Dibarium μ-oxido-bis[pentachloridoruthenate(IV)] decahydrate, Ba
2Ru
2Cl
10O·10H
2O, has been prepared from ruthenium(III) chloride and barium chloride in hydrochloric acid. It crystallizes in the monoclinic system (space group
C2/
c). The structure consists of alternating layers of [Ru
2Cl
10O]
4− and [Ba(H
2O)
7]
2+ complex ions along the
a direction. The O atom bonded to ruthenium occupies the 4
e site, with
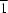
symmetry, while the other atoms occupy general 8
f sites. The overall structure is held together by O—H

O hydrogen bonds and O—H

Cl dipole–dipole interactions.
Supporting information
The title compound was crystallized from a supersaturated hydrochloric acid
solution (50%, 5 ml) prepared using doubly distilled water and a mixture of
ruthenium(III) chloride trihydrate (2.61 g) and dehydrated barium chloride
(2.263 g). Brown plates of Ba2Ru2Cl10O.10H2O were obtained at ambient
temperature by slow evaporation of the solution.
H atoms were fixed by geometric constraints using the HFIX command
(Sheldrick, 1997) and allowed to ride on the attached O atom [this appears to
contradict following sentence]. The O—H distances were restrained to
0.85 (s.u. value?) Å to ensure chemically reasonable geometry, with
Uiso(H) fixed at 1.5Ueq(O). Please check the values in
Table 2; there are minor discrepancies between O—H values here and in
_geom_bond_distance.
Data collection: COLLECT (Nonius, 2002); cell refinement: DIRAX (Duisenberg, 1992); data reduction: EVAL (Nonius, 2002); program(s) used to solve structure: SIR92 (Altomare et al., 1993); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: DIAMOND (Brandenburg, 1998) and ATOMS (Dowty, 1995); software used to prepare material for publication: WinGX (Farrugia, 1999) and PARST (Nardelli, 1995).
dibarium µ-oxo-bis(pentachlororuthenate(IV)) decahydrate
top
Crystal data top
| Ba2Ru2Cl10O·10H2O | F(000) = 1912 |
| Mr = 1027.48 | Dx = 2.78 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 2096 reflections |
| a = 20.9386 (18) Å | θ = 5.0–26.4° |
| b = 8.8654 (7) Å | µ = 5.49 mm−1 |
| c = 16.0560 (15) Å | T = 293 K |
| β = 124.559 (5)° | Plate, brown |
| V = 2454.5 (4) Å3 | 0.25 × 0.13 × 0.03 mm |
| Z = 4 | |
Data collection top
Nonius KappaCCD
diffractometer | 2488 independent reflections |
| Radiation source: fine-focus sealed tube | 2103 reflections with I > 2σ(I) |
| Detector resolution: 9 pixels mm-1 | Rint = 0.05 |
| CCD scans | θmax = 26.4°, θmin = 5° |
Absorption correction: part of the refinement model (ΔF)
(Sheldrick, 1996 or 1990???) | h = −24→26 |
| Tmin = 0.441, Tmax = 0.862 | k = −11→10 |
| 10480 measured reflections | l = −20→18 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.029 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.070 | Only H-atom coordinates refined |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0242P)2 + 9.3433P]
where P = (Fo2 + 2Fc2)/3 |
| 2488 reflections | (Δ/σ)max = 0.001 |
| 144 parameters | Δρmax = 0.53 e Å−3 |
| 15 restraints | Δρmin = −1.28 e Å−3 |
Crystal data top
| Ba2Ru2Cl10O·10H2O | V = 2454.5 (4) Å3 |
| Mr = 1027.48 | Z = 4 |
| Monoclinic, C2/c | Mo Kα radiation |
| a = 20.9386 (18) Å | µ = 5.49 mm−1 |
| b = 8.8654 (7) Å | T = 293 K |
| c = 16.0560 (15) Å | 0.25 × 0.13 × 0.03 mm |
| β = 124.559 (5)° | |
Data collection top
Nonius KappaCCD
diffractometer | 2488 independent reflections |
Absorption correction: part of the refinement model (ΔF)
(Sheldrick, 1996 or 1990???) | 2103 reflections with I > 2σ(I) |
| Tmin = 0.441, Tmax = 0.862 | Rint = 0.05 |
| 10480 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.029 | 15 restraints |
| wR(F2) = 0.070 | Only H-atom coordinates refined |
| S = 1.06 | Δρmax = 0.53 e Å−3 |
| 2488 reflections | Δρmin = −1.28 e Å−3 |
| 144 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Ba1 | 0.190281 (18) | 0.46951 (4) | 0.66934 (2) | 0.03451 (11) | |
| Ru1 | 0.39877 (2) | 0.50729 (4) | 0.65883 (2) | 0.02010 (11) | |
| Cl1 | 0.41487 (7) | 0.55179 (14) | 0.52648 (8) | 0.0317 (3) | |
| Cl2 | 0.37459 (7) | 0.45890 (14) | 0.78403 (8) | 0.0302 (3) | |
| Cl3 | 0.26475 (6) | 0.51664 (14) | 0.53772 (8) | 0.0320 (3) | |
| Cl4 | 0.39331 (7) | 0.24665 (13) | 0.62485 (10) | 0.0352 (3) | |
| Cl5 | 0.39538 (7) | 0.76962 (13) | 0.68603 (10) | 0.0353 (3) | |
| O1 | 0.5 | 0.5020 (4) | 0.75 | 0.0204 (9) | |
| O1W | 0.0670 (3) | 0.2681 (4) | 0.5698 (3) | 0.0488 (10) | |
| H21W | 0.079 (4) | 0.183 (5) | 0.585 (4) | 0.073* | |
| H11W | 0.037 (4) | 0.271 (7) | 0.5060 (10) | 0.073* | |
| O2W | 0.2671 (2) | 0.2783 (4) | 0.8455 (3) | 0.0374 (8) | |
| H12W | 0.304 (3) | 0.323 (7) | 0.893 (4) | 0.056* | |
| H22W | 0.234 (3) | 0.277 (8) | 0.852 (5) | 0.056* | |
| O3W | 0.0683 (3) | 0.5963 (6) | 0.6596 (4) | 0.0580 (12) | |
| H13W | 0.066 (4) | 0.634 (9) | 0.698 (4) | 0.087* | |
| H23W | 0.026 (3) | 0.576 (10) | 0.614 (4) | 0.087* | |
| O4W | 0.2458 (2) | 0.1701 (5) | 0.6492 (3) | 0.0420 (9) | |
| H24W | 0.209 (3) | 0.127 (7) | 0.596 (4) | 0.063* | |
| H14W | 0.281 (3) | 0.177 (8) | 0.645 (5) | 0.063* | |
| O5W | 0.0767 (2) | 0.5701 (5) | 0.4729 (3) | 0.0459 (9) | |
| H15W | 0.043 (4) | 0.616 (8) | 0.472 (5) | 0.069* | |
| H25W | 0.081 (4) | 0.607 (8) | 0.425 (5) | 0.069* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Ba1 | 0.02783 (17) | 0.03410 (18) | 0.03573 (18) | 0.00183 (12) | 0.01453 (14) | 0.00030 (13) |
| Ru1 | 0.01603 (18) | 0.02381 (19) | 0.01740 (18) | −0.00012 (13) | 0.00764 (14) | 0.00005 (14) |
| Cl1 | 0.0326 (6) | 0.0405 (7) | 0.0246 (5) | 0.0010 (5) | 0.0178 (5) | 0.0030 (5) |
| Cl2 | 0.0266 (6) | 0.0404 (6) | 0.0247 (5) | −0.0029 (5) | 0.0153 (5) | 0.0011 (5) |
| Cl3 | 0.0184 (6) | 0.0464 (7) | 0.0237 (5) | 0.0004 (5) | 0.0075 (5) | −0.0041 (5) |
| Cl4 | 0.0372 (7) | 0.0259 (6) | 0.0415 (7) | −0.0035 (5) | 0.0218 (6) | −0.0067 (5) |
| Cl5 | 0.0336 (6) | 0.0288 (6) | 0.0417 (7) | 0.0015 (5) | 0.0202 (6) | 0.0004 (5) |
| O1 | 0.018 (2) | 0.024 (2) | 0.018 (2) | 0 | 0.0085 (17) | 0 |
| O1W | 0.055 (3) | 0.033 (2) | 0.059 (3) | −0.0048 (19) | 0.033 (2) | 0.0003 (19) |
| O2W | 0.040 (2) | 0.041 (2) | 0.0342 (19) | −0.0075 (18) | 0.0227 (18) | −0.0021 (16) |
| O3W | 0.048 (3) | 0.062 (3) | 0.066 (3) | −0.008 (2) | 0.034 (2) | −0.027 (3) |
| O4W | 0.042 (2) | 0.050 (2) | 0.0344 (19) | −0.0087 (19) | 0.0221 (18) | −0.0114 (18) |
| O5W | 0.050 (2) | 0.041 (2) | 0.051 (2) | −0.0005 (19) | 0.030 (2) | 0.0033 (19) |
Geometric parameters (Å, º) top
| Ba1—O3W | 2.713 (4) | Ru1—Cl5 | 2.3755 (15) |
| Ba1—O1W | 2.780 (4) | O1—Ru1ii | 1.7657 (4) |
| Ba1—O5W | 2.809 (4) | O1W—H21W | 0.79 (3) |
| Ba1—O2W | 2.882 (4) | O1W—H11W | 0.846 (10) |
| Ba1—O2Wi | 2.931 (4) | O2W—Ba1iii | 2.931 (4) |
| Ba1—O4W | 2.988 (4) | O2W—H12W | 0.81 (4) |
| Ba1—O4Wi | 3.004 (4) | O2W—H22W | 0.76 (4) |
| Ba1—Cl2 | 3.1986 (12) | O3W—H13W | 0.73 (5) |
| Ba1—Cl3 | 3.2882 (13) | O3W—H23W | 0.78 (4) |
| Ru1—O1 | 1.7657 (4) | O4W—Ba1iii | 3.004 (4) |
| Ru1—Cl3 | 2.3394 (12) | O4W—H24W | 0.85 (3) |
| Ru1—Cl4 | 2.3627 (14) | O4W—H14W | 0.78 (4) |
| Ru1—Cl1 | 2.3665 (12) | O5W—H15W | 0.81 (5) |
| Ru1—Cl2 | 2.3744 (12) | O5W—H25W | 0.88 (5) |
| | | |
| O3W—Ba1—O1W | 72.33 (13) | Cl3—Ru1—Cl4 | 87.97 (4) |
| O3W—Ba1—O5W | 68.55 (14) | O1—Ru1—Cl1 | 91.93 (4) |
| O1W—Ba1—O5W | 68.89 (12) | Cl3—Ru1—Cl1 | 87.84 (4) |
| O3W—Ba1—O2W | 106.46 (14) | Cl4—Ru1—Cl1 | 88.20 (5) |
| O1W—Ba1—O2W | 90.31 (12) | O1—Ru1—Cl2 | 91.45 (4) |
| O5W—Ba1—O2W | 159.20 (12) | Cl3—Ru1—Cl2 | 88.79 (4) |
| O3W—Ba1—O2Wi | 85.88 (14) | Cl4—Ru1—Cl2 | 90.75 (4) |
| O1W—Ba1—O2Wi | 141.23 (12) | Cl1—Ru1—Cl2 | 176.50 (4) |
| O5W—Ba1—O2Wi | 73.44 (11) | O1—Ru1—Cl5 | 91.19 (13) |
| O2W—Ba1—O2Wi | 127.12 (6) | Cl3—Ru1—Cl5 | 88.35 (4) |
| O3W—Ba1—O4W | 140.66 (13) | Cl4—Ru1—Cl5 | 176.30 (5) |
| O1W—Ba1—O4W | 70.02 (12) | Cl1—Ru1—Cl5 | 92.07 (5) |
| O5W—Ba1—O4W | 107.09 (12) | Cl2—Ru1—Cl5 | 88.76 (4) |
| O2W—Ba1—O4W | 63.18 (10) | Ru1—Cl2—Ba1 | 105.92 (4) |
| O2Wi—Ba1—O4W | 131.76 (11) | Ru1—Cl3—Ba1 | 104.10 (4) |
| O3W—Ba1—O4Wi | 72.37 (12) | Ru1—O1—Ru1ii | 176.9 (3) |
| O1W—Ba1—O4Wi | 134.09 (12) | Ba1—O1W—H21W | 115 (5) |
| O5W—Ba1—O4Wi | 121.94 (11) | Ba1—O1W—H11W | 119 (5) |
| O2W—Ba1—O4Wi | 72.71 (11) | H21W—O1W—H11W | 108 (4) |
| O2Wi—Ba1—O4Wi | 62.42 (10) | Ba1—O2W—Ba1iii | 119.74 (12) |
| O4W—Ba1—O4Wi | 130.13 (6) | Ba1—O2W—H12W | 110 (5) |
| O3W—Ba1—Cl2 | 146.54 (10) | Ba1iii—O2W—H12W | 111 (5) |
| O1W—Ba1—Cl2 | 137.37 (9) | Ba1—O2W—H22W | 95 (5) |
| O5W—Ba1—Cl2 | 129.21 (9) | Ba1iii—O2W—H22W | 110 (5) |
| O2W—Ba1—Cl2 | 66.31 (8) | H12W—O2W—H22W | 109 (5) |
| O2Wi—Ba1—Cl2 | 75.56 (8) | Ba1—O3W—H13W | 131 (6) |
| O4W—Ba1—Cl2 | 67.66 (8) | Ba1—O3W—H23W | 119 (6) |
| O4Wi—Ba1—Cl2 | 74.37 (8) | H13W—O3W—H23W | 108 (6) |
| O3W—Ba1—Cl3 | 133.20 (12) | Ba1—O4W—Ba1iii | 114.07 (11) |
| O1W—Ba1—Cl3 | 109.65 (9) | Ba1—O4W—H24W | 108 (5) |
| O5W—Ba1—Cl3 | 69.25 (9) | Ba1iii—O4W—H24W | 111 (5) |
| O2W—Ba1—Cl3 | 120.12 (8) | Ba1—O4W—H14W | 113 (5) |
| O2Wi—Ba1—Cl3 | 63.36 (7) | Ba1iii—O4W—H14W | 101 (5) |
| O4W—Ba1—Cl3 | 71.82 (8) | H24W—O4W—H14W | 109 (5) |
| O4Wi—Ba1—Cl3 | 115.81 (8) | Ba1—O5W—H15W | 111 (5) |
| Cl2—Ba1—Cl3 | 61.10 (3) | Ba1—O5W—H25W | 130 (4) |
| O1—Ru1—Cl3 | 179.48 (13) | H15W—O5W—H25W | 108 (5) |
| O1—Ru1—Cl4 | 92.49 (13) | | |
| Symmetry codes: (i) −x+1/2, y+1/2, −z+3/2; (ii) −x+1, y, −z+3/2; (iii) −x+1/2, y−1/2, −z+3/2. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1W—H11W···O3Wiv | 0.85 (1) | 2.58 (4) | 3.347 (7) | 151 (6) |
| O2W—H12W···Cl1v | 0.82 (6) | 2.36 (6) | 3.176 (4) | 177 (7) |
| O3W—H13W···Cl4i | 0.73 (7) | 2.66 (6) | 3.354 (6) | 161 (7) |
| O4W—H14W···Cl4 | 0.78 (8) | 2.62 (8) | 3.395 (5) | 171 (7) |
| O5W—H15W···O1Wiv | 0.81 (9) | 2.25 (9) | 3.040 (9) | 166 (7) |
| O1W—H21W···Cl2iii | 0.79 (5) | 2.65 (5) | 3.357 (4) | 151 (6) |
| O1W—H21W···Cl1vi | 0.79 (5) | 2.80 (6) | 3.354 (5) | 130 (5) |
| O2W—H22W···Cl5iii | 0.76 (8) | 2.42 (8) | 3.144 (5) | 161 (7) |
| O3W—H23W···O5Wiv | 0.79 (6) | 2.20 (7) | 2.932 (8) | 155 (8) |
| O4W—H24W···Cl1vi | 0.85 (6) | 2.71 (6) | 3.519 (5) | 160 (5) |
| O4W—H24W···Cl3vi | 0.85 (6) | 2.81 (6) | 3.324 (5) | 121 (6) |
| O5W—H25W···Cl5vii | 0.87 (8) | 2.38 (8) | 3.251 (6) | 173 (4) |
| Symmetry codes: (i) −x+1/2, y+1/2, −z+3/2; (iii) −x+1/2, y−1/2, −z+3/2; (iv) −x, −y+1, −z+1; (v) x, −y+1, z+1/2; (vi) −x+1/2, −y+1/2, −z+1; (vii) −x+1/2, −y+3/2, −z+1. |
Experimental details
| Crystal data |
| Chemical formula | Ba2Ru2Cl10O·10H2O |
| Mr | 1027.48 |
| Crystal system, space group | Monoclinic, C2/c |
| Temperature (K) | 293 |
| a, b, c (Å) | 20.9386 (18), 8.8654 (7), 16.0560 (15) |
| β (°) | 124.559 (5) |
| V (Å3) | 2454.5 (4) |
| Z | 4 |
| Radiation type | Mo Kα |
| µ (mm−1) | 5.49 |
| Crystal size (mm) | 0.25 × 0.13 × 0.03 |
| |
| Data collection |
| Diffractometer | Nonius KappaCCD
diffractometer |
| Absorption correction | Part of the refinement model (ΔF)
(Sheldrick, 1996 or 1990???) |
| Tmin, Tmax | 0.441, 0.862 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 10480, 2488, 2103 |
| Rint | 0.05 |
| (sin θ/λ)max (Å−1) | 0.625 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.029, 0.070, 1.06 |
| No. of reflections | 2488 |
| No. of parameters | 144 |
| No. of restraints | 15 |
| H-atom treatment | Only H-atom coordinates refined |
| Δρmax, Δρmin (e Å−3) | 0.53, −1.28 |
Selected geometric parameters (Å, º) top| Ba1—O3W | 2.713 (4) | Ba1—Cl3 | 3.2882 (13) |
| Ba1—O1W | 2.780 (4) | Ru1—O1 | 1.7657 (4) |
| Ba1—O5W | 2.809 (4) | Ru1—Cl3 | 2.3394 (12) |
| Ba1—O2W | 2.882 (4) | Ru1—Cl4 | 2.3627 (14) |
| Ba1—O2Wi | 2.931 (4) | Ru1—Cl1 | 2.3665 (12) |
| Ba1—O4W | 2.988 (4) | Ru1—Cl2 | 2.3744 (12) |
| Ba1—O4Wi | 3.004 (4) | Ru1—Cl5 | 2.3755 (15) |
| Ba1—Cl2 | 3.1986 (12) | | |
| | | |
| O3W—Ba1—O1W | 72.33 (13) | O2W—Ba1—Cl2 | 66.31 (8) |
| O3W—Ba1—O5W | 68.55 (14) | O5W—Ba1—Cl3 | 69.25 (9) |
| O1W—Ba1—O5W | 68.89 (12) | O1—Ru1—Cl3 | 179.48 (13) |
| O3W—Ba1—O2W | 106.46 (14) | O1—Ru1—Cl4 | 92.49 (13) |
| O1W—Ba1—O2W | 90.31 (12) | O1—Ru1—Cl1 | 91.93 (4) |
| O3W—Ba1—O2Wi | 85.88 (14) | O1—Ru1—Cl2 | 91.45 (4) |
| O2W—Ba1—O2Wi | 127.12 (6) | O1—Ru1—Cl5 | 91.19 (13) |
| Symmetry code: (i) −x+1/2, y+1/2, −z+3/2. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1W—H11W···O3Wii | 0.845 (14) | 2.58 (4) | 3.347 (7) | 151 (6) |
| O2W—H12W···Cl1iii | 0.82 (6) | 2.36 (6) | 3.176 (4) | 177 (7) |
| O3W—H13W···Cl4i | 0.73 (7) | 2.66 (6) | 3.354 (6) | 161 (7) |
| O4W—H14W···Cl4 | 0.78 (8) | 2.62 (8) | 3.395 (5) | 171 (7) |
| O5W—H15W···O1Wii | 0.81 (9) | 2.25 (9) | 3.040 (9) | 166 (7) |
| O1W—H21W···Cl2iv | 0.79 (5) | 2.65 (5) | 3.357 (4) | 151 (6) |
| O1W—H21W···Cl1v | 0.79 (5) | 2.80 (6) | 3.354 (5) | 130 (5) |
| O2W—H22W···Cl5iv | 0.76 (8) | 2.42 (8) | 3.144 (5) | 161 (7) |
| O3W—H23W···O5Wii | 0.79 (6) | 2.20 (7) | 2.932 (8) | 155 (8) |
| O4W—H24W···Cl1v | 0.85 (6) | 2.71 (6) | 3.519 (5) | 160 (5) |
| O4W—H24W···Cl3v | 0.85 (6) | 2.81 (6) | 3.324 (5) | 121 (6) |
| O5W—H25W···Cl5vi | 0.87 (8) | 2.38 (8) | 3.251 (6) | 173 (4) |
| Symmetry codes: (i) −x+1/2, y+1/2, −z+3/2; (ii) −x, −y+1, −z+1; (iii) x, −y+1, z+1/2; (iv) −x+1/2, y−1/2, −z+3/2; (v) −x+1/2, −y+1/2, −z+1; (vi) −x+1/2, −y+3/2, −z+1. |
 O hydrogen bonds and O—H
O hydrogen bonds and O—H Cl dipole–dipole interactions.
Cl dipole–dipole interactions.
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2007/08/00/bc3046/bc3046fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2007/08/00/bc3046/bc3046fig2thm.gif)




Previous investigations of MCl2—RuCl3 systems (M = K and Cs) have led to the identification of K4[Ru2Cl10O] (Deloume et al., 1979) and Cs4Ru2Cl10O (Santana Da Silva et al., 1999). We have investigated the hydrated systems MCl2—RuCl3—H2O (M = Mg, Ca and Ba) and, just recently, we have determined the structure of Mg2Ru2Cl10O.16H2O (Boufas et al., 2007). The structural study of the different phases of these systems has been performed in order to compare the cation coordination environments and the number of water molecules, and to study the Ru2Cl10O anionic group and understand the effect of the Mg/Ba substitution.
The asymmetric unit of the title compound contains one [Ru2Cl10O]4- anion, one Ba2+ cation and five water molecules (Fig. 1). The overall structure consists of layers stacked along the c direction, with [Ru2Cl10O]4- dimeric units bridging adjacent sheets that are held together by [Ba(H2O)7]2+ cations through the Cl atoms Cl2 and Cl3 along the (101) plane. As reported for Mg2Ru2Cl10O.16H2O (Boufas et al., 2007), the Ru atom resides in a distorted octahedron involving one bridging O atom [Ru1—O1 bond is 1.7657 (4) Å] and five Cl atoms with an Ru—Cl average distance of 2.3637 Å (Table 1). These distances are similar to those of the anhydrous compounds reported in the literature (1.800 and 2.362 Å for K4[Ru2Cl10O], and 1.791 and 2.357 Å for Cs4Ru2Cl10O] and agree well with those found in Mg2Ru2Cl10O.16H2O (1.7822 and 2.36284 Å). The standard deviations from the values given by the two hydrated compounds differ by about 0.02 and 0.0008 Å. The Ru—O distance in the title compound is the shortest and the Ru—Cl distance is the longest.
The barium cation is surrounded by nine atoms, viz. five O atoms belonging to five water molecules via metal–OH2 bonds and two Cl atoms (Cl2 and Cl3) from the Ru2Cl10O octahedron via metal–Cl bonds. The presence of additional interactions (Ba1—O2Wi and Ba1—O4Wi) leads to ninefold coordination for each Ba atom. The [Ba(H2O)7]2+ cations extend along the b direction in a zigzag fashion, forming layers parallel to the bc plane, and each [Ru2Cl10O]4- anion is surrounded by six [Ba(H2O)7]2+ cations (Fig. 2). The Ba1—Cl2 and Ba1—Cl3 distances (Table 1) are similar to those found in barium dichloride dihydrate (3.0901 and 3.2836 Å; Bochkova et al., 1980) and agree with the sum of the ionic radii of the Ba2+ (1.35 Å) and Cl- ions (1.81 Å) (Shannon, 1976).
The average Ba—Ow distance is 2.8724 Å, similar to that in Ba(OH)I(H2O)4 (2.8425 Å), where Ba is coordinated by only four water molecules (Fromm & Goesmann, 2000), and somewhat longer than that in Ba(C10H4O8)(H2O)5 (2.8094 Å), where Ba is surrounded by five O atoms (Dale et al., 2003). This difference is due to the presence of Ba–carboxylate bonds that reduce the Ba—OH2 distances.
The cations and anions of the title compound are linked into a three-dimensional network by means of O—H···Cl and O—H···O interactions (Table 2 and Fig. 2). The first type links the Ru2Cl10O octahedra to water molecules through all the Cl atoms, with distances of between 2.36 (6) and 2.81 (6) Å. Atoms Cl4 and Cl5 are involved in two hydrogen bonds each, Cl2 and Cl3 establish one hydrogen bond each, and Cl1 is engaged in three interactions. The O2W—H12W···Cl1iii interaction is the strongest, with an O—H···Cl distance of 2.36 (6) Å. Only three O—H···O hydrogen bonds are formed via O3W [2.58 (4) Å], O1W [2.25 (9) Å] and O5W [2.20 (7) Å]. The environment of the [Ru2Cl10O]4- anion contains nine O—H···Cl dipole–dipole interactions between the anion and the water molecules, i.e. this structure displays a lower degree of cohesion than that reported for Mg2Ru2Cl10O.16H2O characterized by 12 O—H···Cl dipole–dipole interactions and five O—H···O hydrogen bonds.
Overall, the various interactions bridge the ruthenate anions within and between layers and form cavities occupied by the [BaCl2(H2O)7] sheets.