.440 (4) |
| N1—H1N | 0.87 (3) | S1—O2 | 1.440 (4) |
| N1—H2N | 0.84 (2) | S1—O2vii | 1.440 (4) |
| C1—C1ii | 1.517 (3) | S1—O2vi | 1.440 (4) |
| C1—H1A | 0.9800 | S1—O2ix | 1.440 (4) |
| C1—H1B | 0.9800 | S1—O2viii | 1.440 (4) |
| | | |
| N1i—Zn1—N1ii | 92.96 (5) | Zn1—N1—H1N | 112.3 (16) |
| N1i—Zn1—N1iii | 92.96 (5) | C1—N1—H2N | 111.4 (17) |
| N1ii—Zn1—N1iii | 92.96 (5) | Zn1—N1—H2N | 110.3 (17) |
| N1i—Zn1—N1iv | 80.33 (7) | H1N—N1—H2N | 108 (2) |
| N1ii—Zn1—N1iv | 170.16 (8) | N1—C1—C1ii | 109.14 (11) |
| N1iii—Zn1—N1iv | 94.55 (8) | N1—C1—H1A | 109.9 |
| N1i—Zn1—N1v | 170.16 (8) | C1ii—C1—H1A | 109.9 |
| N1ii—Zn1—N1v | 94.55 (8) | N1—C1—H1B | 109.9 |
| N1iii—Zn1—N1v | 80.33 (7) | C1ii—C1—H1B | 109.9 |
| N1iv—Zn1—N1v | 92.96 (5) | H1A—C1—H1B | 108.3 |
| N1i—Zn1—N1 | 94.55 (8) | O1vii—S1—O1 | 111.8 (6) |
| N1ii—Zn1—N1 | 80.33 (7) | O1vii—S1—O2 | 110.8 (5) |
| N1iii—Zn1—N1 | 170.16 (8) | O1—S1—O2 | 108.7 (3) |
| N1iv—Zn1—N1 | 92.96 (5) | O1vii—S1—O2vii | 108.7 (3) |
| N1v—Zn1—N1 | 92.96 (5) | O1—S1—O2vii | 110.8 (5) |
| C1—N1—Zn1 | 107.79 (9) | O2—S1—O2vii | 105.9 (7) |
| C1—N1—H1N | 107.2 (16) | | |
| | | |
| N1i—Zn1—N1—C1 | −107.19 (11) | N1v—Zn1—N1—C1 | 79.17 (13) |
| N1ii—Zn1—N1—C1 | −14.95 (7) | Zn1—N1—C1—C1ii | 42.11 (18) |
| N1iv—Zn1—N1—C1 | 172.29 (10) | | |
| Symmetry codes: (i) −y+1, −x+1, −z+1/2; (ii) x, x−y, −z+1/2; (iii) −x+y+1, y, −z+1/2; (iv) −x+y+1, −x+1, z; (v) −y+1, x−y, z; (vi) x, x−y+1, −z+1/2; (vii) −x+y, y, −z+1/2; (viii) −x+y, −x+1, z; (ix) −y+1, x−y+1, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1 | 0.87 (3) | 1.98 (3) | 2.835 (5) | 169 (2) |
| N1—H2N···O2x | 0.84 (2) | 2.18 (2) | 3.019 (6) | 177 (2) |
| Symmetry code: (x) x−y+1, x, −z. |
Crystal data top
| C6H24N6Zn·O4S | Dx = 1.690 Mg m−3 |
| Mr = 341.74 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, P31c | Cell parameters from 10365 reflections |
| Hall symbol: -P 3 2c | θ = 2.1–35.0° |
| a = 8.9855 (2) Å | µ = 2.00 mm−1 |
| c = 9.6043 (1) Å | T = 260 K |
| V = 671.55 (3) Å3 | Needle, colourless |
| Z = 2 | 0.30 × 0.09 × 0.09 mm |
| F(000) = 360 | |
Data collection top
Nonius KappaCCD
diffractometer | 694 independent reflections |
| Radiation source: rotating anode | 653 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.029 |
| φ and ω scans | θmax = 30.5°, θmin = 2.6° |
Absorption correction: analytical
SADABS-2008/1 (Sheldrick, 2008b) | h = −12→12 |
| Tmin = 0.621, Tmax = 0.874 | k = −12→12 |
| 11110 measured reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.024 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.063 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0373P)2 + 0.3022P]
where P = (Fo2 + 2Fc2)/3 |
| 694 reflections | (Δ/σ)max < 0.001 |
| 49 parameters | Δρmax = 0.66 e Å−3 |
| 16 restraints | Δρmin = −0.29 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Zn1 | 0.6667 | 0.3333 | 0.2500 | 0.02039 (13) | |
| N1 | 0.45185 (18) | 0.30916 (19) | 0.12549 (14) | 0.0302 (3) | |
| H1N | 0.432 (3) | 0.393 (4) | 0.136 (3) | 0.048 (6)* | |
| H2N | 0.471 (3) | 0.303 (3) | 0.038 (2) | 0.045 (6)* | |
| C1 | 0.2958 (2) | 0.1531 (2) | 0.17122 (18) | 0.0343 (3) | |
| H1A | 0.2927 | 0.0525 | 0.1311 | 0.041* | |
| H1B | 0.1948 | 0.1563 | 0.1399 | 0.041* | |
| S1 | 0.3333 | 0.6667 | 0.2500 | 0.02050 (17) | |
| O1 | 0.3538 (12) | 0.5637 (9) | 0.1465 (5) | 0.096 (3) | 0.33333 |
| O2 | 0.2838 (11) | 0.7780 (10) | 0.1822 (6) | 0.115 (4) | 0.33333 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Zn1 | 0.02054 (15) | 0.02054 (15) | 0.02010 (19) | 0.01027 (7) | 0.000 | 0.000 |
| N1 | 0.0281 (6) | 0.0368 (7) | 0.0268 (6) | 0.0170 (5) | −0.0035 (5) | 0.0027 (5) |
| C1 | 0.0232 (6) | 0.0385 (8) | 0.0365 (8) | 0.0118 (6) | −0.0063 (6) | −0.0058 (6) |
| S1 | 0.0207 (2) | 0.0207 (2) | 0.0201 (3) | 0.01036 (11) | 0.000 | 0.000 |
| O1 | 0.202 (10) | 0.112 (6) | 0.042 (3) | 0.131 (7) | 0.013 (4) | −0.007 (3) |
| O2 | 0.233 (11) | 0.158 (8) | 0.058 (4) | 0.175 (9) | 0.018 (5) | 0.029 (4) |
Geometric parameters (Å, º) top
| Zn1—N1i | 2.1871 (13) | S1—O1vi | 1.432 (4) |
| Zn1—N1ii | 2.1871 (13) | S1—O1 | 1.432 (4) |
| Zn1—N1iii | 2.1871 (13) | S1—O1iv | 1.432 (4) |
| Zn1—N1 | 2.1872 (13) | S1—O1vii | 1.432 (4) |
| Zn1—N1iv | 2.1872 (13) | S1—O1viii | 1.432 (4) |
| Zn1—N1v | 2.1872 (13) | S1—O1ix | 1.432 (4) |
| N1—C1 | 1.469 (2) | S1—O2vii | 1.438 (5) |
| N1—H1N | 0.86 (3) | S1—O2ix | 1.438 (5) |
| N1—H2N | 0.87 (2) | S1—O2viii | 1.438 (5) |
| C1—C1iii | 1.516 (3) | S1—O2iv | 1.438 (5) |
| C1—H1A | 0.9700 | S1—O2 | 1.438 (5) |
| C1—H1B | 0.9700 | S1—O2vi | 1.438 (5) |
| | | |
| N1i—Zn1—N1ii | 80.31 (7) | Zn1—N1—H1N | 114.1 (18) |
| N1i—Zn1—N1iii | 92.96 (5) | C1—N1—H2N | 110.9 (16) |
| N1ii—Zn1—N1iii | 94.58 (8) | Zn1—N1—H2N | 110.3 (17) |
| N1i—Zn1—N1 | 170.13 (8) | H1N—N1—H2N | 107 (2) |
| N1ii—Zn1—N1 | 92.96 (5) | N1—C1—C1iii | 109.13 (11) |
| N1iii—Zn1—N1 | 80.31 (7) | N1—C1—H1A | 109.9 |
| N1i—Zn1—N1iv | 92.96 (5) | C1iii—C1—H1A | 109.9 |
| N1ii—Zn1—N1iv | 170.13 (8) | N1—C1—H1B | 109.9 |
| N1iii—Zn1—N1iv | 92.96 (5) | C1iii—C1—H1B | 109.9 |
| N1—Zn1—N1iv | 94.58 (8) | H1A—C1—H1B | 108.3 |
| N1i—Zn1—N1v | 94.58 (8) | O1vi—S1—O1 | 111.9 (6) |
| N1ii—Zn1—N1v | 92.96 (5) | O1vi—S1—O2 | 110.8 (5) |
| N1iii—Zn1—N1v | 170.13 (8) | O1—S1—O2 | 108.7 (3) |
| N1—Zn1—N1v | 92.96 (5) | O1vi—S1—O2vi | 108.7 (3) |
| N1iv—Zn1—N1v | 80.31 (7) | O1—S1—O2vi | 110.8 (5) |
| C1—N1—Zn1 | 107.76 (10) | O2—S1—O2vi | 105.9 (7) |
| C1—N1—H1N | 106.6 (17) | | |
| | | |
| N1ii—Zn1—N1—C1 | 79.17 (13) | N1v—Zn1—N1—C1 | 172.29 (10) |
| N1iii—Zn1—N1—C1 | −14.98 (8) | Zn1—N1—C1—C1iii | 42.19 (18) |
| N1iv—Zn1—N1—C1 | −107.20 (11) | | |
| Symmetry codes: (i) −x+y+1, y, −z+1/2; (ii) −y+1, x−y, z; (iii) x, x−y, −z+1/2; (iv) −y+1, −x+1, −z+1/2; (v) −x+y+1, −x+1, z; (vi) −x+y, y, −z+1/2; (vii) x, x−y+1, −z+1/2; (viii) −x+y, −x+1, z; (ix) −y+1, x−y+1, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1 | 0.86 (3) | 1.99 (3) | 2.839 (5) | 172 (3) |
| N1—H2N···O2x | 0.87 (2) | 2.16 (2) | 3.022 (6) | 177 (2) |
| Symmetry code: (x) x−y+1, x, −z. |
Crystal data top
| C6H24N6Zn·O4S | Dx = 1.686 Mg m−3 |
| Mr = 341.74 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, P31c | Cell parameters from 10072 reflections |
| Hall symbol: -P 3 2c | θ = 2.1–34.9° |
| a = 8.9929 (2) Å | µ = 2.00 mm−1 |
| c = 9.6101 (2) Å | T = 290 K |
| V = 673.07 (3) Å3 | Needle, colourless |
| Z = 2 | 0.30 × 0.09 × 0.09 mm |
| F(000) = 360 | |
Data collection top
Nonius KappaCCD
diffractometer | 696 independent reflections |
| Radiation source: rotating anode | 647 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.030 |
| φ and ω scans | θmax = 30.5°, θmin = 2.6° |
Absorption correction: analytical
SADABS-2008/1 (Sheldrick, 2008b) | h = −12→12 |
| Tmin = 0.627, Tmax = 0.872 | k = −12→12 |
| 11155 measured reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.023 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.064 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0402P)2 + 0.1924P]
where P = (Fo2 + 2Fc2)/3 |
| 696 reflections | (Δ/σ)max < 0.001 |
| 49 parameters | Δρmax = 0.62 e Å−3 |
| 16 restraints | Δρmin = −0.29 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Zn1 | 0.6667 | 0.3333 | 0.2500 | 0.02240 (13) | |
| N1 | 0.45193 (17) | 0.30908 (19) | 0.12575 (14) | 0.0328 (3) | |
| H1N | 0.432 (3) | 0.392 (4) | 0.136 (3) | 0.053 (6)* | |
| H2N | 0.469 (3) | 0.304 (3) | 0.038 (2) | 0.050 (6)* | |
| C1 | 0.2960 (2) | 0.1532 (2) | 0.17146 (18) | 0.0375 (3) | |
| H1A | 0.2929 | 0.0527 | 0.1314 | 0.045* | |
| H1B | 0.1951 | 0.1564 | 0.1402 | 0.045* | |
| S1 | 0.3333 | 0.6667 | 0.2500 | 0.02259 (16) | |
| O1 | 0.3543 (12) | 0.5644 (9) | 0.1464 (5) | 0.100 (3) | 0.33333 |
| O2 | 0.2843 (11) | 0.7782 (10) | 0.1825 (6) | 0.119 (4) | 0.33333 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Zn1 | 0.02248 (15) | 0.02248 (15) | 0.02224 (19) | 0.01124 (7) | 0.000 | 0.000 |
| N1 | 0.0302 (6) | 0.0399 (7) | 0.0295 (6) | 0.0185 (5) | −0.0038 (5) | 0.0024 (5) |
| C1 | 0.0255 (6) | 0.0417 (8) | 0.0407 (8) | 0.0134 (6) | −0.0072 (6) | −0.0070 (6) |
| S1 | 0.0230 (2) | 0.0230 (2) | 0.0218 (3) | 0.01150 (11) | 0.000 | 0.000 |
| O1 | 0.206 (10) | 0.116 (6) | 0.045 (3) | 0.131 (7) | 0.015 (4) | −0.008 (3) |
| O2 | 0.239 (11) | 0.159 (8) | 0.063 (4) | 0.177 (9) | 0.020 (5) | 0.028 (4) |
Geometric parameters (Å, º) top
| Zn1—N1i | 2.1866 (13) | S1—O1v | 1.430 (4) |
| Zn1—N1ii | 2.1866 (13) | S1—O1vi | 1.430 (4) |
| Zn1—N1iii | 2.1866 (13) | S1—O1 | 1.430 (4) |
| Zn1—N1 | 2.1866 (13) | S1—O1vii | 1.430 (4) |
| Zn1—N1iv | 2.1866 (13) | S1—O1viii | 1.430 (4) |
| Zn1—N1v | 2.1867 (13) | S1—O1ix | 1.430 (4) |
| N1—C1 | 1.469 (2) | S1—O2vii | 1.436 (5) |
| N1—H1N | 0.86 (3) | S1—O2 | 1.436 (5) |
| N1—H2N | 0.86 (2) | S1—O2ix | 1.436 (5) |
| C1—C1iii | 1.512 (3) | S1—O2vi | 1.436 (5) |
| C1—H1A | 0.9700 | S1—O2viii | 1.436 (5) |
| C1—H1B | 0.9700 | S1—O2v | 1.436 (5) |
| | | |
| N1i—Zn1—N1ii | 80.22 (7) | Zn1—N1—H1N | 114.3 (18) |
| N1i—Zn1—N1iii | 93.02 (5) | C1—N1—H2N | 110.8 (16) |
| N1ii—Zn1—N1iii | 94.56 (8) | Zn1—N1—H2N | 111.4 (17) |
| N1i—Zn1—N1 | 170.09 (8) | H1N—N1—H2N | 106 (2) |
| N1ii—Zn1—N1 | 93.02 (5) | N1—C1—C1iii | 109.14 (11) |
| N1iii—Zn1—N1 | 80.22 (7) | N1—C1—H1A | 109.9 |
| N1i—Zn1—N1iv | 94.56 (8) | C1iii—C1—H1A | 109.9 |
| N1ii—Zn1—N1iv | 93.02 (5) | N1—C1—H1B | 109.9 |
| N1iii—Zn1—N1iv | 170.09 (8) | C1iii—C1—H1B | 109.9 |
| N1—Zn1—N1iv | 93.02 (5) | H1A—C1—H1B | 108.3 |
| N1i—Zn1—N1v | 93.02 (5) | O1vi—S1—O1 | 112.3 (6) |
| N1ii—Zn1—N1v | 170.09 (8) | O1vi—S1—O2 | 110.7 (5) |
| N1iii—Zn1—N1v | 93.02 (5) | O1—S1—O2 | 108.7 (3) |
| N1—Zn1—N1v | 94.56 (8) | O1vi—S1—O2vi | 108.7 (3) |
| N1iv—Zn1—N1v | 80.22 (7) | O1—S1—O2vi | 110.7 (5) |
| C1—N1—Zn1 | 107.80 (10) | O2—S1—O2vi | 105.6 (7) |
| C1—N1—H1N | 106.5 (18) | | |
| | | |
| N1ii—Zn1—N1—C1 | 79.15 (13) | N1v—Zn1—N1—C1 | −107.24 (11) |
| N1iii—Zn1—N1—C1 | −14.96 (8) | Zn1—N1—C1—C1iii | 42.21 (18) |
| N1iv—Zn1—N1—C1 | 172.34 (10) | | |
| Symmetry codes: (i) −x+y+1, y, −z+1/2; (ii) −y+1, x−y, z; (iii) x, x−y, −z+1/2; (iv) −x+y+1, −x+1, z; (v) −y+1, −x+1, −z+1/2; (vi) −x+y, y, −z+1/2; (vii) x, x−y+1, −z+1/2; (viii) −y+1, x−y+1, z; (ix) −x+y, −x+1, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1 | 0.86 (3) | 1.99 (3) | 2.846 (6) | 172 (3) |
| N1—H2N···O2x | 0.86 (2) | 2.17 (2) | 3.029 (6) | 178 (2) |
| Symmetry code: (x) x−y+1, x, −z. |
Crystal data top
| C6H24CuN6·O4S | Dx = 1.704 Mg m−3 |
| Mr = 339.91 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, P31c | Cell parameters from 9767 reflections |
| Hall symbol: -P 3 2c | θ = 2.1–35.0° |
| a = 8.9387 (1) Å | µ = 1.82 mm−1 |
| c = 9.5766 (1) Å | T = 190 K |
| V = 662.66 (2) Å3 | Plate, blue |
| Z = 2 | 0.33 × 0.15 × 0.06 mm |
| F(000) = 358 | |
Data collection top
Nonius KappaCCD
diffractometer | 683 independent reflections |
| Radiation source: rotating anode | 644 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.025 |
| φ and ω scans | θmax = 30.5°, θmin = 2.6° |
Absorption correction: analytical
SADABS-2008/1 (Sheldrick, 2008b) | h = −12→12 |
| Tmin = 0.580, Tmax = 0.940 | k = −12→12 |
| 9708 measured reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.022 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.056 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.11 | w = 1/[σ2(Fo2) + (0.0277P)2 + 0.3168P]
where P = (Fo2 + 2Fc2)/3 |
| 683 reflections | (Δ/σ)max < 0.001 |
| 49 parameters | Δρmax = 0.45 e Å−3 |
| 16 restraints | Δρmin = −0.28 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Cu1 | 0.6667 | 0.3333 | 0.2500 | 0.01628 (11) | |
| N1 | 0.45572 (19) | 0.31150 (19) | 0.12709 (15) | 0.0333 (3) | |
| H1N | 0.435 (3) | 0.390 (3) | 0.135 (2) | 0.039 (6)* | |
| H2N | 0.471 (3) | 0.305 (3) | 0.042 (2) | 0.050 (6)* | |
| C1 | 0.29734 (18) | 0.1540 (2) | 0.17112 (16) | 0.0298 (3) | |
| H1A | 0.2940 | 0.0509 | 0.1297 | 0.036* | |
| H1B | 0.1943 | 0.1583 | 0.1390 | 0.036* | |
| S1 | 0.3333 | 0.6667 | 0.2500 | 0.01729 (15) | |
| O1 | 0.3523 (10) | 0.5611 (8) | 0.1455 (4) | 0.081 (2) | 0.33333 |
| O2 | 0.2834 (10) | 0.7788 (9) | 0.1815 (6) | 0.104 (3) | 0.33333 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cu1 | 0.01773 (13) | 0.01773 (13) | 0.01339 (17) | 0.00886 (7) | 0.000 | 0.000 |
| N1 | 0.0369 (7) | 0.0313 (6) | 0.0302 (6) | 0.0159 (6) | 0.0085 (5) | 0.0030 (5) |
| C1 | 0.0216 (6) | 0.0342 (7) | 0.0292 (7) | 0.0105 (5) | −0.0037 (5) | −0.0036 (5) |
| S1 | 0.0185 (2) | 0.0185 (2) | 0.0150 (3) | 0.00923 (10) | 0.000 | 0.000 |
| O1 | 0.170 (7) | 0.102 (5) | 0.032 (2) | 0.114 (5) | −0.003 (3) | −0.016 (3) |
| O2 | 0.210 (9) | 0.133 (6) | 0.058 (3) | 0.154 (7) | 0.013 (4) | 0.024 (4) |
Geometric parameters (Å, º) top
| Cu1—N1 | 2.1473 (16) | S1—O1vi | 1.442 (4) |
| Cu1—N1i | 2.1473 (16) | S1—O1vii | 1.442 (4) |
| Cu1—N1ii | 2.1473 (16) | S1—O1viii | 1.442 (4) |
| Cu1—N1iii | 2.1473 (16) | S1—O1 | 1.442 (4) |
| Cu1—N1iv | 2.1473 (16) | S1—O1v | 1.442 (4) |
| Cu1—N1v | 2.1473 (16) | S1—O1ix | 1.442 (4) |
| N1—C1 | 1.473 (2) | S1—O2vii | 1.443 (4) |
| N1—H1N | 0.81 (3) | S1—O2vi | 1.443 (4) |
| N1—H2N | 0.83 (2) | S1—O2 | 1.443 (4) |
| C1—C1iv | 1.514 (3) | S1—O2ix | 1.443 (4) |
| C1—H1A | 0.9900 | S1—O2v | 1.443 (4) |
| C1—H1B | 0.9900 | S1—O2viii | 1.443 (4) |
| | | |
| N1—Cu1—N1i | 92.83 (5) | Cu1—N1—H1N | 115.8 (16) |
| N1—Cu1—N1ii | 92.82 (5) | C1—N1—H2N | 109.0 (17) |
| N1i—Cu1—N1ii | 92.83 (5) | Cu1—N1—H2N | 111.8 (18) |
| N1—Cu1—N1iii | 170.97 (8) | H1N—N1—H2N | 106 (2) |
| N1i—Cu1—N1iii | 80.99 (7) | N1—C1—C1iv | 108.40 (10) |
| N1ii—Cu1—N1iii | 94.04 (8) | N1—C1—H1A | 110.0 |
| N1—Cu1—N1iv | 80.99 (7) | C1iv—C1—H1A | 110.0 |
| N1i—Cu1—N1iv | 94.04 (8) | N1—C1—H1B | 110.0 |
| N1ii—Cu1—N1iv | 170.97 (8) | C1iv—C1—H1B | 110.0 |
| N1iii—Cu1—N1iv | 92.82 (5) | H1A—C1—H1B | 108.4 |
| N1—Cu1—N1v | 94.04 (8) | O1viii—S1—O1 | 111.0 (5) |
| N1i—Cu1—N1v | 170.97 (8) | O1viii—S1—O2 | 111.3 (4) |
| N1ii—Cu1—N1v | 80.99 (7) | O1—S1—O2 | 108.5 (3) |
| N1iii—Cu1—N1v | 92.82 (5) | O1viii—S1—O2viii | 108.5 (3) |
| N1iv—Cu1—N1v | 92.82 (5) | O1—S1—O2viii | 111.3 (4) |
| C1—N1—Cu1 | 108.09 (10) | O2—S1—O2viii | 106.0 (6) |
| C1—N1—H1N | 105.9 (16) | | |
| | | |
| N1i—Cu1—N1—C1 | 78.55 (13) | N1v—Cu1—N1—C1 | −107.32 (11) |
| N1ii—Cu1—N1—C1 | 171.52 (10) | Cu1—N1—C1—C1iv | 42.31 (17) |
| N1iv—Cu1—N1—C1 | −15.09 (7) | | |
| Symmetry codes: (i) −y+1, x−y, z; (ii) −x+y+1, −x+1, z; (iii) −x+y+1, y, −z+1/2; (iv) x, x−y, −z+1/2; (v) −y+1, −x+1, −z+1/2; (vi) −y+1, x−y+1, z; (vii) −x+y, −x+1, z; (viii) −x+y, y, −z+1/2; (ix) x, x−y+1, −z+1/2. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1 | 0.81 (3) | 2.01 (3) | 2.816 (5) | 172 (2) |
| N1—H2N···O2x | 0.83 (2) | 2.19 (2) | 3.017 (6) | 179 (2) |
| Symmetry code: (x) x−y+1, x, −z. |
Crystal data top
| C6H24CuN6·O4S | Dx = 1.700 Mg m−3 |
| Mr = 339.91 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, P31c | Cell parameters from 9618 reflections |
| Hall symbol: -P 3 2c | θ = 2.1–35.0° |
| a = 8.9445 (1) Å | µ = 1.82 mm−1 |
| c = 9.5826 (1) Å | T = 210 K |
| V = 663.94 (2) Å3 | Plate, blue |
| Z = 2 | 0.33 × 0.15 × 0.06 mm |
| F(000) = 358 | |
Data collection top
Nonius KappaCCD
diffractometer | 685 independent reflections |
| Radiation source: rotating anode | 637 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.026 |
| φ and ω scans | θmax = 30.5°, θmin = 2.6° |
Absorption correction: analytical
SADABS-2008/1 (Sheldrick, 2008b) | h = −12→12 |
| Tmin = 0.581, Tmax = 0.940 | k = −12→12 |
| 9739 measured reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.022 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.057 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.09 | w = 1/[σ2(Fo2) + (0.0302P)2 + 0.2851P]
where P = (Fo2 + 2Fc2)/3 |
| 685 reflections | (Δ/σ)max < 0.001 |
| 49 parameters | Δρmax = 0.46 e Å−3 |
| 16 restraints | Δρmin = −0.26 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Cu1 | 0.6667 | 0.3333 | 0.2500 | 0.01760 (12) | |
| N1 | 0.4559 (2) | 0.31159 (19) | 0.12711 (15) | 0.0352 (3) | |
| H1N | 0.435 (3) | 0.390 (3) | 0.135 (2) | 0.047 (6)* | |
| H2N | 0.468 (3) | 0.304 (3) | 0.042 (2) | 0.051 (6)* | |
| C1 | 0.29761 (19) | 0.1542 (2) | 0.17121 (16) | 0.0319 (3) | |
| H1A | 0.2942 | 0.0522 | 0.1302 | 0.038* | |
| H1B | 0.1956 | 0.1584 | 0.1394 | 0.038* | |
| S1 | 0.3333 | 0.6667 | 0.2500 | 0.01877 (15) | |
| O1 | 0.3531 (11) | 0.5622 (9) | 0.1458 (5) | 0.087 (2) | 0.33333 |
| O2 | 0.2842 (11) | 0.7787 (9) | 0.1813 (6) | 0.107 (3) | 0.33333 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cu1 | 0.01900 (14) | 0.01900 (14) | 0.01480 (17) | 0.00950 (7) | 0.000 | 0.000 |
| N1 | 0.0385 (7) | 0.0341 (7) | 0.0318 (6) | 0.0173 (6) | 0.0084 (5) | 0.0036 (5) |
| C1 | 0.0240 (6) | 0.0358 (7) | 0.0319 (7) | 0.0120 (5) | −0.0045 (5) | −0.0041 (6) |
| S1 | 0.0200 (2) | 0.0200 (2) | 0.0163 (3) | 0.01000 (10) | 0.000 | 0.000 |
| O1 | 0.177 (8) | 0.108 (5) | 0.037 (2) | 0.117 (6) | −0.002 (3) | −0.018 (3) |
| O2 | 0.212 (10) | 0.139 (7) | 0.064 (3) | 0.157 (8) | 0.018 (5) | 0.028 (4) |
Geometric parameters (Å, º) top
| Cu1—N1i | 2.1479 (16) | S1—O1ii | 1.438 (4) |
| Cu1—N1ii | 2.1479 (16) | S1—O1vi | 1.438 (4) |
| Cu1—N1iii | 2.1479 (16) | S1—O1vii | 1.438 (4) |
| Cu1—N1 | 2.1479 (16) | S1—O1 | 1.438 (4) |
| Cu1—N1iv | 2.1479 (16) | S1—O1viii | 1.438 (4) |
| Cu1—N1v | 2.1479 (16) | S1—O1ix | 1.438 (4) |
| N1—C1 | 1.474 (2) | S1—O2 | 1.439 (4) |
| N1—H1N | 0.81 (3) | S1—O2vii | 1.439 (4) |
| N1—H2N | 0.83 (2) | S1—O2ii | 1.439 (4) |
| C1—C1iii | 1.513 (3) | S1—O2vi | 1.439 (4) |
| C1—H1A | 0.9800 | S1—O2ix | 1.439 (4) |
| C1—H1B | 0.9800 | S1—O2viii | 1.439 (4) |
| | | |
| N1i—Cu1—N1ii | 92.82 (5) | Cu1—N1—H1N | 115.9 (17) |
| N1i—Cu1—N1iii | 92.82 (5) | C1—N1—H2N | 108.0 (17) |
| N1ii—Cu1—N1iii | 92.82 (5) | Cu1—N1—H2N | 113.4 (18) |
| N1i—Cu1—N1 | 171.00 (8) | H1N—N1—H2N | 105 (2) |
| N1ii—Cu1—N1 | 94.02 (8) | N1—C1—C1iii | 108.45 (11) |
| N1iii—Cu1—N1 | 81.02 (7) | N1—C1—H1A | 110.0 |
| N1i—Cu1—N1iv | 94.02 (8) | C1iii—C1—H1A | 110.0 |
| N1ii—Cu1—N1iv | 81.02 (7) | N1—C1—H1B | 110.0 |
| N1iii—Cu1—N1iv | 171.00 (8) | C1iii—C1—H1B | 110.0 |
| N1—Cu1—N1iv | 92.82 (5) | H1A—C1—H1B | 108.4 |
| N1i—Cu1—N1v | 81.02 (7) | O1vii—S1—O1 | 111.5 (6) |
| N1ii—Cu1—N1v | 171.00 (8) | O1vii—S1—O2 | 111.4 (5) |
| N1iii—Cu1—N1v | 94.02 (8) | O1—S1—O2 | 108.3 (3) |
| N1—Cu1—N1v | 92.82 (5) | O2—S1—O2vii | 105.8 (7) |
| N1iv—Cu1—N1v | 92.82 (5) | O1vii—S1—O2vii | 108.3 (3) |
| C1—N1—Cu1 | 108.04 (10) | O1—S1—O2vii | 111.4 (5) |
| C1—N1—H1N | 105.8 (17) | | |
| | | |
| N1ii—Cu1—N1—C1 | −107.29 (11) | N1v—Cu1—N1—C1 | 78.56 (13) |
| N1iii—Cu1—N1—C1 | −15.07 (7) | Cu1—N1—C1—C1iii | 42.28 (17) |
| N1iv—Cu1—N1—C1 | 171.52 (10) | | |
| Symmetry codes: (i) −x+y+1, y, −z+1/2; (ii) −y+1, −x+1, −z+1/2; (iii) x, x−y, −z+1/2; (iv) −x+y+1, −x+1, z; (v) −y+1, x−y, z; (vi) x, x−y+1, −z+1/2; (vii) −x+y, y, −z+1/2; (viii) −y+1, x−y+1, z; (ix) −x+y, −x+1, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1 | 0.81 (3) | 2.01 (3) | 2.821 (5) | 172 (2) |
| N1—H2N···O2x | 0.83 (2) | 2.19 (2) | 3.017 (6) | 179 (2) |
| Symmetry code: (x) x−y+1, x, −z. |
Crystal data top
| C6H24CuN6·O4S | Dx = 1.697 Mg m−3 |
| Mr = 339.91 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, P31c | Cell parameters from 9445 reflections |
| Hall symbol: -P 3 2c | θ = 2.1–35.0° |
| a = 8.9513 (1) Å | µ = 1.82 mm−1 |
| c = 9.5876 (1) Å | T = 230 K |
| V = 665.29 (2) Å3 | Plate, blue |
| Z = 2 | 0.33 × 0.15 × 0.06 mm |
| F(000) = 358 | |
Data collection top
Nonius KappaCCD
diffractometer | 687 independent reflections |
| Radiation source: rotating anode | 642 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.027 |
| φ and ω scans | θmax = 30.5°, θmin = 2.6° |
Absorption correction: analytical
SADABS-2008/1 (Sheldrick, 2008b) | h = −12→12 |
| Tmin = 0.578, Tmax = 0.937 | k = −12→12 |
| 9736 measured reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.023 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.060 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.12 | w = 1/[σ2(Fo2) + (0.0315P)2 + 0.2737P]
where P = (Fo2 + 2Fc2)/3 |
| 687 reflections | (Δ/σ)max < 0.001 |
| 49 parameters | Δρmax = 0.51 e Å−3 |
| 16 restraints | Δρmin = −0.28 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Cu1 | 0.6667 | 0.3333 | 0.2500 | 0.01892 (12) | |
| N1 | 0.4561 (2) | 0.3115 (2) | 0.12728 (16) | 0.0369 (3) | |
| H1N | 0.437 (3) | 0.390 (4) | 0.135 (3) | 0.048 (7)* | |
| H2N | 0.469 (3) | 0.304 (3) | 0.043 (3) | 0.052 (7)* | |
| C1 | 0.2978 (2) | 0.1543 (2) | 0.17130 (17) | 0.0339 (3) | |
| H1A | 0.2944 | 0.0523 | 0.1303 | 0.041* | |
| H1B | 0.1960 | 0.1586 | 0.1395 | 0.041* | |
| S1 | 0.3333 | 0.6667 | 0.2500 | 0.02010 (16) | |
| O1 | 0.3538 (11) | 0.5626 (9) | 0.1460 (5) | 0.091 (3) | 0.33333 |
| O2 | 0.2842 (11) | 0.7787 (10) | 0.1816 (6) | 0.113 (4) | 0.33333 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cu1 | 0.02039 (15) | 0.02039 (15) | 0.01598 (18) | 0.01020 (7) | 0.000 | 0.000 |
| N1 | 0.0410 (8) | 0.0362 (7) | 0.0327 (7) | 0.0186 (6) | 0.0079 (6) | 0.0036 (5) |
| C1 | 0.0248 (7) | 0.0386 (8) | 0.0340 (7) | 0.0126 (6) | −0.0049 (5) | −0.0044 (6) |
| S1 | 0.0214 (2) | 0.0214 (2) | 0.0176 (3) | 0.01069 (11) | 0.000 | 0.000 |
| O1 | 0.187 (9) | 0.112 (5) | 0.040 (3) | 0.123 (6) | 0.002 (4) | −0.016 (3) |
| O2 | 0.225 (11) | 0.142 (7) | 0.069 (4) | 0.164 (8) | 0.016 (5) | 0.029 (4) |
Geometric parameters (Å, º) top
| Cu1—N1i | 2.1466 (17) | S1—O1vi | 1.437 (4) |
| Cu1—N1ii | 2.1466 (17) | S1—O1vii | 1.437 (4) |
| Cu1—N1iii | 2.1467 (17) | S1—O1viii | 1.437 (4) |
| Cu1—N1 | 2.1467 (17) | S1—O1ii | 1.437 (4) |
| Cu1—N1iv | 2.1467 (17) | S1—O1ix | 1.437 (4) |
| Cu1—N1v | 2.1467 (17) | S1—O1 | 1.437 (4) |
| N1—C1 | 1.474 (2) | S1—O2vi | 1.439 (4) |
| N1—H1N | 0.80 (3) | S1—O2vii | 1.439 (4) |
| N1—H2N | 0.83 (2) | S1—O2 | 1.439 (4) |
| C1—C1iii | 1.512 (3) | S1—O2ix | 1.439 (4) |
| C1—H1A | 0.9800 | S1—O2viii | 1.439 (4) |
| C1—H1B | 0.9800 | S1—O2ii | 1.439 (4) |
| | | |
| N1i—Cu1—N1ii | 80.98 (8) | Cu1—N1—H1N | 115.6 (18) |
| N1i—Cu1—N1iii | 170.96 (9) | C1—N1—H2N | 108.0 (18) |
| N1ii—Cu1—N1iii | 92.83 (6) | Cu1—N1—H2N | 113.1 (19) |
| N1i—Cu1—N1 | 92.83 (6) | H1N—N1—H2N | 105 (3) |
| N1ii—Cu1—N1 | 94.04 (9) | N1—C1—C1iii | 108.42 (11) |
| N1iii—Cu1—N1 | 80.98 (8) | N1—C1—H1A | 110.0 |
| N1i—Cu1—N1iv | 92.83 (6) | C1iii—C1—H1A | 110.0 |
| N1ii—Cu1—N1iv | 170.96 (9) | N1—C1—H1B | 110.0 |
| N1iii—Cu1—N1iv | 94.04 (9) | C1iii—C1—H1B | 110.0 |
| N1—Cu1—N1iv | 92.83 (6) | H1A—C1—H1B | 108.4 |
| N1i—Cu1—N1v | 94.04 (9) | O1ix—S1—O1 | 111.7 (6) |
| N1ii—Cu1—N1v | 92.83 (6) | O1ix—S1—O2 | 111.1 (5) |
| N1iii—Cu1—N1v | 92.83 (6) | O1—S1—O2 | 108.5 (3) |
| N1—Cu1—N1v | 170.96 (9) | O1ix—S1—O2ix | 108.5 (3) |
| N1iv—Cu1—N1v | 80.98 (8) | O1—S1—O2ix | 111.1 (5) |
| C1—N1—Cu1 | 108.12 (11) | O2—S1—O2ix | 105.7 (7) |
| C1—N1—H1N | 106.4 (18) | | |
| | | |
| N1i—Cu1—N1—C1 | 171.56 (11) | N1iv—Cu1—N1—C1 | 78.58 (14) |
| N1ii—Cu1—N1—C1 | −107.29 (12) | Cu1—N1—C1—C1iii | 42.25 (18) |
| N1iii—Cu1—N1—C1 | −15.06 (8) | | |
| Symmetry codes: (i) −x+y+1, −x+1, z; (ii) −y+1, −x+1, −z+1/2; (iii) x, x−y, −z+1/2; (iv) −y+1, x−y, z; (v) −x+y+1, y, −z+1/2; (vi) −x+y, −x+1, z; (vii) −y+1, x−y+1, z; (viii) x, x−y+1, −z+1/2; (ix) −x+y, y, −z+1/2. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1 | 0.80 (3) | 2.03 (3) | 2.825 (6) | 172 (3) |
| N1—H2N···O2x | 0.83 (2) | 2.20 (3) | 3.022 (6) | 179 (2) |
| Symmetry code: (x) x−y+1, x, −z. |
Crystal data top
| C6H24CuN6·O4S | Dx = 1.694 Mg m−3 |
| Mr = 339.91 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, P31c | Cell parameters from 9343 reflections |
| Hall symbol: -P 3 2c | θ = 2.1–35.0° |
| a = 8.9572 (1) Å | µ = 1.81 mm−1 |
| c = 9.5926 (1) Å | T = 250 K |
| V = 666.52 (2) Å3 | Plate, blue |
| Z = 2 | 0.33 × 0.15 × 0.06 mm |
| F(000) = 358 | |
Data collection top
Nonius KappaCCD
diffractometer | 687 independent reflections |
| Radiation source: rotating anode | 638 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.027 |
| φ and ω scans | θmax = 30.5°, θmin = 2.6° |
Absorption correction: analytical
SADABS-2008/1 (Sheldrick, 2008b) | h = −12→12 |
| Tmin = 0.579, Tmax = 0.947 | k = −12→12 |
| 9785 measured reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.024 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.061 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.10 | w = 1/[σ2(Fo2) + (0.031P)2 + 0.3143P]
where P = (Fo2 + 2Fc2)/3 |
| 687 reflections | (Δ/σ)max < 0.001 |
| 49 parameters | Δρmax = 0.52 e Å−3 |
| 16 restraints | Δρmin = −0.29 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Cu1 | 0.6667 | 0.3333 | 0.2500 | 0.02025 (13) | |
| N1 | 0.4560 (2) | 0.3115 (2) | 0.12733 (17) | 0.0385 (3) | |
| H1N | 0.436 (3) | 0.390 (4) | 0.135 (3) | 0.048 (7)* | |
| H2N | 0.469 (3) | 0.304 (3) | 0.043 (3) | 0.056 (7)* | |
| C1 | 0.2980 (2) | 0.1544 (2) | 0.17132 (18) | 0.0361 (3) | |
| H1A | 0.2946 | 0.0526 | 0.1303 | 0.043* | |
| H1B | 0.1962 | 0.1587 | 0.1396 | 0.043* | |
| S1 | 0.3333 | 0.6667 | 0.2500 | 0.02154 (16) | |
| O1 | 0.3551 (12) | 0.5636 (9) | 0.1464 (5) | 0.096 (3) | 0.33333 |
| O2 | 0.2847 (11) | 0.7790 (10) | 0.1813 (7) | 0.116 (4) | 0.33333 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cu1 | 0.02168 (15) | 0.02168 (15) | 0.01737 (19) | 0.01084 (7) | 0.000 | 0.000 |
| N1 | 0.0427 (8) | 0.0376 (7) | 0.0347 (7) | 0.0198 (6) | 0.0076 (6) | 0.0034 (6) |
| C1 | 0.0266 (7) | 0.0402 (8) | 0.0370 (8) | 0.0134 (6) | −0.0056 (6) | −0.0050 (6) |
| S1 | 0.0227 (2) | 0.0227 (2) | 0.0193 (3) | 0.01133 (11) | 0.000 | 0.000 |
| O1 | 0.191 (9) | 0.119 (6) | 0.045 (3) | 0.127 (7) | 0.001 (4) | −0.019 (3) |
| O2 | 0.222 (11) | 0.145 (8) | 0.075 (4) | 0.163 (9) | 0.019 (5) | 0.032 (5) |
Geometric parameters (Å, º) top
| Cu1—N1i | 2.1479 (17) | S1—O1vi | 1.434 (4) |
| Cu1—N1ii | 2.1479 (17) | S1—O1vii | 1.434 (4) |
| Cu1—N1 | 2.1479 (17) | S1—O1 | 1.434 (4) |
| Cu1—N1iii | 2.1479 (17) | S1—O1viii | 1.434 (4) |
| Cu1—N1iv | 2.1479 (17) | S1—O1iii | 1.434 (4) |
| Cu1—N1v | 2.1479 (17) | S1—O1ix | 1.434 (4) |
| N1—C1 | 1.473 (2) | S1—O2viii | 1.440 (5) |
| N1—H1N | 0.81 (3) | S1—O2ix | 1.440 (5) |
| N1—H2N | 0.83 (3) | S1—O2 | 1.440 (5) |
| C1—C1i | 1.513 (3) | S1—O2vii | 1.440 (5) |
| C1—H1A | 0.9800 | S1—O2vi | 1.440 (5) |
| C1—H1B | 0.9800 | S1—O2iii | 1.440 (5) |
| | | |
| N1i—Cu1—N1ii | 94.01 (9) | Cu1—N1—H1N | 116.1 (18) |
| N1i—Cu1—N1 | 80.96 (8) | C1—N1—H2N | 108.4 (18) |
| N1ii—Cu1—N1 | 92.85 (6) | Cu1—N1—H2N | 113.0 (19) |
| N1i—Cu1—N1iii | 92.85 (6) | H1N—N1—H2N | 105 (3) |
| N1ii—Cu1—N1iii | 170.97 (9) | N1—C1—C1i | 108.44 (12) |
| N1—Cu1—N1iii | 94.01 (9) | N1—C1—H1A | 110.0 |
| N1i—Cu1—N1iv | 92.85 (6) | C1i—C1—H1A | 110.0 |
| N1ii—Cu1—N1iv | 80.96 (8) | N1—C1—H1B | 110.0 |
| N1—Cu1—N1iv | 170.97 (9) | C1i—C1—H1B | 110.0 |
| N1iii—Cu1—N1iv | 92.85 (6) | H1A—C1—H1B | 108.4 |
| N1i—Cu1—N1v | 170.97 (9) | O1vi—S1—O1 | 112.3 (6) |
| N1ii—Cu1—N1v | 92.85 (6) | O1vi—S1—O2 | 110.8 (5) |
| N1—Cu1—N1v | 92.85 (6) | O1—S1—O2 | 108.6 (3) |
| N1iii—Cu1—N1v | 80.96 (8) | O1vi—S1—O2vi | 108.6 (3) |
| N1iv—Cu1—N1v | 94.01 (9) | O1—S1—O2vi | 110.8 (5) |
| C1—N1—Cu1 | 108.10 (11) | O2—S1—O2vi | 105.6 (7) |
| C1—N1—H1N | 106.3 (18) | | |
| | | |
| N1i—Cu1—N1—C1 | −15.06 (8) | N1v—Cu1—N1—C1 | 171.55 (11) |
| N1ii—Cu1—N1—C1 | 78.55 (15) | Cu1—N1—C1—C1i | 42.26 (19) |
| N1iii—Cu1—N1—C1 | −107.32 (12) | | |
| Symmetry codes: (i) x, x−y, −z+1/2; (ii) −y+1, x−y, z; (iii) −y+1, −x+1, −z+1/2; (iv) −x+y+1, y, −z+1/2; (v) −x+y+1, −x+1, z; (vi) −x+y, y, −z+1/2; (vii) −x+y, −x+1, z; (viii) x, x−y+1, −z+1/2; (ix) −y+1, x−y+1, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1 | 0.81 (3) | 2.02 (3) | 2.827 (6) | 173 (3) |
| N1—H2N···O2x | 0.83 (3) | 2.19 (3) | 3.022 (7) | 180 (3) |
| Symmetry code: (x) x−y+1, x, −z. |
Crystal data top
| C6H24CuN6·O4S | Dx = 1.689 Mg m−3 |
| Mr = 339.91 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, P31c | Cell parameters from 8734 reflections |
| Hall symbol: -P 3 2c | θ = 2.1–35.0° |
| a = 8.9664 (1) Å | µ = 1.81 mm−1 |
| c = 9.5996 (1) Å | T = 270 K |
| V = 668.38 (2) Å3 | Plate, blue |
| Z = 2 | 0.33 × 0.15 × 0.06 mm |
| F(000) = 358 | |
Data collection top
Nonius KappaCCD
diffractometer | 690 independent reflections |
| Radiation source: rotating anode | 639 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.027 |
| φ and ω scans | θmax = 30.5°, θmin = 2.6° |
Absorption correction: analytical
SADABS-2008/1 (Sheldrick, 2008b) | h = −12→12 |
| Tmin = 0.578, Tmax = 0.947 | k = −12→12 |
| 9819 measured reflections | l = −13→13 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.025 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.065 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.12 | w = 1/[σ2(Fo2) + (0.0364P)2 + 0.2214P]
where P = (Fo2 + 2Fc2)/3 |
| 690 reflections | (Δ/σ)max < 0.001 |
| 49 parameters | Δρmax = 0.56 e Å−3 |
| 16 restraints | Δρmin = −0.35 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Cu1 | 0.6667 | 0.3333 | 0.2500 | 0.02174 (13) | |
| N1 | 0.4561 (2) | 0.3115 (2) | 0.12731 (17) | 0.0405 (3) | |
| H1N | 0.434 (4) | 0.389 (4) | 0.134 (3) | 0.054 (7)* | |
| H2N | 0.469 (4) | 0.304 (3) | 0.042 (3) | 0.057 (7)* | |
| C1 | 0.2983 (2) | 0.1545 (2) | 0.17148 (18) | 0.0387 (4) | |
| H1A | 0.2951 | 0.0537 | 0.1310 | 0.046* | |
| H1B | 0.1976 | 0.1585 | 0.1401 | 0.046* | |
| S1 | 0.3333 | 0.6667 | 0.2500 | 0.02308 (17) | |
| O1 | 0.3561 (12) | 0.5645 (10) | 0.1468 (6) | 0.102 (3) | 0.33333 |
| O2 | 0.2845 (11) | 0.7783 (10) | 0.1814 (7) | 0.121 (4) | 0.33333 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cu1 | 0.02328 (15) | 0.02328 (15) | 0.01866 (19) | 0.01164 (8) | 0.000 | 0.000 |
| N1 | 0.0443 (8) | 0.0405 (8) | 0.0364 (7) | 0.0210 (6) | 0.0076 (6) | 0.0038 (6) |
| C1 | 0.0290 (7) | 0.0438 (9) | 0.0398 (8) | 0.0155 (6) | −0.0060 (6) | −0.0052 (7) |
| S1 | 0.0242 (2) | 0.0242 (2) | 0.0208 (3) | 0.01211 (11) | 0.000 | 0.000 |
| O1 | 0.201 (10) | 0.126 (6) | 0.050 (3) | 0.134 (7) | 0.003 (4) | −0.019 (4) |
| O2 | 0.226 (11) | 0.156 (8) | 0.081 (4) | 0.171 (9) | 0.020 (6) | 0.035 (5) |
Geometric parameters (Å, º) top
| Cu1—N1i | 2.1495 (18) | S1—O1vi | 1.431 (5) |
| Cu1—N1 | 2.1495 (18) | S1—O1vii | 1.431 (5) |
| Cu1—N1ii | 2.1495 (18) | S1—O1viii | 1.431 (5) |
| Cu1—N1iii | 2.1495 (18) | S1—O1 | 1.431 (5) |
| Cu1—N1iv | 2.1495 (18) | S1—O1ix | 1.431 (5) |
| Cu1—N1v | 2.1495 (18) | S1—O1i | 1.431 (5) |
| N1—C1 | 1.474 (2) | S1—O2ix | 1.437 (5) |
| N1—H1N | 0.82 (3) | S1—O2vii | 1.437 (5) |
| N1—H2N | 0.84 (3) | S1—O2vi | 1.437 (5) |
| C1—C1ii | 1.511 (4) | S1—O2 | 1.437 (5) |
| C1—H1A | 0.9700 | S1—O2viii | 1.437 (5) |
| C1—H1B | 0.9700 | S1—O2i | 1.437 (5) |
| | | |
| N1i—Cu1—N1 | 94.03 (9) | Cu1—N1—H1N | 117.2 (19) |
| N1i—Cu1—N1ii | 92.85 (6) | C1—N1—H2N | 108.1 (18) |
| N1—Cu1—N1ii | 80.96 (8) | Cu1—N1—H2N | 113.0 (19) |
| N1i—Cu1—N1iii | 80.96 (8) | H1N—N1—H2N | 105 (3) |
| N1—Cu1—N1iii | 92.85 (6) | N1—C1—C1ii | 108.48 (12) |
| N1ii—Cu1—N1iii | 170.97 (9) | N1—C1—H1A | 110.0 |
| N1i—Cu1—N1iv | 170.96 (9) | C1ii—C1—H1A | 110.0 |
| N1—Cu1—N1iv | 92.85 (6) | N1—C1—H1B | 110.0 |
| N1ii—Cu1—N1iv | 94.02 (9) | C1ii—C1—H1B | 110.0 |
| N1iii—Cu1—N1iv | 92.84 (6) | H1A—C1—H1B | 108.4 |
| N1i—Cu1—N1v | 92.85 (6) | O1viii—S1—O1 | 112.7 (6) |
| N1—Cu1—N1v | 170.96 (9) | O1viii—S1—O2 | 110.5 (5) |
| N1ii—Cu1—N1v | 92.84 (6) | O1—S1—O2 | 108.6 (3) |
| N1iii—Cu1—N1v | 94.02 (9) | O1viii—S1—O2viii | 108.6 (3) |
| N1iv—Cu1—N1v | 80.96 (8) | O1—S1—O2viii | 110.5 (5) |
| C1—N1—Cu1 | 108.01 (11) | O2—S1—O2viii | 105.8 (7) |
| C1—N1—H1N | 105.2 (19) | | |
| | | |
| N1i—Cu1—N1—C1 | −107.30 (12) | N1iv—Cu1—N1—C1 | 78.57 (15) |
| N1ii—Cu1—N1—C1 | −15.06 (8) | Cu1—N1—C1—C1ii | 42.33 (19) |
| N1iii—Cu1—N1—C1 | 171.56 (11) | | |
| Symmetry codes: (i) −y+1, −x+1, −z+1/2; (ii) x, x−y, −z+1/2; (iii) −x+y+1, −x+1, z; (iv) −y+1, x−y, z; (v) −x+y+1, y, −z+1/2; (vi) −x+y, −x+1, z; (vii) x, x−y+1, −z+1/2; (viii) −x+y, y, −z+1/2; (ix) −y+1, x−y+1, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1 | 0.82 (3) | 2.01 (3) | 2.832 (6) | 175 (3) |
| N1—H2N···O2x | 0.84 (3) | 2.19 (3) | 3.025 (7) | 180 (3) |
| Symmetry code: (x) x−y+1, x, −z. |

 Subscribe to Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials
Subscribe to Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
support@iucr.org for assistance.

 Subscribe to Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials
Subscribe to Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials
 journal menu
journal menu









 Subscribe to Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials
Subscribe to Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials


