In the crystal of the title compound, C
20H
42O, the molecules are packed in layers parallel to the (100) plane. The alkyl chains are parallel to the [30
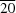
] direction and these molecular chains are hydrogen-bonded into chains parallel to the
c axis. All C—C bonds of the alkyl chain show an antiperiplanar (
trans) conformation, with a slight deviation from the ideal value (180°) in the C—C bonds close to the hydrogen bonds. The length of the alkyl chain is 27.92 (2) Å and the tilt angle is 59.7 (2)°.
Supporting information
CCDC reference: 142763
Crystals of (I) were obtained by slowly evaporating saturated solutions of the compound in 4-chlorotoluene at room temperature.
Intensities were collected to θ = 30°. (hkl) reflections with θ > 26° were omitted due to peak overlapping problems (asymmetric background) and to a low intensity value. The positions of 41 H atoms linked to C atoms were geometrically calculated. The H atom linked to O was located from a difference map and found to be on a disordered site. An occupancy factor of 0.5 was assigned according to the peak heights in the difference map. Each O atom is close to two further O atoms (related by an inversion centre and a twofold axis) at a distance appropriate to a hydrogen bond, so an ordered hydrogen-bond scheme is incompatible with the space group symmetry. An overall isotropic displacement parameter for the H atoms was used in the refinement and all H atoms were refined using a riding model. The anisotropic displacement parameters for the O atom revealed a possible disorder in the localization of this atom. The maximum and minimum principal axes of the displacement ellipsoids are 0.131 (1) and 0.015 (1) Å2, respectively. The direction of that corresponding to the maximum eigenvalue is practically perpendicular to the C—O bond.
Data collection: CAD-4-PC (Kretschmar, 1996); cell refinement: CAD-4-PC; data reduction: CFEO (Solans, 1978); program(s) used to solve structure: SHELXS97 (Sheldrick, 1990); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: ORTEP (Johnson, 1965; Brueggermann & Schmid, 1990); software used to prepare material for publication: PLATON (Spek, 1990).
γ-form of n-Eicosanol
top
Crystal data top
| C20H42O | F(000) = 1360 |
| Mr = 298.54 | Dx = 0.982 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71069 Å |
| a = 91.116 (17) Å | Cell parameters from 25 reflections |
| b = 4.932 (4) Å | θ = 8–15° |
| c = 8.997 (10) Å | µ = 0.06 mm−1 |
| β = 93.01 (4)° | T = 293 K |
| V = 4038 (6) Å3 | Lozenge, colourless |
| Z = 8 | 0.6 × 0.6 × 0.2 mm |
Data collection top
Enraf-Nonius CAD-4
diffractometer | Rint = 0.023 |
| Radiation source: fine-focus sealed tube | θmax = 26.0°, θmin = 2.2° |
| Graphite monochromator | h = −110→112 |
| ω–2θ scans | k = 0→6 |
| 4539 measured reflections | l = 0→10 |
| 3932 independent reflections | 3 standard reflections every 120 min |
| 2007 reflections with I > 2σ(I) | intensity decay: none |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: mixed |
| wR(F2) = 0.159 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.98 | Calculated w = 1/[σ2(Fo2) + (0.0879P)2]
where P = (Fo2 + 2Fc2)/3 |
| 3932 reflections | (Δ/σ)max = 0.009 |
| 198 parameters | Δρmax = 0.15 e Å−3 |
| 2 restraints | Δρmin = −0.12 e Å−3 |
Crystal data top
| C20H42O | V = 4038 (6) Å3 |
| Mr = 298.54 | Z = 8 |
| Monoclinic, C2/c | Mo Kα radiation |
| a = 91.116 (17) Å | µ = 0.06 mm−1 |
| b = 4.932 (4) Å | T = 293 K |
| c = 8.997 (10) Å | 0.6 × 0.6 × 0.2 mm |
| β = 93.01 (4)° | |
Data collection top
Enraf-Nonius CAD-4
diffractometer | Rint = 0.023 |
| 4539 measured reflections | 3 standard reflections every 120 min |
| 3932 independent reflections | intensity decay: none |
| 2007 reflections with I > 2σ(I) | |
Refinement top
| R[F2 > 2σ(F2)] = 0.041 | 2 restraints |
| wR(F2) = 0.159 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.98 | Δρmax = 0.15 e Å−3 |
| 3932 reflections | Δρmin = −0.12 e Å−3 |
| 198 parameters | |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| O | 0.005323 (9) | 0.3433 (2) | 0.11346 (8) | 0.0658 (4) | |
| C1 | 0.018551 (13) | 0.2041 (3) | 0.09344 (13) | 0.0583 (4) | |
| H1 | 0.0164 | 0.0138 | 0.0757 | 0.0989 (16)* | |
| H1A | 0.0248 | 0.2171 | 0.1840 | 0.0989 (16)* | |
| C2 | 0.026676 (12) | 0.3140 (2) | −0.03390 (12) | 0.0499 (3) | |
| H2 | 0.0280 | 0.5074 | −0.0194 | 0.0989 (16)* | |
| H2A | 0.0206 | 0.2891 | −0.1249 | 0.0989 (16)* | |
| C3 | 0.041483 (12) | 0.1857 (2) | −0.05364 (11) | 0.0407 (3) | |
| H3 | 0.0401 | −0.0072 | −0.0702 | 0.0989 (16)* | |
| H3A | 0.0475 | 0.2080 | 0.0377 | 0.0989 (16)* | |
| C4 | 0.049606 (11) | 0.3033 (2) | −0.18150 (11) | 0.0376 (3) | |
| H4 | 0.0437 | 0.2750 | −0.2731 | 0.0989 (16)* | |
| H4A | 0.0506 | 0.4974 | −0.1667 | 0.0989 (16)* | |
| C5 | 0.064749 (11) | 0.1846 (2) | −0.19995 (11) | 0.0400 (3) | |
| H5 | 0.0638 | −0.0094 | −0.2153 | 0.0989 (16)* | |
| H5A | 0.0707 | 0.2127 | −0.1085 | 0.0989 (16)* | |
| C6 | 0.072729 (10) | 0.3052 (2) | −0.32812 (11) | 0.0366 (3) | |
| H6 | 0.0668 | 0.2768 | −0.4195 | 0.0989 (16)* | |
| H6A | 0.0736 | 0.4993 | −0.3128 | 0.0989 (16)* | |
| C7 | 0.087904 (11) | 0.1878 (2) | −0.34692 (11) | 0.0398 (3) | |
| H7 | 0.0870 | −0.0063 | −0.3619 | 0.0989 (16)* | |
| H7A | 0.0938 | 0.2166 | −0.2556 | 0.0989 (16)* | |
| C8 | 0.095919 (11) | 0.3082 (2) | −0.47584 (11) | 0.0386 (3) | |
| H8 | 0.0900 | 0.2794 | −0.5672 | 0.0989 (16)* | |
| H8A | 0.0968 | 0.5024 | −0.4609 | 0.0989 (16)* | |
| C9 | 0.111082 (11) | 0.1903 (2) | −0.49424 (12) | 0.0411 (3) | |
| H9 | 0.1102 | −0.0040 | −0.5087 | 0.0989 (16)* | |
| H9A | 0.1170 | 0.2199 | −0.4031 | 0.0989 (16)* | |
| C10 | 0.119043 (12) | 0.3101 (2) | −0.62378 (11) | 0.0407 (3) | |
| H10 | 0.1131 | 0.2814 | −0.7149 | 0.0989 (16)* | |
| H10A | 0.1200 | 0.5043 | −0.6090 | 0.0989 (16)* | |
| C11 | 0.134116 (12) | 0.1918 (2) | −0.64282 (12) | 0.0428 (3) | |
| H11 | 0.1331 | −0.0025 | −0.6572 | 0.0989 (16)* | |
| H11A | 0.1401 | 0.2209 | −0.5518 | 0.0989 (16)* | |
| C12 | 0.142101 (12) | 0.3098 (2) | −0.77209 (11) | 0.0435 (3) | |
| H12 | 0.1361 | 0.2808 | −0.8631 | 0.0989 (16)* | |
| H12A | 0.1431 | 0.5040 | −0.7576 | 0.0989 (16)* | |
| C13 | 0.157136 (12) | 0.1919 (2) | −0.79133 (12) | 0.0454 (3) | |
| H13 | 0.1562 | −0.0023 | −0.8057 | 0.0989 (16)* | |
| H13A | 0.1631 | 0.2210 | −0.7003 | 0.0989 (16)* | |
| C14 | 0.165162 (13) | 0.3092 (2) | −0.92044 (12) | 0.0454 (3) | |
| H14 | 0.1592 | 0.2790 | −1.0117 | 0.0989 (16)* | |
| H14A | 0.1661 | 0.5036 | −0.9065 | 0.0989 (16)* | |
| C15 | 0.180218 (13) | 0.1918 (2) | −0.93831 (12) | 0.0499 (3) | |
| H15 | 0.1793 | −0.0025 | −0.9524 | 0.0989 (16)* | |
| H15A | 0.1861 | 0.2215 | −0.8468 | 0.0989 (16)* | |
| C16 | 0.188386 (13) | 0.3090 (2) | −1.06745 (12) | 0.0495 (3) | |
| H16 | 0.1826 | 0.2769 | −1.1592 | 0.0989 (16)* | |
| H16A | 0.1892 | 0.5037 | −1.0542 | 0.0989 (16)* | |
| C17 | 0.203576 (13) | 0.1924 (2) | −1.08302 (13) | 0.0543 (4) | |
| H17 | 0.2027 | −0.0024 | −1.0957 | 0.0989 (16)* | |
| H17A | 0.2094 | 0.2254 | −0.9915 | 0.0989 (16)* | |
| C18 | 0.211663 (14) | 0.3085 (2) | −1.21240 (12) | 0.0545 (4) | |
| H18 | 0.2059 | 0.2733 | −1.3042 | 0.0989 (16)* | |
| H18A | 0.2124 | 0.5036 | −1.2004 | 0.0989 (16)* | |
| C19 | 0.226915 (15) | 0.1938 (3) | −1.22607 (14) | 0.0720 (4) | |
| H19 | 0.2262 | −0.0016 | −1.2363 | 0.0989 (16)* | |
| H19A | 0.2327 | 0.2314 | −1.1347 | 0.0989 (16)* | |
| C20 | 0.235048 (16) | 0.3051 (3) | −1.35654 (14) | 0.0863 (5) | |
| H20 | 0.2446 | 0.2228 | −1.3577 | 0.0989 (16)* | |
| H20A | 0.2361 | 0.4979 | −1.3462 | 0.0989 (16)* | |
| H20B | 0.2296 | 0.2647 | −1.4479 | 0.0989 (16)* | |
| H1O | −0.0004 (3) | 0.313 (4) | 0.204 (2) | 0.0989 (16)* | 0.50 |
| H1O' | −0.0010 (3) | 0.376 (4) | 0.021 (2) | 0.0989 (16)* | 0.50 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O | 0.0457 (5) | 0.1042 (8) | 0.0502 (5) | 0.0385 (5) | 0.0294 (4) | 0.0157 (6) |
| C1 | 0.0350 (6) | 0.0849 (9) | 0.0573 (7) | 0.0157 (6) | 0.0256 (5) | 0.0133 (6) |
| C2 | 0.0318 (6) | 0.0754 (8) | 0.0441 (6) | 0.0077 (6) | 0.0183 (5) | 0.0016 (6) |
| C3 | 0.0341 (6) | 0.0487 (7) | 0.0408 (6) | 0.0048 (5) | 0.0146 (4) | −0.0033 (5) |
| C4 | 0.0213 (5) | 0.0517 (7) | 0.0414 (6) | 0.0051 (5) | 0.0167 (4) | 0.0023 (5) |
| C5 | 0.0296 (6) | 0.0530 (7) | 0.0390 (6) | 0.0006 (5) | 0.0157 (4) | −0.0035 (5) |
| C6 | 0.0198 (5) | 0.0514 (7) | 0.0402 (6) | 0.0065 (5) | 0.0165 (4) | 0.0015 (5) |
| C7 | 0.0282 (6) | 0.0497 (7) | 0.0430 (6) | −0.0003 (5) | 0.0180 (5) | −0.0007 (5) |
| C8 | 0.0228 (5) | 0.0526 (7) | 0.0420 (6) | 0.0040 (5) | 0.0186 (4) | 0.0048 (5) |
| C9 | 0.0298 (6) | 0.0521 (7) | 0.0432 (6) | 0.0017 (5) | 0.0192 (5) | 0.0021 (5) |
| C10 | 0.0283 (6) | 0.0523 (7) | 0.0434 (6) | 0.0030 (5) | 0.0204 (4) | 0.0034 (5) |
| C11 | 0.0366 (7) | 0.0503 (7) | 0.0434 (6) | −0.0003 (5) | 0.0205 (5) | −0.0010 (5) |
| C12 | 0.0341 (6) | 0.0547 (7) | 0.0438 (6) | 0.0028 (5) | 0.0227 (5) | 0.0040 (5) |
| C13 | 0.0398 (7) | 0.0517 (7) | 0.0468 (6) | −0.0030 (5) | 0.0216 (5) | 0.0023 (5) |
| C14 | 0.0382 (6) | 0.0532 (7) | 0.0470 (6) | 0.0045 (5) | 0.0224 (5) | 0.0051 (5) |
| C15 | 0.0438 (7) | 0.0580 (7) | 0.0506 (6) | −0.0019 (6) | 0.0267 (5) | 0.0001 (6) |
| C16 | 0.0409 (7) | 0.0619 (8) | 0.0479 (6) | 0.0020 (6) | 0.0250 (5) | 0.0076 (6) |
| C17 | 0.0489 (7) | 0.0637 (8) | 0.0534 (7) | 0.0007 (6) | 0.0312 (6) | 0.0023 (6) |
| C18 | 0.0570 (8) | 0.0607 (8) | 0.0485 (6) | −0.0021 (6) | 0.0291 (6) | 0.0049 (6) |
| C19 | 0.0774 (10) | 0.0815 (10) | 0.0605 (7) | 0.0073 (8) | 0.0354 (7) | 0.0069 (7) |
| C20 | 0.0819 (11) | 0.1154 (12) | 0.0653 (8) | −0.0090 (9) | 0.0394 (8) | 0.0000 (8) |
Geometric parameters (Å, º) top
| O—C1 | 1.407 (1) | C10—H10A | 0.9700 |
| O—H1O | 1.001 (10) | C11—C12 | 1.520 (2) |
| O—H1O' | 1.000 (10) | C11—H11 | 0.9700 |
| C1—C2 | 1.498 (2) | C11—H11A | 0.9700 |
| C1—H1 | 0.9700 | C12—C13 | 1.507 (1) |
| C1—H1A | 0.9700 | C12—H12 | 0.9700 |
| C2—C3 | 1.509 (1) | C12—H12A | 0.9700 |
| C2—H2 | 0.9700 | C13—C14 | 1.519 (2) |
| C2—H2A | 0.9700 | C13—H13 | 0.9700 |
| C3—C4 | 1.516 (2) | C13—H13A | 0.9700 |
| C3—H3 | 0.9700 | C14—C15 | 1.506 (1) |
| C3—H3A | 0.9700 | C14—H14 | 0.9700 |
| C4—C5 | 1.516 (1) | C14—H14A | 0.9700 |
| C4—H4 | 0.9700 | C15—C16 | 1.526 (2) |
| C4—H4A | 0.9700 | C15—H15 | 0.9700 |
| C5—C6 | 1.517 (2) | C15—H15A | 0.9700 |
| C5—H5 | 0.9700 | C16—C17 | 1.512 (2) |
| C5—H5A | 0.9700 | C16—H16 | 0.9700 |
| C6—C7 | 1.517 (1) | C16—H16A | 0.9700 |
| C6—H6 | 0.9700 | C17—C18 | 1.522 (2) |
| C6—H6A | 0.9700 | C17—H17 | 0.9700 |
| C7—C8 | 1.523 (2) | C17—H17A | 0.9700 |
| C7—H7 | 0.9700 | C18—C19 | 1.511 (2) |
| C7—H7A | 0.9700 | C18—H18 | 0.9700 |
| C8—C9 | 1.516 (1) | C18—H18A | 0.9700 |
| C8—H8 | 0.9700 | C19—C20 | 1.523 (2) |
| C8—H8A | 0.9700 | C19—H19 | 0.9700 |
| C9—C10 | 1.524 (2) | C19—H19A | 0.9700 |
| C9—H9 | 0.9700 | C20—H20 | 0.9600 |
| C9—H9A | 0.9700 | C20—H20A | 0.9600 |
| C10—C11 | 1.510 (1) | C20—H20B | 0.9600 |
| C10—H10 | 0.9700 | | |
| | | |
| C1—O—H1O | 121.3 (15) | H10—C10—H10A | 107.6 |
| C1—O—H1O' | 115.7 (17) | C10—C11—C12 | 114.4 (1) |
| H1O—O—H1O' | 113 (2) | C10—C11—H11 | 108.7 |
| O—C1—C2 | 112.5 (1) | C12—C11—H11 | 108.7 |
| O—C1—H1 | 109.1 | C10—C11—H11A | 108.7 |
| C2—C1—H1 | 109.1 | C12—C11—H11A | 108.7 |
| O—C1—H1A | 109.1 | H11—C11—H11A | 107.6 |
| C2—C1—H1A | 109.1 | C13—C12—C11 | 114.5 (1) |
| H1—C1—H1A | 107.8 | C13—C12—H12 | 108.6 |
| C1—C2—C3 | 114.8 (1) | C11—C12—H12 | 108.6 |
| C1—C2—H2 | 108.6 | C13—C12—H12A | 108.6 |
| C3—C2—H2 | 108.6 | C11—C12—H12A | 108.6 |
| C1—C2—H2A | 108.6 | H12—C12—H12A | 107.6 |
| C3—C2—H2A | 108.6 | C12—C13—C14 | 114.7 (1) |
| H2—C2—H2A | 107.6 | C12—C13—H13 | 108.6 |
| C2—C3—C4 | 113.8 (1) | C14—C13—H13 | 108.6 |
| C2—C3—H3 | 108.8 | C12—C13—H13A | 108.6 |
| C4—C3—H3 | 108.8 | C14—C13—H13A | 108.6 |
| C2—C3—H3A | 108.8 | H13—C13—H13A | 107.6 |
| C4—C3—H3A | 108.8 | C15—C14—C13 | 114.4 (1) |
| H3—C3—H3A | 107.7 | C15—C14—H14 | 108.7 |
| C5—C4—C3 | 114.8 (1) | C13—C14—H14 | 108.7 |
| C5—C4—H4 | 108.6 | C15—C14—H14A | 108.7 |
| C3—C4—H4 | 108.6 | C13—C14—H14A | 108.7 |
| C5—C4—H4A | 108.6 | H14—C14—H14A | 107.6 |
| C3—C4—H4A | 108.6 | C14—C15—C16 | 114.8 (1) |
| H4—C4—H4A | 107.6 | C14—C15—H15 | 108.6 |
| C4—C5—C6 | 114.0 (1) | C16—C15—H15 | 108.6 |
| C4—C5—H5 | 108.7 | C14—C15—H15A | 108.6 |
| C6—C5—H5 | 108.7 | C16—C15—H15A | 108.6 |
| C4—C5—H5A | 108.7 | H15—C15—H15A | 107.5 |
| C6—C5—H5A | 108.7 | C17—C16—C15 | 114.3 (1) |
| H5—C5—H5A | 107.6 | C17—C16—H16 | 108.7 |
| C7—C6—C5 | 114.3 (1) | C15—C16—H16 | 108.7 |
| C7—C6—H6 | 108.7 | C17—C16—H16A | 108.7 |
| C5—C6—H6 | 108.7 | C15—C16—H16A | 108.7 |
| C7—C6—H6A | 108.7 | H16—C16—H16A | 107.6 |
| C5—C6—H6A | 108.7 | C16—C17—C18 | 114.2 (1) |
| H6—C6—H6A | 107.6 | C16—C17—H17 | 108.7 |
| C6—C7—C8 | 114.4 (1) | C18—C17—H17 | 108.7 |
| C6—C7—H7 | 108.7 | C16—C17—H17A | 108.7 |
| C8—C7—H7 | 108.7 | C18—C17—H17A | 108.7 |
| C6—C7—H7A | 108.7 | H17—C17—H17A | 107.6 |
| C8—C7—H7A | 108.7 | C19—C18—C17 | 113.9 (1) |
| H7—C7—H7A | 107.6 | C19—C18—H18 | 108.8 |
| C9—C8—C7 | 114.2 (1) | C17—C18—H18 | 108.8 |
| C9—C8—H8 | 108.7 | C19—C18—H18A | 108.8 |
| C7—C8—H8 | 108.7 | C17—C18—H18A | 108.8 |
| C9—C8—H8A | 108.7 | H18—C18—H18A | 107.7 |
| C7—C8—H8A | 108.7 | C18—C19—C20 | 114.5 (1) |
| H8—C8—H8A | 107.6 | C18—C19—H19 | 108.6 |
| C8—C9—C10 | 114.1 (1) | C20—C19—H19 | 108.6 |
| C8—C9—H9 | 108.7 | C18—C19—H19A | 108.6 |
| C10—C9—H9 | 108.7 | C20—C19—H19A | 108.6 |
| C8—C9—H9A | 108.7 | H19—C19—H19A | 107.6 |
| C10—C9—H9A | 108.7 | C19—C20—H20 | 109.5 |
| H9—C9—H9A | 107.6 | C19—C20—H20A | 109.5 |
| C11—C10—C9 | 114.1 (1) | H20—C20—H20A | 109.5 |
| C11—C10—H10 | 108.7 | C19—C20—H20B | 109.5 |
| C9—C10—H10 | 108.7 | H20—C20—H20B | 109.5 |
| C11—C10—H10A | 108.7 | H20A—C20—H20B | 109.5 |
| C9—C10—H10A | 108.7 | | |
| | | |
| O—C1—C2—C3 | 175.77 (9) | C9—C10—C11—C12 | 179.84 (8) |
| C1—C2—C3—C4 | −179.04 (9) | C10—C11—C12—C13 | −179.97 (8) |
| C2—C3—C4—C5 | 177.86 (8) | C11—C12—C13—C14 | 179.98 (8) |
| C3—C4—C5—C6 | −179.85 (8) | C12—C13—C14—C15 | 179.60 (8) |
| C4—C5—C6—C7 | 179.85 (8) | C13—C14—C15—C16 | −179.83 (8) |
| C5—C6—C7—C8 | 179.86 (8) | C14—C15—C16—C17 | 179.19 (9) |
| C6—C7—C8—C9 | −179.99 (8) | C15—C16—C17—C18 | 179.68 (9) |
| C7—C8—C9—C10 | 179.71 (8) | C16—C17—C18—C19 | 179.23 (9) |
| C8—C9—C10—C11 | −179.68 (8) | C17—C18—C19—C20 | 179.12 (10) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O—H1O···Oi | 1.00 (2) | 1.73 (2) | 2.688 (3) | 159 (2) |
| O—H1O′···Oii | 1.00 (2) | 1.87 (2) | 2.700 (3) | 139 (2) |
| Symmetry codes: (i) −x, y, −z+1/2; (ii) −x, −y+1, −z. |
Experimental details
| Crystal data |
| Chemical formula | C20H42O |
| Mr | 298.54 |
| Crystal system, space group | Monoclinic, C2/c |
| Temperature (K) | 293 |
| a, b, c (Å) | 91.116 (17), 4.932 (4), 8.997 (10) |
| β (°) | 93.01 (4) |
| V (Å3) | 4038 (6) |
| Z | 8 |
| Radiation type | Mo Kα |
| µ (mm−1) | 0.06 |
| Crystal size (mm) | 0.6 × 0.6 × 0.2 |
| |
| Data collection |
| Diffractometer | Enraf-Nonius CAD-4
diffractometer |
| Absorption correction | – |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 4539, 3932, 2007 |
| Rint | 0.023 |
| (sin θ/λ)max (Å−1) | 0.617 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.041, 0.159, 0.98 |
| No. of reflections | 3932 |
| No. of parameters | 198 |
| No. of restraints | 2 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.15, −0.12 |
Selected geometric parameters (Å, º) top| O—C1 | 1.407 (1) | C1—C2 | 1.498 (2) |
| | | |
| O—C1—C2 | 112.5 (1) | | |
| | | |
| O—C1—C2—C3 | 175.77 (9) | C2—C3—C4—C5 | 177.86 (8) |
| C1—C2—C3—C4 | −179.04 (9) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O—H1O···Oi | 1.00 (2) | 1.73 (2) | 2.688 (3) | 159 (2) |
| O—H1O'···Oii | 1.00 (2) | 1.87 (2) | 2.700 (3) | 139 (2) |
| Symmetry codes: (i) −x, y, −z+1/2; (ii) −x, −y+1, −z. |

 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2000/02/00/na1435/na1435fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2000/02/00/na1435/na1435fig2thm.gif)




The n-alkanols (normal fatty alcohols CnH2n+1OH) show a complicated polymorphic behaviour, only partially characterized to date. The principle of a common structural description is summed up by Small (1986). This description is based on the assumption that the presence of long alkanol chains packed in a regular and parallel crystallographic array would contribute a small periodic structure within the real unit cell that defines the repeating unit of the whole structure. For a given mode of alkanol chain packing, this small internal periodic structure describes the three-dimensional relationship between equivalent positions of translationally related positions within a single alkanol chain and between its lateral neighbours. This three-dimensional subcell is defined by three axes: cs for the translation between equivalent positions within a chain and as and bs for the lateral translations. In the title compound, (I), cs = 2.553 Å and as and bs are given by the rectangular planar subcell, 4.889 × 6.032 Å.
When the long axis of the molecule tilts over the short axis of the rectangular planar subcell (4.889 Å), the modification is called γ, and if the long axis of the molecule tilts over the long axis of the rectangular subcell (6.032 Å), the modification is called β. The γ-modifications are typically exhibited by even-numbered n-alkanols. They are subdivided into several forms γi, depending on the degree of tilting of the molecules with respect to the molecular layers (Precht, 1974). The structure of two such crystals, n-hexadecanol (Abrahamsson et al., 1960) and n-octadecanol (Seto, 1962; Fujimoto et al., 1985), have already been determined. They were proved to be of the γ4 form and were reported in the space group A2/a. The cell transformation between the conventional and the reported cell is aR = c, bR = −b and cR = a - nc, where n is 5 for n-octadecanol and 6 for n-hexadecanol. For the rest of the series, as well as for other γi forms, powder diffraction patterns are indexed by extrapolation, using a constant orthorhombic subcell connected with the –CH2– repetition in the structure (Precht, 1974, 1976). In spite of the consistency of the subcell description, a complete and unambiguous indexing of the patterns becomes difficult as the length of the chain increases.
The present study is part of our work dedicated to n-alkanol mixed crystals. The investigation of the crystal structure of (I) was undertaken in order to support powder diffraction analyses.
The average C—C bond length is 1.515 (7) Å. All C—C bonds show an anti-periplanar (trans) conformation. The C2—C3—C4—C5 [177.86 (8)°], C1—C2—C3—C4 [−179.04 (9)°] and O—C1—C2—C3 [175.77 (9)°] torsion angles all deviate significantly from the ideal 180°. This is explained by the steric hindrance produced by the hydrogen bond. The chains are parallel to [3020] direction, while they are parallel to [106] in n-octadecanol and [3017] in n-hexadecanol if the conventional cell is used. Precht (1974) classified the three compounds in the [106] group.
The length of the alkyl chain is 27.92 (2) Å. This value is obtained by dividing the modulus of the [3020] vector by 12, the number of chains in this period; the value is 27.08 Å if Precht's aproximation is used. The corresponding chain lengths are 24.9 for n-octadecanol and 21.6 Å for n-hexadecanol. The chain length for (I) can also be obtained in Å from the equation l = 1.246 n + 3, where n is the number of C atoms. The long axis of the molecule in (I) tilts 59.7 (2)° over the short axis (4.889 Å) of the orthorhombic subcell defined by Precht (1974), so the structure corresponds to the γ4 form according to the classification of Precht.
A rigid body treatment of the atomic displacement parameters for the whole chain has been carried out, using the EKRT program (Craven et al., 1992). The inertial axes are defined as follows: the first is normal to the plane of the chain, the second is in the chain plane and perpendicular to the long axis of the molecule and the third is the long axis of the molecule. The principal values of the translational tensor are 0.043, 0.051 and 0.019 Å2, where the first two principal axes of the translational tensor are the first two inertial axes rotated 60° around the third inertial axis and the third is parallel to the long axis of the molecule. The principal values of the librational tensor are 0.4, 1.0 and 97.2°2, where the first two principal axes of the librational tensor are the first two inertial axes rotated 74° around the third inertial axis and the third is parallel to the long axis of the molecule. These results could explain the formation of gauche conformers in the phases at high temperature.
The alkyl chains are hydrogen-bonded, producing chains parallel to the c axis. The direction of the alkyl chains and the direction of the hydrogen bond produce a layered structure, with the layers parallel to the (100) plane. This fact explains the pseudo-rhombohedral morphology of the crystals with two more-developed opposite faces, which are (100). The short diagonal of these rhombic faces is the direction of the b axis. The angles between the edges of the main face are close to 120 and 60°.